Abstract
RNA-binding proteins act at various stages of gene expression to regulate and fine-tune patterns of mRNA accumulation. One protein in this class is Drosophila Su(s), a nuclear protein that has been previously shown to inhibit the accumulation of mutant transcripts by an unknown mechanism. Here, we have identified several additional RNAs that are downregulated by Su(s). These Su(s) targets include cryptic wild-type transcripts from the developmentally regulated Sgs4 and ng1 genes, noncoding RNAs derived from tandemly repeated αβ/αγ elements within an Hsp70 locus, and aberrant transcripts induced by Hsp70 promoter transgenes inserted at ectopic sites. We used the αβ RNAs to investigate the mechanism of Su(s) function and obtained evidence that these transcripts are degraded by the nuclear exosome and that Su(s) promotes this process. Furthermore, we showed that the RNA binding domains of Su(s) are important for this effect and mapped the sequences involved to a 267-nucleotide region of an αβ element. Taken together, these results suggest that Su(s) binds to certain nascent transcripts and stimulates their degradation by the nuclear exosome.
RNA-binding proteins participate in many aspects of eukaryotic gene expression, including transcription, pre-mRNA splicing and polyadenylation, mRNA export from the nucleus, localization, translation, and degradation (18, 33, 36). Proteins in this class have been shown to perform inhibitory and stimulatory roles in these processes. For example, the binding of poly(A) binding protein to the 3′ end of eukaryotic mRNAs prevents their degradation by ribonucleases, whereas other RNA-binding proteins promote the rapid degradation of certain transcripts (24, 41). Although numerous RNA-binding proteins have been well characterized, the functions of many others are not as well understood. Thus, much remains to be learned about the roles that RNA-binding proteins play in shaping the patterns of mRNA accumulation, especially in multicellular organisms.
Su(s) of Drosophila is a 144-kDa nuclear RNA-binding protein (34, 50) that negatively regulates the levels of its mRNA targets. Molecular genetic studies have shown that Su(s) inhibits the accumulation of RNA from several mutant alleles with transposon insertions in the 5′ transcribed region (14, 17, 26). These mutant pre-mRNAs contain antisense transposon sequences which are inefficiently spliced from the transcripts. In su(s)+ flies, the amount of RNA that accumulates from these mutant alleles is very low, and it increases in an su(s) mutant. A previous analysis of one allele suggested that the position of the transposon insertion relative to splicing signals determines whether or not the RNA is targeted by the Su(s) pathway (15). However, the mechanism by which Su(s) regulates RNA accumulation has not been previously established. The prior studies were consistent with possible roles for this protein in pre-mRNA splicing, RNA degradation, or transcription (15, 17, 27), and direct evidence for a role in any one of these processes has been lacking.
Two different types of RNA-binding domains have been identified within Su(s). Two arginine-rich motifs mediate RNA binding in vitro (48). In addition, two tandem CCCH zinc fingers of Su(s) are similar to motifs found in several proteins, including the RNA destabilization protein Tis11 and the polyadenylation factor subunit CPSF30. The CCCH zinc fingers of Tis11 bind AU-rich instability elements in the 3′ untranslated region (UTR) of its target mRNAs (6). Both the arginine-rich motifs and the zinc fingers are important for the inhibitory activity of Su(s) (27). The RNA-binding activity of Su(s) together with the observation that this protein localizes to distinct sites on Drosophila larval salivary gland polytene chromosomes (34) suggest that Su(s) associates with nascent transcripts and acts cotranscriptionally.
The polytene chromosomes of larval salivary glands have been used to examine the global distribution of numerous Drosophila proteins that participate in nuclear RNA metabolism (e.g., 3, 4, 7, 43). A visual indicator of the developmental stage of these chromosomes is the pattern of puffs, i.e., decondensed euchromatic regions, at the sites of highly transcribed genes. Changes in the puffing pattern during the last 6 to 8 h of larval development reflect transitions in gene expression in response to a pulse of the steroid hormone ecdysone. Ashburner defined 11 distinct puff stages between the beginning of the wandering larval phase and puparium formation (5). Under heat shock conditions, the developmental puffs regress, and a distinct set of puffs forms at loci that encode heat shock proteins.
The goals of this study were to identify additional Su(s) RNA targets and to explore further the mechanism of Su(s) function. Toward these ends, we used polytene chromosome immunofluorescence analysis to compare the chromosomal localization of Su(s) with polymerase II (Pol II) or the hnRNP protein Hrp40 at specific puff stages and during heat shock. These experiments showed that Su(s) localizes to a subset of sites where Pol II is bound. After identifying several of the sites, we determined that Su(s) negatively regulates the accumulation of transcripts at these loci, and we used the RNAs from one locus to explore the mechanism involved. This analysis revealed that the nuclear exosome degrades these transcripts and indicate that degradation is enhanced by the presence of Su(s). We also showed that this inhibitory effect depends on the RNA binding domains of Su(s) and sequences within the transcribed region of the RNA. Together, these findings indicate that Su(s) interacts with RNA sequences in nascent transcripts and stimulates RNA degradation by the nuclear exosome.
MATERIALS AND METHODS
Fly stocks.
The null mutant su(s)R39 has a deletion of the entire su(s) coding region (unpublished data). The wild-type stock used for RNA analysis was yw, the progenitor of su(s)R39. Transformant lines carrying the su(s) wild-type, zinc finger, and arginine-rich motif mutant transgenes in the su(s)R39 background were generated previously (27). In the arginine-rich motif mutant, sequences encoding amino acids (aa) 151 to 168 and aa 269 to 294 have been deleted. The zinc finger mutant has missense mutations that alter Cys or His residues at aa 350, aa 374, and aa 378. Oregon-R and yw were used as the wild-type stocks for immunofluorescence analysis. Deficiency stocks were obtained from the Bloomington Stock Center, except for the Df(1)N5419 and Df(1)N79f alleles, which were obtained from S. Artavanis-Tsakonas. The Kochi mutant was obtained from A. Hofmann. A transformant line containing an Hsp70 promoter transgene inserted at 23A was obtained from the Bloomington Stock Center (stock no. 10221). A transformant line carrying an Hsp70-LacZ reporter gene (z-243-2) was obtained from J. Lis.
Heat shock treatments.
Third-instar larvae or adult males were heat shocked as follows. Individual larvae or sets of five adult flies were placed in microcentrifuge tubes that contained a small hole in the top for gas exchange and a small piece of wet filter paper to prevent dehydration. Larvae used for the polytene chromosome analysis were heat shocked in a water bath at 37°C for 20 min, and the salivary glands were dissected immediately. The adult flies were heat shocked in a water bath (32°C or 37°C) or maintained at room temperature (25°C) for 15 to 30 min. The flies were quickly frozen on dry ice and stored at −70°C.
Staging of larvae.
To generate synchronized populations of larvae, females were transferred to fresh egg-laying plates for 2 h to preclear them of any fertilized eggs that they might be holding. After this procedure had been repeated twice, the females were allowed to lay eggs on fresh plates for 2 h. These plates were incubated at 25°C for 24 h, and 50 newly hatched first-instar larvae were transferred to a vial containing freshly prepared food. Tegosept was omitted from the food because it delays larval development. All of the larvae in the same experiment were reared on the same batch of food. At the appropriate times, larvae were harvested, quickly frozen, and used for RNA analysis.
Polytene chromosome immunofluorescence analysis.
Salivary glands were dissected from wandering third-instar larvae. The procedures used for preparing chromosome squashes and antibody staining have been described previously (27). DAPI staining was performed as described by Weeks et al. (51). The rabbit anti-Su(s) antibody was raised against Su(s) amino acids 648 to 808 (34). Goat anti-Pol IIa and anti-Pol IIo were provided by A. Greenleaf (51). These antibodies were directed against unphosphorylated and CTDK-1 phosphorylated C-terminal domain (CTD) repeats of Drosophila Pol II. Monoclonal Pol II antibodies H5 (anti-Ser2∼P) and H14 (anti-Ser5∼P) were obtained from Covance. H5 and goat anti-Pol IIo give identical immunofluorescence banding patterns (unpublished observations), and thus, both of these antibodies preferentially recognize Ser2∼P. Monoclonal antibody 8G6 (anti-Hrp40) was obtained from G. Dreyfuss (32). The following antibody dilutions were used: anti-Su(s), 1:150; anti-Pol IIa and anti-Pol IIo, 1:50; H5, 1:10; H14, 1:50; and 8G6, 1:800. Cy2- or Cy3-conjugated secondary antibodies were used at dilutions recommended by the supplier (Jackson ImmunoResearch Laboratories, Inc.). Digital images were taken with a Zeiss laser-scanning confocal microscope and processed using Photoshop software.
RNA analysis.
Total RNA was isolated as described previously (13). Northern blots and antisense RNA probes were prepared as described previously (15). The plasmid clones used to generate the antisense RNA probes were as follows. The LacZ probe was made from a recombinant plasmid created by cloning a 1,035-nucleotide (nt) fragment of the pPelican-LacZ vector (GenBank accession no. AF242360; nt 561 to 1596) into pCRII-TOPO. The αβ probe was prepared from pAB1γ (provided by S. Lakhotia) and consisted of a 961-nt fragment (GenBank accession no. AE014297.2; nt 8300427 to 8299466). A 226-nt Hsp70 probe was prepared from pHsp70 (provided by J. Lis). The rp49 probe was prepared from pGRP49 (39). Other probes were prepared from RE38782 (CG9894, Drosophila Genomics Resource Center), pGEMSgs4 (provided by S. Beckendorf), and the ng1 clone RE05239 (ResGen). RNA levels were quantitated using a Storm 800 PhosphorImager with ImageQuant5.2 analysis software (Molecular Dymanics).
Nuclear run-on experiments were performed as described by So and Roshbash (46) with the following modifications. Groups of 20 adult females (2 to 5 days old) were heat shocked or maintained at room temperature for 20 min and placed immediately on ice prior to homogenization. Radioactively labeled run-on RNA was hybridized to antisense RNA probes bound to a Hybond-N (GE Healthcare) membrane. The αβ and Hsp70 probes (961 nt and 226 nt, respectively) were the same as those used in the Northern analysis. The histone H4 plasmid (provided by R. Duronio) was used to synthesize a 575-nt probe (GenBank accession no. AE014134.5; nt 21425167 to 21424592). One microgram of each probe was slot blotted onto the nylon membrane. The hybridization reactions were performed overnight at 65°C.
The Invitrogen 3′ rapid amplification of cDNA ends (RACE) system was used to determine the 3′ ends of αβ RNAs. The oligo(dT)-containing universal primer with an adaptor sequence (AP; Invitrogen) was used for cDNA synthesis. An αβ-specific upstream primer (primer 4; [see Fig. 5C]) and the complementary universal primer AUAP (Invitrogen) were used for the PCR. Circular reverse transcriptase PCR (cRT-PCR) analysis was used to define the 5′ and 3′ RNA ends and was performed as previously described (12). Briefly, 10 μg of total RNA was treated with oligo(dT) and RNase H to remove all but a few nucleotides of the poly(A) tail, decapped with tobacco acid pyrophosphatase, and circularized with RNA ligase. The circular RNA was reverse transcribed into cDNA, and an aliquot of the cDNA was amplified by PCR. The primers used in the cDNA synthesis and PCRs were derived from the αβ region and are shown in Fig. 5C. Primer 1 was used in the reverse transcription reaction, whereas primers 2 and 3 or primers 2 and 5 were used for the PCR amplification of cDNAs. Thirty cycles of PCR were performed. The PCR products were examined by agarose gel electrophoresis, cloned into the pCRII-TOPO vector, and sequenced.
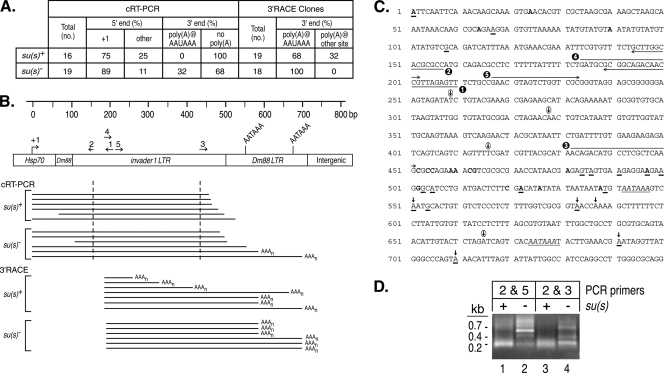
FIG. 5.
cRT-PCR and 3′ RACE experiments suggest that Su(s) promotes the degradation of αβ RNAs. (A) Summary of αβ cDNA clone sequence data. The RNA used in these experiments was from su(s)+ and su(s)− flies heat shocked at 32°C. (B) Schematic map of a portion of an αγ element. Numbered arrows above the map indicate the primers used in this analysis. Horizontal lines beneath the map indicate the types of cDNAs recovered in these experiments. The cDNA clones obtained by cRT-PCR contained sequences upstream and downstream of the dashed vertical lines. Poly(A) tails at the 3′ ends are indicated by AAAn. (C) Sequences of the first 750 bp of an αγ element. Transcription starts at +1. Primers used for cRT-PCR and 3′ RACE are indicated by the numbered arrows. Polyadenylation signals AATAAA are italicized and underlined. Bold vertical arrows above the sequences indicate canonical polyadenylation sites detected in cRT-PCR and 3′ RACE experiments. Circled vertical arrows indicate unusual polyadenylation sites detected only in 3′ RACE of su(s)+ RNA. The 5′ and 3′ ends of su(s)+ cDNA clones generated by cRT-PCR are shown in bold. The 5′ ends of these cDNAs were at positions 1, 24, 64, and 90; the 3′ ends were at positions 452, 454, 455, 459, 460, 462, 463, 481, 497, 502, 520, 522, 526, and 538. The 5′ and 3′ ends of su(s)− cDNA clones obtained by cRT-PCR are underlined. The 5′ ends were at positions 1, 67, and 108; the 3′ ends were at positions 484, 486, 491, 497, 500, 503, 505, 522, 539, 551, 554, 583, 691, and 710. (D) Ethidium bromide-stained agarose gel of cRT-PCR products prior to cloning. The PCR primers used are shown in panel C. Similarly sized fragments were seen with both primer sets from the su(s)+ samples. This is probably because most of the cDNAs were amplified from RNAs with intact 5′ ends and extensively degraded 3′ ends. Thus, many cDNAs were truncated within a short distance of the downstream primer (primer 3 or 5). In contrast, longer cDNAs were amplified from su(s)− RNA.
RNAi analysis.
DNA templates for double-stranded RNA (dsRNA) synthesis were PCR amplified from either cDNA clones or genomic DNA and consisted of the following segments of mRNA sequences: Su(s), nt 3855 to 4652; Rrp6, nt 1971 to 2518; Dis3, nt 1283 to 1799; Pcm, nt 1546 to 2446; Rat1, nt 107 to 1088; Trf4-1, nt 1345 to 1820; and Trf4-2, nt 517 to 983. The MEGAscript RNA interference (RNAi) kit (Ambion) was used to synthesize dsRNA from these templates. The LacZ template, provided with the kit, was used to prepare control dsRNA. Drosophila S2 cells (2 × 106 cells/well) were treated with 30 μg of dsRNA in a well of a six-well plate for 4 days as described previously (16), after which the cells from each well were divided into two equal portions. One set of samples was maintained at room temperature, and the other set was heat shocked by incubation at 32°C for 20 min. The cells were chilled and pelleted by centrifugation, and RNA was isolated using TRIzol reagent (Invitrogen). The RNA samples were analyzed by Northern blotting as described above.
Inverse PCR.
The position of the Hsp70 promoter transgene insertion in Bloomington Stock no. 10221 was determined using the protocol described on the Berkeley Drosophila Genome Project's website (http://www.fruitfly.org/about/methods/inverse.pcr.html). Genomic DNA was prepared from adults and digested with Sau3AI, and the resulting fragments were circularized in a DNA ligation reaction. Subsequently, the primers Pry1 and Pry4 were used to PCR amplify the fragment containing the transgene insertion. The fragment was cloned into the pCRII-TOPO vector and sequenced.
Transgenic DNA construct generation.
Various segments of an αγ element (see Fig. 7A) were cloned into the pPelican-LacZ transformation vector (Drosophila Genomics Research Center). The DNA fragments were PCR amplified from yw genomic DNA, and during this process, EcoRI and KpnI restriction sites were added to the 5′ and 3′ ends of the αγ segments, respectively. The primers used for the PCR amplification were γ5′ (5′-ACGAATTCCCGCATAACAAAGCGCTTCG-3′) and one of the following: αß-A (5′-GCGGTACCCCTTCTGCGCTTGTTTATTTGC-3′), αß-B (5′-GAGGTACCATGACAGTTGTTCTAGTCCGC-3′), αß-C (5′-CTGGTACCGTCGAAGAGTCATCAGGATGC-3′), or αß-D (5′-ATGGTACCCAGTCTGCGCGCTCTCAGC-3′). The fragments were cloned into the EcoRI and KpnI sites of the vector, upstream of the LacZ coding region. The resulting reporter gene constructs in pPelican are flanked by insulator elements, which prevent position-dependent effects on the expression of transgenes inserted at different genomic positions. The plasmids were injected into w1118 embryos at the Duke Model System Genomics facility. Transformant lines were identified and homozygous stocks were established using standard genetic crosses.
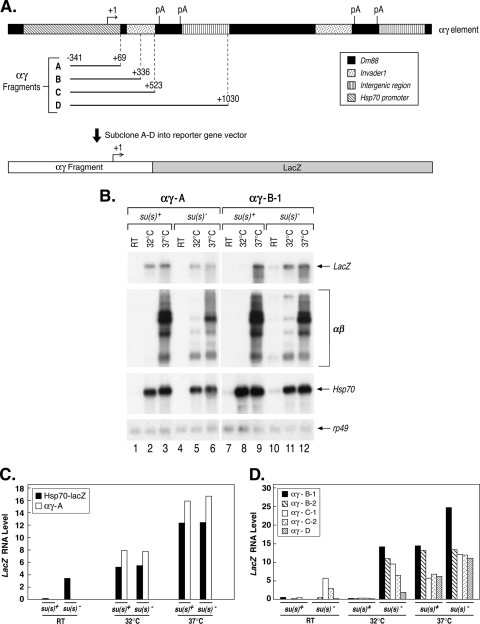
FIG. 7.
An Su(s) target site is found within the first 267 nt of αβ sequences. (A) Map of an αγ element, which consists of Hsp70 promoter and 5′ UTR sequences joined to αβ sequences. Four fragments (αγ A to D) were cloned into a LacZ reporter vector and transformed into flies. The numbers at the boundaries of the cloned fragments are relative to the Hsp70 transcription start site. pA, polyadenylation signal. (B) Representative Northern blots of RNA from two transformant lines after heat shock induction of the transgenes. Probes are indicated on the right. RT, control. (C) Graph illustrating the results of the Northern analysis of two heat shock-inducible LacZ reporter transgenes lacking αβ sequences. The RNAs produced by these transgenes are not regulated by Su(s). (D) Graph illustrating the results of the Northern blot analysis of experiments with three heat shock-inducible LacZ reporter transgenes containing variable amounts of αβ sequences. Two different transformant lines containing the αγ-B and αγ-C constructs were analyzed. Su(s) downregulates the RNAs produced by these transgenes at 32°C. The results shown in panels C and D are from two different experiments. LacZ RNA levels were normalized with rp49.
RESULTS
Su(s) localizes to a subset of RNA Pol II-associated sites on polytene chromosomes.
Su(s) was originally shown to downregulate the accumulation of transcripts produced by mutant alleles of several genes with transposon insertions in the transcribed region. To broaden our understanding of the types of transcripts that Su(s) regulates, we sought to identify other Su(s) targets. Thus, we performed immunofluorescence analysis to examine the global distribution of Su(s) on third-instar larval polytene chromosomes at specific puff stages and under heat shock conditions. Chromosomes were probed with antibodies that detect Su(s) and either Pol II or Hrp40, a member of the hnRNP A/B family that binds to nascent transcripts (31, 38).
Pol II that is bound to chromosomes can be present in various phosphorylation states because multiple sites within the CTD of the largest Pol II subunit are dynamically phosphorylated during transcription. The CTD consists of tandem copies of a seven-amino-acid repeat, which has the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7. Phosphorylation occurs primarily at two sites within this repeat, i.e., position 2 (Ser2∼P) and position 5 (Ser5∼P). Ser5 is phosphorylated during the early elongation phase, whereas Ser2 phosphorylation occurs subsequently. The predominant elongating polymerase is Pol IISer2∼P for some genes, whereas Pol IISer2∼P Ser5∼P performs elongation at other genes (40). Since various mRNA processing factors associate with specific phosphorylated forms of the CTD, these modifications are important for cotranscriptional RNA processing (21). Thus, the phosphorylation state of Pol II at sites where Su(s) is bound can be informative about whether active transcription and efficient RNA processing are likely to be occurring at those sites. The Pol II antibodies that we used detect unphosphorylated (anti-Pol IIa), Ser2∼P (H5), or Ser5∼P (H14) CTD repeats. In addition, we used an anti-Pol IIo antibody, which, as we determined in double-labeling experiments, produces the same immunofluorescence pattern as H5 (unpublished data). Thus, anti-Pol IIo also preferentially recognizes Ser2∼P.
While Pol II was present at all of the Su(s)-associated sites in these experiments, this was a subset of the loci that were bound by Pol II (Fig. 1 and 2). For example, at puff stage 7, Ser2∼P and Hrp40 were widely distributed at many sites in very similar patterns. Su(s) was also found at a number of these same sites during this stage, but little or no Su(s) was detected at the early ecdysone-induced puffs at 74EF and 75B or the late puff at 78C (Fig. 1A to B), and these are among the strongest Ser2∼P and Hrp40 sites.
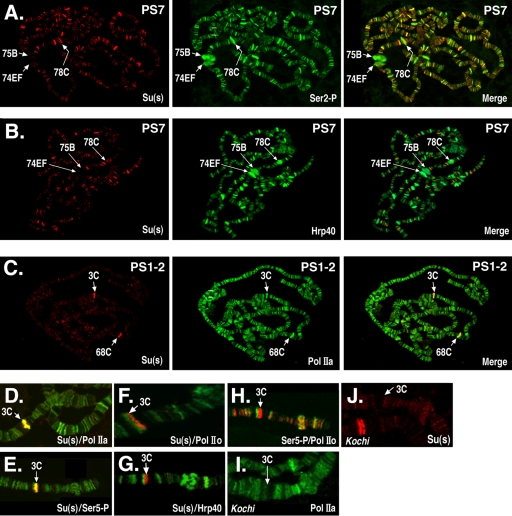
FIG. 1.
Su(s) localizes to a subset of Pol II-associated sites. The polytene chromosome distributions of Su(s) and either Pol II or Hrp40 were compared by immunofluorescence analysis. (A to C) Global Su(s) distribution at specific puff stages (PS). Arrows indicate the cytological positions of several prominent puffs. Chromosomes were probed with antibodies that recognize Su(s) (red) and either Pol II or Hrp40 (green). Sites of colocalization appear yellow in merged images. (D to J) Distribution of Su(s), various forms of Pol II, and Hrp40 at the 3C locus during PS 1 to 2. Genotypes: wild type (D to H); Kochi mutant with Sgs4 enhancer deletion (I and J). Panels D to H show merged images of chromosomes probed with the antibodies indicated in the lower right corner. The chromosomes shown in panels I and J were probed with a single antibody. Antibody signals are shown in the following colors: Su(s), red (D to H and J); Ser5∼P, green (E) and red (H); Pol IIa, green (D and I); Hrp40, green (G); and Pol IIo, green (F and H). Su(s) colocalizes at 3C with Pol IIa and Ser5∼P but not Ser2∼P or Hrp40. Pol IIa and Su(s) are absent from 3C in the Kochi mutant.
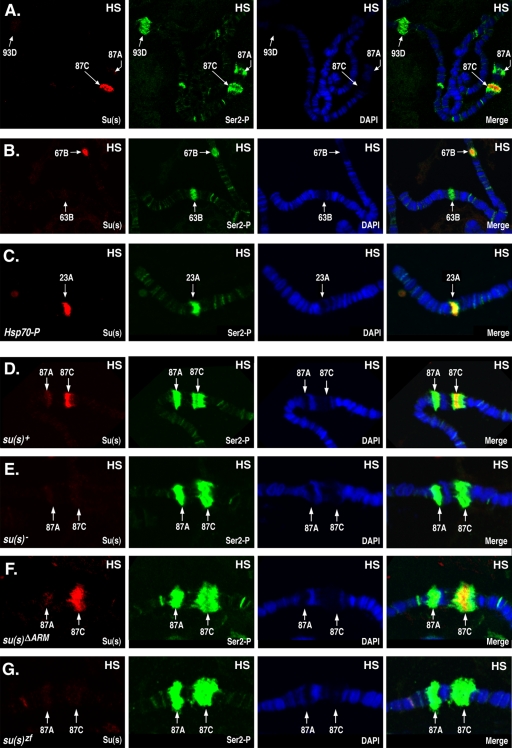
FIG. 2.
Su(s) localizes to a subset of heat shock (HS) puffs. Chromosomes from heat-shocked (37°C) larvae were probed to detect Su(s) (red) and Ser2∼P (green) and stained with DAPI (4′,6-diamidino-2-phenylindole; blue). Arrows indicate the cytological positions of heat shock loci. Chromosomes were obtained from the following stocks: su(s)+ (A, B, and D); su(s)+ transformant line with an Hsp70 promoter insertion at 23A (C); su(s) null mutant (E); and transformant lines carrying su(s)ΔARM and su(s)ZF mutant transgenes, respectively, in the su(s) null mutant background (F and G). Su(s) was detected at the endogenous 87C and 67B heat shock puffs and at the site of an ectopic Hsp70 promoter insertion at 23A. Su(s) was present at 87C in the su(s)ΔARM mutant but was absent from this region in the su(s) null and su(s)zf mutants.
In chromosomes of early wandering larvae (puff stage 1 to 2), a strong Su(s) signal was consistently detected at two sites, i.e., the intermoult puffs at 3C and 68C (Fig. 1C), but not at the other six intermoult puffs (data not shown). Both 3C and 68C contain clusters of salivary gland-specific genes. The examination of numerous images revealed that Su(s) colocalizes with a high level of Pol IIa at these sites, and the levels of Ser2∼P and Hrp40 were usually relatively low (Fig. 1C and data not shown). We focused on the 3C region to examine this in more detail (Fig. 1D to H). The strong Su(s) site at 3C coincided with strong Pol IIa and Ser5∼P sites (Fig. 1D to E). On the other hand, the Su(s) site could be resolved from the Hrp40 and Ser2∼P (anti-Pol IIo) site in this region (Fig. 1F to H). These results indicate that hypophosphorylated Pol IISer5∼P is the predominant form of Pol II present at the site in the 3C region where Su(s) is bound. Furthermore, the low levels of Ser2∼P, which is required for cotranscriptional polyadenylation (21), and Hrp40 suggest that mature mRNAs are not being generated at this site during this stage.
We examined the chromosomal distribution of Su(s) during heat shock, and under these conditions, a very strong Su(s) signal was detected at the 87C heat shock puff (Fig. 2A and D). Although Su(s) and Ser2∼P overlapped at this site, Su(s) was restricted to a subregion within the broader Ser2∼P signal. A moderate Su(s) signal was frequently detected at the 67B heat shock locus (Fig. 2B), which, like 87C, contains a cluster of heat shock protein (Hsp) genes. Su(s) also localized to the 23A region during heat shock in a fly stock (45) that carries a transgenic Hsp70 promoter fragment at that site (Fig. 2C). On the other hand, little or no Su(s) was detected at the other endogenous heat shock loci, e.g., 87A, 63B, and 93D (Fig. 2A, B, and D).
Su(s) inhibits the aberrant temporal accumulation of wild-type Sgs4 and ng1 RNA.
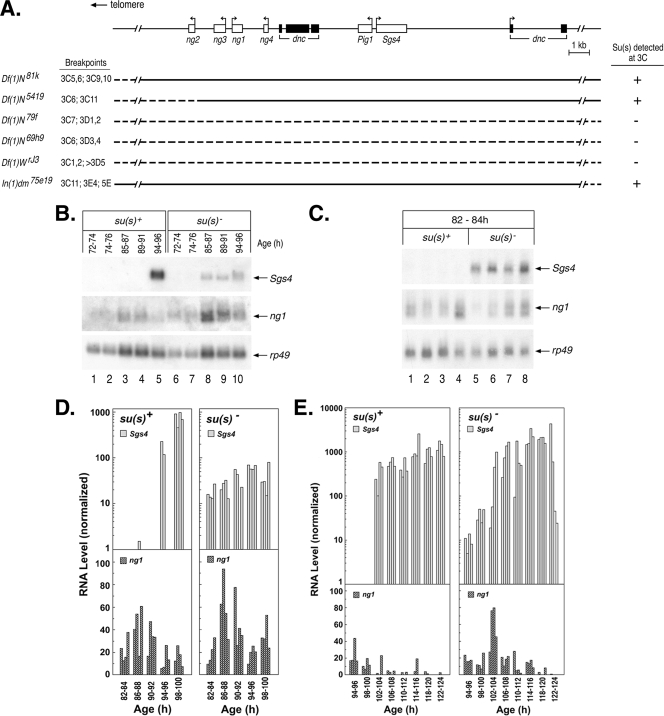
To determine how Su(s) affects the RNA transcripts at sites where it is bound, we performed molecular analysis on genes at a few sites where the identities of the relevant genes could be easily determined. One of the focal points of our molecular analysis was the 3C region of the X chromosome. Using existing deficiency stocks, we mapped the Su(s) localization site to the cytological interval 3C10 to 3C11 (Fig. 3A). This region contains a cluster of intronless salivary gland-specific genes (ng1, ng2, ng3, ng4, Pig1, and Sgs4) embedded within two introns of dunce, a large and complex gene with multiple transcription start sites. Sgs4 is expected to be expressed at the time when Su(s) was observed in this region. To determine if the transcription of Sgs4 is required for the localization of Su(s) at this site, we analyzed chromosomes of the Kochi stock, which has a 51-bp deletion that removes Sgs4 regulatory sequences (−305 to −356) and eliminates Sgs4 mRNA (29, 35). This deletion resulted in the loss of the Su(s) and Pol IIa signals at 3C (Fig. 1I and J). Thus, either the Sgs4 cis-regulatory region or the transcription that it induces is necessary for the recruitment of Su(s) to this locus.
FIG. 3.
Su(s) prevents aberrant temporal accumulation of wild-type Sgs4 and ng1 transcripts. (A) Schematic map of genes in the 3C10 to 3C11 region. Immunofluorescence analysis of deficiency stocks confirmed that Su(s) localization requires sequences within this genomic interval. Dashed lines indicate sequences deleted in each deficiency stock. (B, C) Representative developmental Northern blots of total RNA isolated from various stages (h after egg laying) of su(s)+ and su(s)− larvae. Probes used are shown on the right. Each lane contains RNA from multiple larvae (B) or a single larva (C). (D, E) Graphical representation of developmental Northern blots of RNA from individual larvae at various times during the early (D) and late (E) third-instar stages. Sgs4 and ng1 RNA signals were normalized to rp49. The data shown in panel C are included in panel D. The development of both the su(s)+ and su(s)− larvae in the experiment for which the results are shown in panel E was delayed relative to that of the larvae in the experiments for which the results are shown in panels B and D, i.e., Sgs4 induction occurred at 102 to 104 h instead of 94 to 96 h. This effect was probably related to the batch of larval food used in this experiment.
Since the results described above suggested that Sgs4 may be a regulatory target of Su(s), we next examined how the expression of Sgs4 is affected by the loss of su(s) function. The wild-type temporal patterns of Sgs4 and ng gene expression have been previously characterized (2, 13). This analysis has revealed a precise developmental switch in the expression of Sgs4 and three of the ng genes (ng1 to ng3). The ng genes are active at an earlier phase of the third instar than Sgs4, and Sgs4 is activated precisely at the point when ng1 to ng3 mRNA levels decrease. Thus, normally there is little or no overlap between the expression of Sgs4 and these ng genes.
To determine if the expression pattern is affected in an su(s) mutant, we performed developmental Northern blot analysis on total RNA isolated from carefully staged su(s)+ and su(s) null mutant larvae and initially examined Sgs4 and ng1 mRNA levels in samples prepared from pools of multiple larvae harvested at various intervals during the first day of the third instar (72 to 96 h; Fig. 3B). In the mutant larvae, Sgs4 mRNA began accumulating earlier than usual, during the time when ng1 RNA was expressed. To rule out the possibility that these larvae were not well synchronized, i.e., a few older larvae were present in the pool of young larvae, we subsequently analyzed RNA prepared from individual larvae (four per time point), collected at 2-h intervals. In these experiments, Sgs4 mRNA consistently accumulated prematurely in the su(s) mutant larvae (Fig. 3C to E). Furthermore, whereas Sgs4 mRNA rapidly increased to a high level in the wild-type larvae, in the su(s) mutant the time required to reach the maximal RNA level was longer than normal (Fig. 3D and E). A subtle difference was also seen in ng1 RNA accumulation between wild-type and su(s) mutant larvae (Fig. 3E). In su(s)+ larvae, the amount of ng1 RNA decreased before Sgs4 was activated and was barely detectable during the time of Sgs4 RNA accumulation. However, in the su(s) mutant, ng1 RNA was detected during this time with a second peak occurring precisely at the time that Sgs4 RNA levels increased (Fig. 3E). Thus, Su(s) also plays a role in preventing ng1 RNA accumulation when Sgs4 is active.
In summary, although the general expression patterns for these RNAs are similar in su(s)+ and su(s) mutant backgrounds, the finer details of this pattern are disrupted in su(s) mutants. Our results indicate that Su(s) plays a role in preventing the inappropriate accumulation of both Sgs4 and ng1 mRNA and maintaining distinct temporal expression profiles for these genes. Furthermore, Su(s) promotes the rapid accumulation of Sgs4 mRNA.
Su(s) inhibits the accumulation of RNAs transcribed from repetitive αβ and αγ elements in the vicinity of Hsp70 genes during a mild heat shock.
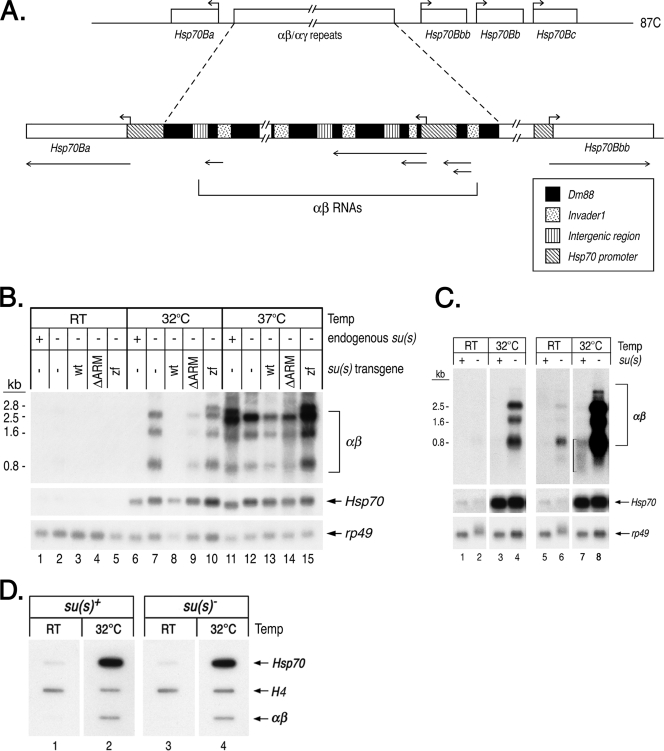
The 87C region where Su(s) localizes during heat shock is the site of four Hsp70 genes (Fig. 4A), and two other Hsp70 genes are located at 87A. While the 5′ regulatory and coding sequences of all Drosophila Hsp70 genes are highly conserved, a high level of Su(s) was observed at 87C but not at 87A during heat shock (Fig. 2A and D). A distinctive feature of the Hsp70 gene cluster at 87C is that two of the four Hsp70 genes in this region are separated by an approximately 38-kb cluster of tandemly repeated transposon fragments. This is the largest euchromatic cluster of transposons in the Drosophila genome, and it appears to have been created by multiple insertion and duplication events (25). Each repeat unit consists of a long terminal repeat (LTR) of the retrotransposon invader1, nested within a larger remnant of the retrotransposon Dm88 (Fig. 4A). Lis et al. (30) originally identified these repeated sequences and named them αβ elements. A subset of the repeats (the αγ elements) contains a duplicated copy of the Hsp70 promoter (a γ element) fused to an αβ repeat (20). These elements produce several noncoding, polyadenylated RNAs (αβ RNAs) during a 37°C heat shock. Based on their genomic position, we suspected that these might be the Su(s) targets. To determine if Su(s) regulates the accumulation of αβ transcripts, we used Northern blot analysis to examine RNA isolated from wild-type and su(s) mutant flies after a 32°C or 37°C heat shock. At 32°C, αβ transcripts accumulated in the su(s) mutant but not in the su(s)+ flies (Fig. 4B, lanes 6 and 7, and C, lanes 3 and 4). This indicates that Su(s) blocks αβ RNA accumulation at this temperature. At 37°C, αβ RNA accumulated in both su(s)+ and su(s) mutant flies (Fig. 4B, lanes 11 and 12). Thus, although Su(s) associates with the αβ region at 37°C (Fig. 2), the Su(s) regulatory pathway does not function efficiently at this temperature. This is not particularly surprising since several RNA metabolic processes, e.g., pre-mRNA splicing and mRNA degradation, are disrupted by heat shocking flies at 37°C (8, 54). In contrast to the effect on αβ RNA, the level of Hsp70 RNA was apparently unaffected by the absence of Su(s) (Fig. 4B to C). Since the heat shock response is relatively simple to induce and it occurs in all cell types, we focused most of our mechanistic studies on the αβ RNAs.
FIG. 4.
Su(s) inhibits the accumulation of noncoding αβ RNAs from the 87C region during a mild heat shock. (A) Schematic map of transcription units in the 87C region, which contains several Hsp70 genes and a cluster of tandemly repeated αβ/αγ elements. The αβ elements are segments of transposons invader1 and Dm88, whereas a γ element is a duplicated Hsp70 promoter fragment joined to αβ sequences. Hsp70 transcription start sites are indicated by the right-angled arrows above the map. Regions that give rise to αβ and Hsp70 RNAs are indicated by lines with arrows beneath the map. (B, C) Northern blots of RNA from heat-shocked and control (RT) flies. The abundance of αβ RNA is substantially lower than Hsp70 RNA; thus, blots probed to detect αβ RNAs were exposed for longer times. (B) RNA from su(s)+, the su(s) mutant, and flies carrying the su(s) transgenes indicated in the null mutant background. (C) RNA from su(s)+ and the su(s) null mutant. Two different exposures of the blot probed for αβ RNA are shown in lanes 1 to 4 and lanes 5 to 8. The bracket in lane 7 indicates the residual, heterogenous-sized αβ transcripts seen in su(s)+ RNA at 32°C. (D) Nuclear run-on experiment comparing the amount of elongating Pol II in the αβ/αγ region after a 32°C heat shock in wild-type and su(s) mutant adult flies. Internal control histone H4 genes are transcribed in the presence and absence of heat shock. Probes in panels B to D are indicated on the right. This analysis indicates that Su(s) does not regulate the transcription of the αβ/αγ region. wt, wild type.
The RNA-binding domains of Su(s) mediate its association with the αβ chromosomal region and the inhibition of αβ RNA accumulation.
We previously made Su(s) derivatives with alterations in the RNA-binding domains and generated transformant lines that stably express the mutant proteins in the su(s) null mutant background (27). To test whether these domains are required for the association of Su(s) with the 87C region, we performed immunofluorescence analysis on chromosomes from transgenic larvae expressing Su(s) derivatives with either a deletion of both arginine-rich motifs [Su(s)ΔARM] or with both zinc fingers mutated [Su(s)ZF]. We found that Su(s)ΔARM localized to the 87C region during heat shock, but Su(s)ZF did not (Fig. 2F and G). Thus, the zinc fingers are necessary for the stable association of Su(s) with this chromosomal region. The arginine-rich motifs do not appear to be required for this interaction. However, since this analysis is not quantitative, we cannot rule out the possibility that the interaction of Su(s)ΔARM with this locus is somewhat less stable than normal.
We used Northern blot analysis to determine if Su(s)ΔARM and Su(s)ZF were impaired in their ability to inhibit αβ RNA accumulation. This analysis showed that flies carrying the su(s)zf transgene accumulate a similar level of αβ RNA at 32°C as the su(s) null mutant (Fig. 4B, compare lanes 7 and 10). In the su(s)ΔARM mutant, the level of αβ RNA was higher than that observed in flies expressing wild-type Su(s) but not as high as in the null mutant (Fig. 4B, compare lane 9 to lanes 6 to 8). Based on multiple replicates of this experiment, we estimate that su(s)ΔARM flies accumulate 20 to 50% as much αβ RNA as the null mutant, whereas αβ RNA is undetectable in flies carrying a wild-type su(s) transgene (data not shown). Thus, the inhibitory effect of Su(s) on αβ RNA levels depends on the zinc fingers and, to a lesser extent, the arginine-rich motifs. At 37°C, the wild-type and mutant proteins were ineffective at inhibiting the accumulation of these transcripts (Fig. 4B, lanes 11 to 15).
Nuclear run-on and αβ cDNA analyses suggest that Su(s) promotes the degradation of αβ RNA.
Su(s) might inhibit RNA accumulation either by blocking transcription or by promoting the degradation of its RNA targets. To determine if Su(s) regulates transcription, we performed nuclear run-on analysis to compare the level of actively engaged RNA polymerases on αβ/αγ elements in su(s)+ and su(s) mutant flies during heat shock. We found that nuclei prepared from su(s)+ and su(s) mutant flies after a 32°C heat shock produce the same amount of αβ run-on RNA (Fig. 4D). This indicates that the transcription of αβ/αγ elements occurs at the same level in the presence and absence of Su(s). The amount of αβ run-on RNA was substantially lower than the amount of Hsp70 run-on RNA (Fig. 4D). Similar differences in αβ and Hsp70 RNA signals were observed in our Northern analysis. This may be because the genome contains fewer αγ elements (αβ repeats with an Hsp70 promoter upstream) than Hsp70 genes, i.e., one or two versus six, respectively. However, αβ and αγ elements may also be transcribed at a lower rate than Hsp70 genes.
Since the nuclear run-on results indicate that Su(s) does not regulate transcription, next we investigated whether Su(s) promotes the degradation of αβ RNA. The residual, heterogeneous-sized αβ transcripts detected in su(s)+ RNA at 32°C suggested that degradation was occurring (Fig. 4C, lane 7). To get an initial indication of whether this was the case, we generated cDNA clones of αβ RNAs from su(s)+ and su(s)− flies after a 32°C heat shock. Intact αβ RNAs were expected to start at the Hsp70 (γ) transcription initiation site and to be polyadenylated a short distance downstream of one of the consensus polyadenylation signals in the Dm88 LTR, centered at approximately +545 and +675 (Fig. 5B and C). These sequences are repeated further downstream at about +2140 and +2270 (data not shown). On the other hand, degraded transcripts were expected to be truncated from one or both of these ends.
One approach used to generate the cDNAs was cRT-PCR (12). This involved the circularization of total RNA to ligate the 5′ and 3′ ends of individual mRNA molecules, reverse transcription across the site where the ends are joined, and PCR amplification of cDNA fragments that span the joined ends. The sequencing of these cDNA clones provided information about the 5′ and 3′ mRNA ends. For cDNA synthesis, we used a primer located about 200 nt downstream of the Hsp70 (γ) transcription start site (primer 1; Fig. 5B and C), and we used two different primer pairs (primers 2 and 5 or primers 2 and 3; Fig. 5B and C) in the PCRs. The sizes of the PCR products obtained from the su(s)+ and su(s)− samples were different, as expected (Fig. 5D). Short PCR fragments (∼200 bp) were the predominant products produced from su(s)+ RNA with both sets of primers, whereas short as well as longer discrete cDNA fragments were produced from su(s)− RNA. We cloned and sequenced su(s)+ and su(s)− cDNA clones from the PCRs with primers 2 and 3. The 5′ ends of most of the clones from both backgrounds corresponded to the Hsp70 (γ) transcription start site (Fig. 5A to C). Thus, the 5′ ends were intact in most of the cDNA clones. The other 5′ ends were probably generated by degradation that occurred in vivo or during the purification of the RNA. On the other hand, all of the su(s)+ αβ cDNAs were truncated at the 3′ end at various sites upstream of the consensus polyadenylation signals. About one-third of the su(s)− αβ cDNAs were polyadenylated at the expected position downstream of the consensus sites, whereas others lacked a poly(A) tail and ended upstream of these sites (Fig. 5A to C). These results suggest that the αβ transcripts are preferentially degraded in the 3′→5′ direction and that this is more efficient in the presence of Su(s). Since the PCR conditions were not quantitative, this analysis probably overestimates the extent of degradation that occurs in the absence of Su(s) because shorter fragments are preferentially amplified. However, the detection of 3′ truncated αβ RNA in the mutant suggests that there may be an alternative, though less efficient, mechanism for degrading these RNAs in the absence of Su(s).
We also performed 3′ RACE to identify the 3′ ends of these RNAs. In this case, an oligo(dT)-containing primer with a unique adaptor sequence on the end was used for cDNA synthesis. Subsequently, an αβ primer (primer 4; Fig. 5B and C) and a complementary adaptor primer were used in the PCR. Thus, all of the cDNA clones isolated by this approach were derived from polyadenylated RNAs. All of the su(s)− and most of the su(s)+ αβ cDNA clones isolated after a 32°C heat shock were polyadenylated downstream of consensus polyadenylation signals. In addition, several su(s)+αβ cDNAs were polyadenylated at sites without a consensus poly(A) signal at the expected position upstream (Fig. 5A to C). These transcripts were truncated at several different 3′ endpoints and, thus, also appear to be degradation intermediates. Thus, the 3′ RACE analysis also indicates that αβ transcripts are subject to 3′→5′ RNA degradation in su(s)+ flies.
RNAi experiments reveal that the nuclear exosome degrades αβ RNA.
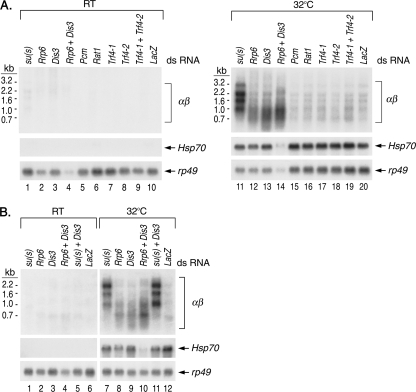
If Su(s) promotes the degradation of αβ RNA, then the depletion of the protein(s) involved should produce an effect similar to that of the depletion of Su(s), i.e., result in a higher level of these transcripts. Thus, we treated cultured Drosophila cells (S2 cell line) with dsRNA to reduce, by the RNAi pathway, the level of Su(s) and the Drosophila homologues of several RNA degradation proteins. The proteins tested included Rrp6 and Dis3 (or Rrp44), two subunits of the nuclear exosome, which is a complex of 3′→5′ exonucleases (23). Rrp6 is unique to the nuclear exosome, and Drosophila Dis3 appears to be predominantly nuclear (19). We also tested Trf4-1 and Trf4-2, which are homologous to the TRAMP subunits Trf4 and Trf5. These proteins stimulate the nuclear exosome by polyadenylating degradation intermediates (28) and generate RNAs with poly(A) tails at unusual positions like those seen in our 3′ RACE analysis. Finally, we tested two exonucleases that degrade RNA in the 5′→3′ direction, i.e., the cytoplasmic protein Pcm (or Xrn1) and its nuclear counterpart Rat1.
After being treated with dsRNA, cells were heat shocked at 32°C or maintained at room temperature. Then RNA was isolated and analyzed on Northern blots (Fig. 6). There were several interesting observations in these experiments. First, the treatment of cells with su(s) dsRNA increased αβ RNA accumulation, thereby confirming that the Su(s) regulatory pathway is functional in these cells (Fig. 6A, lane 11, and B, lane 7). Second, treatment with Rrp6 or Dis3 dsRNA alone partially increased αβ RNA levels, and this effect was enhanced by simultaneous treatment with Rrp6 and Dis3 dsRNAs (Fig. 6A, lanes 12 to 14, and B, lanes 8 to 10). The depletion of exosome components did not increase the level of the longer αβ RNAs to the extent observed when Su(s) was depleted, and the smaller fragments were more heterogeneous in length. These observations suggest that nuclear exosome function is reduced but not eliminated by treatment with Rrp6 and Dis3 dsRNAs. We did not attempt to reduce this activity further because the nuclear exosome is essential for cell viability. Nevertheless, these results indicate that the nuclear exosome degrades αβ RNA. A third interesting finding was that simultaneous treatment with su(s) and Dis3 dsRNA produced a similar effect as su(s) dsRNA treatment alone (Fig. 6B, compare lanes 7, 9, and 11). This is consistent with the notion that Su(s) functions in the same pathway as the exosome. The level of αβ RNA was slightly higher when both Su(s) and Dis3 were depleted. This is probably because dsRNA treatment reduces, but does not completely eliminate, the activity of these proteins. Thus, the activity of residual Su(s) is likely to be further reduced when nuclear exosome function is partially impaired. The other dsRNA treatments, which targeted Trf4 proteins, 5′→3′ exonucleases, or a decapping protein that acts in the 5′→3′ degradation pathway (data not shown), did not significantly increase αβ RNA above the negative control LacZ. Thus, TRAMP and 5′→3′ degradation RNA enzymes do not appear to be involved in regulating αβ RNA levels. However, since it is not possible at present to evaluate the effectiveness of the dsRNA treatment in reducing the activity of these proteins, this is not a definitive conclusion.
FIG. 6.
RNAi depletion experiments indicate that αβ transcripts are degraded by the nuclear exosome. Shown are Northern blots of RNA isolated from control (RT) or Drosophila S2 cells heat shocked at 32°C after treatment with the dsRNAs indicated above the lanes to deplete Su(s) and several RNA degradation proteins. LacZ dsRNA was used to control for the effects of dsRNA treatment. Probes are indicated on the right. The Northern blots in panel A were from different gels but were processed together. Normalization with rp49 indicates that about one-third as much RNA was loaded in lane 14 as lane 11. Thus, the depletion of Rrp6 and Dis3 may have a more pronounced effect than the αβ signal intensity in lane 14 indicates. The samples for which the results are shown in panel B were from a different experiment.
Su(s)-mediated inhibition depends on sequences located within the first 267 nt of αβ RNA.
To map the αβ sequences that mediate the inhibitory activity of Su(s), we generated four transgenic LacZ reporter constructs that contained various segments of an αγ element, which consists of an Hsp70 promoter joined by an in vivo duplication event to αβ sequences (Fig. 7A). All four reporter constructs contained the Hsp70 (γ) promoter and 5′ UTR fragment (−341 to +69) and thus were expected to transcribe LacZ sequences during heat shock. A control construct (αγ-A) contained no repetitive αβ sequences. The three others contained the first 267 bp, 454 bp, or 961 bp of αβ sequences immediately downstream of +69 (αγ-B, αγ-C, and αγ-D, respectively). In addition, we obtained an existing transformant line containing an Hsp70-LacZ reporter gene with an Hsp70 promoter fragment extending from −194 to +265 (44).
We examined the expression of all five transgenes in wild-type and su(s) mutant adults during heat shock by Northern blot analysis. The blots were sequentially probed to detect LacZ-containing RNA produced by the transgenes as well as the endogenous αβ, Hsp70, and rp49 transcripts (Fig. 7B and data not shown). Graphical representations of the results of the LacZ RNA analysis are shown in Fig. 7C and D. In these experiments, LacZ RNA accumulated at the same level at 32° in su(s)+ and su(s)− flies carrying the control transgenes Hsp70-LacZ or αγ-A (Fig. 7B, lanes 2 and 5, and C). In contrast, LacZ RNA was absent in su(s)+ flies carrying the αγ-B, αγ-C, or αγ-D transgene (Fig. 7B, lane 8, and D). At 37°C LacZ RNA accumulated in all of the transformant lines. Since the Su(s) regulatory pathway does not function efficiently at 37°C, the accumulation of LacZ RNA at this temperature confirms that the transgenes were present in the flies. In summary, these results indicate that Su(s)-mediated inhibition is not determined by the Hsp70 promoter but, instead, depends on αβ sequences. Since the shortest construct that is regulated by Su(s) contains the first 267 bp of the αβ region, an Su(s) target site must be present within this fragment, which consists of 39 bp of Dm88 and 237 bp of invader1 LTR sequences. These observations are consistent with a model in which the binding of Su(s) to this region causes these transcripts to be targeted for degradation. However, additional experiments, beyond the scope of this study, will be required to test this hypothesis.
Su(s) inhibits the accumulation of an aberrant RNA that is induced by an ectopic Hsp70 promoter insertion in the 23A region of the second chromosome.
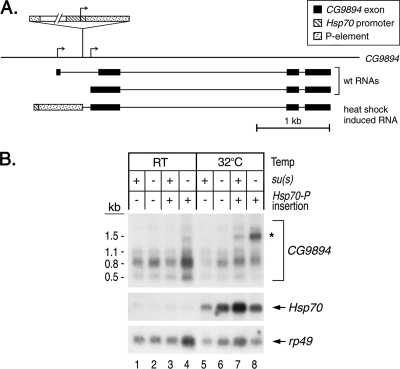
The experiment in Fig. 2C showed that Su(s) is recruited to chromosomal band 23A in a fly stock that has a transgenic Hsp70 promoter and a 5′ UTR fragment without any downstream coding sequences (i.e., −194 to +65) at that site (45). During heat shock, transcripts initiating at this ectopic promoter are expected to extend into the downstream genomic region and produce an aberrant transcript. To determine if Su(s) regulates the accumulation of this transcript, we identified the precise position of the insertion and then used this information to identify a probe capable of detecting transcripts from this region. Using inverse PCR, we mapped the transgene insertion to a site in the 5′ region of CG9894, a gene with two transcription start sites. The transgene is located within the first intron of the longest CG9894 pre-mRNA, and it is 101 bp upstream of the second transcription start site (Fig. 8A). An RNA that begins at the Hsp70 transcription start site and extends through CG9894 is predicted to be about 0.7 kb larger than the CG9894 RNA that initiates downstream of the insertion site.
FIG. 8.
Su(s) inhibits the accumulation of an aberrant transcript induced by an ectopic Hsp70 promoter (Hsp70-P) insertion in CG9894 at 23A. (A) Schematic map illustrating the position of the insertion in CG9894 and the RNAs predicted to be made from this region. Flies carrying the ectopic Hsp70-P insertion are expected to transcribe a novel RNA during heat shock. (B) Northern blot of RNA from heat-shocked and control (RT) flies carrying this transgene. Probes are indicated on the right. At 32°C, a 1.5-kb CG9894 transcript (indicated by the asterisk) accumulates at a higher level in su(s) mutants carrying the Hsp70-P insertion than in su(s)+ flies. RT, room temperature.
We performed genetic crosses to generate males carrying either an su(s)+ or su(s) null allele on the X chromosome and one copy of the Hsp70 promoter insertion at CG9894. These males and control males lacking this insertion were heat shocked at 32°C or maintained at room temperature, and RNA from these flies was analyzed by Northern blotting. Although the Drosophila genome sequence database (Flybase) indicates that CG9894 encodes two transcripts (Fig. 8A), we detected three RNAs, the major transcript being 0.8 kb, in flies lacking the transgene (Fig. 8, lanes 1, 2, 5, and 6). In heat-shocked flies with the insertion at CG9894, a larger 1.5-kb transcript was observed (Fig. 8, lanes 7 and 8). This RNA accumulated at a substantially higher level (at least fourfold) in the su(s) mutant. Based on our other results, it is likely that Su(s) binds to this aberrant transcript and stimulates its degradation. Thus, the activity of Su(s) prevents this insertion from inducing an aberrant mRNA, most likely generated by read through from the ectopic promoter.
DISCUSSION
Su(s) promotes RNA degradation.
The identification of the machinery required for destroying aberrant transcripts in animal cells is a key challenge. Previous studies by our lab and others had shown that Su(s) downregulates the level of several aberrant mutant RNAs, but the mechanism involved was unclear. In this report, we have presented evidence that Su(s) inhibits RNA accumulation by a degradation mechanism that involves the nuclear exosome. Since the Saccharomyces cerevisiae nuclear exosome performs a key role in nuclear RNA quality control (or surveillance) (23), this suggests that Su(s) also functions in this process. The types of transcripts that Su(s) regulates, e.g., mutant RNAs with insertions that disrupt RNA processing, cryptic Sgs4 RNA, and repetitive αβ transcripts, are similar to some of the types of transcripts that are degraded by the yeast nuclear exosome. For example, in yeast, this complex degrades incompletely processed transcripts (9, 10), cryptic intergenic transcripts (53), and cryptic repetitive transcripts arising from regions of repressed chromatin (22). In addition, the nuclear exosome regulates the processing or accumulation of a subset of normal pre-mRNAs (11, 42) and both normal and aberrant rRNA and tRNA (52).
Su(s) may be an RNA-binding cofactor of the nuclear exosome.
We showed that RNA-binding domains, primarily the CCCH zinc fingers, mediate the interaction of Su(s) with the αγ/αβ region and are required for the downregulation of αβ RNA. Furthermore, our reporter gene assay defined a 267-nt segment of the αβ transcribed region that is sufficient to target RNA to the Su(s) pathway. Thus, although we have not demonstrated that Su(s) interacts directly with these sequences, these observations suggest that Su(s) binding stimulates RNA degradation by the exosome. There are several possible ways that this could happen. Perhaps in some instances Su(s) functions as a nuclear exosome cofactor like the yeast RNA-binding protein Nrd1/Nab3 (47, 49). Vasiljeva and Buratowski (49) proposed that Nrd1/Nab3 binds to RNA sequences that are normally removed during processing, and when processing is inefficient, the association of Nrd1/Nab3 with the pre-mRNA activates degradation by the exosome. Thus, the fate of a nuclear pre-mRNA might be determined by competition between normal processing and quality control reactions. Alternatively, Su(s) might stimulate degradation by inhibiting a step in 3′ end formation. For example, since RNAs with short poly(A) tails are good substrates for the exosome (see reference 24), it is conceivable that Su(s) binding to pre-mRNA inhibits poly(A) addition at the 3′ end by canonical poly(A) polymerase. This, in turn, might trigger the degradation of the transcript by the nuclear exosome. This model could explain why the degradation intermediates detected in our cDNA analysis appear to be progressively truncated at the 3′ end from sites near the consensus polyadenylation signals (Fig. 5B).
Su(s)-mediated RNA surveillance targets transposon-containing and wild-type coding RNAs.
Su(s) target sites are apparently found in multiple transposable elements: sense strand Dm88/invader1 LTR sequences (this study), antisense 412 LTR sequences (14, 15), and antisense P element sequences (17). The activity of Su(s) prevents the accumulation of several types of transposon-associated transcripts, including mutant transcripts that cannot be efficiently processed due to transposon sequences in the pre-mRNA, transcripts arising from transposons (αβ/αγ elements) inserted in the vicinity of actively transcribed genes, and possibly read-through transcripts that initiate in a transposon and extend into an adjacent gene (Fig. 8). This mechanism for targeting transposon-containing RNAs for degradation could be important in minimizing the deleterious effects of transposon insertions.
Su(s)-mediated regulation also ensures precise control of the expression of wild-type coding RNAs, i.e., it prevents Sgs4 (and ng1) RNA from accumulating at an inappropriate time when nearby genes are being expressed. It is unclear how cryptic Sgs4 and ng1 transcripts, produced at the wrong time, could be unstable, while the same transcripts produced at the appropriate stages are stable, but the model described above can explain how this might happen. For example, perhaps Su(s) binds to Sgs4 pre-mRNA sequences in the 3′ region that are rapidly removed when polyadenylation is efficient. The transcripts produced at the wrong time might be inefficiently polyadenylated, e.g., because Pol II is not properly phosphorylated. Prior studies have shown that efficient, transcription-coupled polyadenylation depends on CTD phosphorylation at Ser2 (1, 37), and Su(s) colocalizes with hypophosphorylated Pol IISer5∼P in the Sgs4 region. Thus, perhaps a form of Pol II that does not promote polyadenylation performs the cryptic transcription of Sgs4 and ng1.
We suspect that the rate of Sgs4 mRNA synthesis is low at the time when the levels of Su(s) and hypophosphorylated Pol IISer5∼P are high in this region. The Su(s) and Pol IIa signals at the 3C locus were very strong with chromosomes of both prewandering and early wandering larvae. Under our growth conditions, larvae typically begin wandering at a time (112 to 114 h) when ng genes are repressed and Sgs4 RNA is approaching the maximal level. Thus, the rate of Sgs4 mRNA synthesis is expected to be relatively low during this time. Perhaps Su(s) plays a role in shutting down the production of mRNA from this region or in preventing the synthesis of cryptic Sgs4 transcripts when the downstream dunce exons are being transcribed (Fig. 3A).
Together, these results indicate that Su(s) participates in several important processes that ensure that aberrant RNAs are degraded and that prevent functional genes from inappropriately producing mRNA. Future studies will hopefully provide additional mechanistic insights into the details of these processes.
Acknowledgments
We thank the following individuals for providing materials used in this study: S. Artavanis-Tsakonas, A. Hofmann, and J. Lis for fly stocks; A. Greenleaf and G. Dreyfuss for antibodies; and S. Beckendorf, R. Duronio, S. Lakhotia, and J. Lis for plasmids. We thank A. Hutchins, M. Peifer, and the reviewers for providing helpful comments on the manuscript.
This work was supported by grants MCB-0111821, MCB-0417019, and MCB-0517103 from the National Science Foundation.
Footnotes
Published ahead of print on 17 August 2009.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 2.Andres, A. J., J. C. Fletcher, F. D. Karim, and C. S. Thummel. 1993. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev. Biol. 160:388-404. [DOI] [PubMed] [Google Scholar]
- 3.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrulis, E. D., J. Werner, A. Nazarian, H. Erdjument-Bromage, P. Tempst, and J. T. Lis. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420:837-841. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner, M. 1978. Puffing of polytene chromosomes, p. 316-376. In M. Ashburner and T. R. F. Wright (ed.), The genetics and biology of Drosophila, vol. 2b. Academic Press, London, United Kingdom. [Google Scholar]
- 6.Blackshear, P. J. 2002. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30:945-952. [DOI] [PubMed] [Google Scholar]
- 7.Boehm, A. K., A. Saunders, J. Werner, and J. T. Lis. 2003. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 23:7628-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonisch, C., C. Temme, B. Moritz, and E. Wahle. 2007. Degradation of hsp70 and other mRNAs in Drosophila via the 5′-3′ pathway and its regulation by heat shock. J. Biol. Chem. 282:21818-21828. [DOI] [PubMed] [Google Scholar]
- 9.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765-775. [DOI] [PubMed] [Google Scholar]
- 10.Burkard, K. T., and J. S. Butler. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 20:604-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciais, D., M. T. Bohnsack, and D. Tollervey. 2008. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res. 36:3075-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couttet, P., M. Fromont-Racine, D. Steel, R. Pictet, and T. Grange. 1997. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. USA 94:5628-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Avino, P. P., S. Crispi, L. C. Polito, and M. Furia. 1995. The role of the BR-C locus on the expression of genes located at the ecdysone-regulated 3C puff of Drosophila melanogaster. Mech. Dev. 49:161-171. [DOI] [PubMed] [Google Scholar]
- 14.Fridell, R. A., A. M. Pret, and L. L. Searles. 1990. A retrotransposon 412 insertion within an exon of the Drosophila melanogaster vermilion gene is spliced from the precursor RNA. Genes Dev. 4:559-566. [DOI] [PubMed] [Google Scholar]
- 15.Fridell, R. A., and L. L. Searles. 1994. Evidence for a role of the Drosophila melanogaster suppressor of sable gene in the pre-mRNA splicing pathway. Mol. Cell. Biol. 14:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatfield, D., L. Unterholzner, F. D. Ciccarelli, P. Bork, and E. Izaurralde. 2003. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geyer, P. K., A. J. Chien, V. G. Corces, and M. M. Green. 1991. Mutations in the su(s) gene affect RNA processing in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 88:7116-7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glisovic, T., J. L. Bachorik, J. Yong, and G. Dreyfuss. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582:1977-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, A. C., D. L. Kiss, and E. D. Andrulis. 2006. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol. Biol. Cell 17:1399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackett, R. W., and J. T. Lis. 1981. DNA sequence analysis reveals extensive homologies of regions preceding hsp70 and alphabeta heat shock genes in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 78:6196-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirose, Y., and Y. Ohkuma. 2007. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J. Biochem. 141:601-608. [DOI] [PubMed] [Google Scholar]
- 22.Houseley, J., K. Kotovic, A. El Hage, and D. Tollervey. 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 26:4996-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houseley, J., J. LaCava, and D. Tollervey. 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7:529-539. [DOI] [PubMed] [Google Scholar]
- 24.Houseley, J., and D. Tollervey. 2009. The many pathways of RNA degradation. Cell 136:763-776. [DOI] [PubMed] [Google Scholar]
- 25.Kaminker, J. S., C. M. Bergman, B. Kronmiller, J. Carlson, R. Svirskas, S. Patel, E. Frise, D. A. Wheeler, S. E. Lewis, G. M. Rubin, M. Ashburner, and S. E. Celniker. 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3:RESEARCH0084. [DOI] [PMC free article] [PubMed]
- 26.Kim, N., J. Kim, D. Park, C. Rosen, D. Dorsett, and J. Yim. 1996. Structure and expression of wild-type and suppressible alleles of the Drosophila purple gene. Genetics 142:1157-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuan, Y. S., P. Brewer-Jensen, and L. L. Searles. 2004. Suppressor of sable, a putative RNA-processing protein, functions at the level of transcription. Mol. Cell. Biol. 24:3734-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713-724. [DOI] [PubMed] [Google Scholar]
- 29.Lehmann, M., and G. Korge. 1995. Ecdysone regulation of the Drosophila Sgs-4 gene is mediated by the synergistic action of ecdysone receptor and SEBP 3. EMBO J. 14:716-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lis, J. T., D. Ish-Horowicz, and S. M. Pinchin. 1981. Genomic organization and transcription of the alpha beta heat shock DNA in Drosophila melanogaster. Nucleic Acids Res. 9:5297-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matunis, E. L., R. Kelley, and G. Dreyfuss. 1994. Essential role for a heterogeneous nuclear ribonucleoprotein (hnRNP) in oogenesis: hrp40 is absent from the germ line in the dorsoventral mutant squid. Proc. Natl. Acad. Sci. USA 91:2781-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matunis, E. L., M. J. Matunis, and G. Dreyfuss. 1993. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J. Cell Biol. 121:219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore, M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514-1518. [DOI] [PubMed] [Google Scholar]
- 34.Murray, M. V., M. A. Turnage, K. J. Williamson, W. R. Steinhauer, and L. L. Searles. 1997. The Drosophila suppressor of sable protein binds to RNA and associates with a subset of polytene chromosome bands. Mol. Cell. Biol. 17:2291-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muskavitch, M. A., and D. S. Hogness. 1980. Molecular analysis of a gene in a developmentally regulated puff of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 77:7362-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narita, T., Y. Yamaguchi, K. Yano, S. Sugimoto, S. Chanarat, T. Wada, D. K. Kim, J. Hasegawa, M. Omori, N. Inukai, M. Endoh, T. Yamada, and H. Handa. 2003. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 23:1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13:55-65. [DOI] [PubMed] [Google Scholar]
- 38.Norvell, A., R. L. Kelley, K. Wehr, and T. Schupbach. 1999. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 13:864-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connell, P. O., and M. Rosbash. 1984. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 12:5495-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phatnani, H. P., and A. L. Greenleaf. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20:2922-2936. [DOI] [PubMed] [Google Scholar]
- 41.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth, K. M., M. K. Wolf, M. Rossi, and J. S. Butler. 2005. The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol. Cell. Biol. 25:1577-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saunders, A., J. Werner, E. D. Andrulis, T. Nakayama, S. Hirose, D. Reinberg, and J. T. Lis. 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301:1094-1096. [DOI] [PubMed] [Google Scholar]
- 44.Simon, J. A., and J. T. Lis. 1987. A germline transformation analysis reveals flexibility in the organization of heat shock consensus elements. Nucleic Acids Res. 15:2971-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon, J. A., C. A. Sutton, R. B. Lobell, R. L. Glaser, and J. T. Lis. 1985. Determinants of heat shock-induced chromosome puffing. Cell 40:805-817. [DOI] [PubMed] [Google Scholar]
- 46.So, W. V., and M. Rosbash. 1997. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 16:7146-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiebaut, M., E. Kisseleva-Romanova, M. Rougemaille, J. Boulay, and D. Libri. 2006. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell 23:853-864. [DOI] [PubMed] [Google Scholar]
- 48.Turnage, M. A., P. Brewer-Jensen, W. L. Bai, and L. L. Searles. 2000. Arginine-rich regions mediate the RNA binding and regulatory activities of the protein encoded by the Drosophila melanogaster suppressor of sable gene. Mol. Cell. Biol. 20:8198-8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasiljeva, L., and S. Buratowski. 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell 21:239-248. [DOI] [PubMed] [Google Scholar]
- 50.Voelker, R. A., W. Gibson, J. P. Graves, J. F. Sterling, and M. T. Eisenberg. 1991. The Drosophila suppressor of sable gene encodes a polypeptide with regions similar to those of RNA-binding proteins. Mol. Cell. Biol. 11:894-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weeks, J. R., S. E. Hardin, J. Shen, J. M. Lee, and A. L. Greenleaf. 1993. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 7:2329-2344. [DOI] [PubMed] [Google Scholar]
- 52.Wolin, S. L., and K. M. Reinisch. 2006. The Ro 60 kDa autoantigen comes into focus: interpreting epitope mapping experiments on the basis of structure. Autoimmun. Rev. 5:367-372. [DOI] [PubMed] [Google Scholar]
- 53.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725-737. [DOI] [PubMed] [Google Scholar]
- 54.Yost, H. J., and S. Lindquist. 1986. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell 45:185-193. [DOI] [PubMed] [Google Scholar]