Abstract
In all telomerases, the template region of the RNA subunit contains a region of telomere homology that is longer than the unit telomeric repeat. This allows a newly synthesized telomeric repeat to translocate back to the 3′ end of the template prior to a second round of telomeric repeat synthesis. In the yeast Kluyveromyces lactis, the telomerase RNA (Ter1) template has 30 nucleotides of perfect homology to the 25-bp telomeric repeat. Here we provide strong evidence that three additional nucleotides at positions −2 through −4 present on the 3′ side of the template form base-pairing interactions with telomeric DNA. Mutation of these bases can lead to opposite effects on telomere length depending on the sequence permutation of the template in a manner consistent with whether the mutation increases or decreases the base-pairing potential with the telomere. Additionally, mutations in the −2 and −3 positions that restore base-pairing potential can suppress corresponding sequence changes in the telomeric repeat. Finally, multiple other yeast species were found to also have telomerase RNAs that encode relatively long 7- to 10-nucleotide domains predicted to base pair, often with imperfect pairing, with telomeric DNA. We further demonstrate that K. lactis telomeric fragments produce banded patterns with a 25-bp periodicity. This indicates that K. lactis telomeres have preferred termination points within the 25-bp telomeric repeat.
Telomeres are DNA and protein complexes present at the ends of eukaryotic chromosomes that function to protect chromosome ends from terminal sequence losses and fusions (3, 36). Telomeric DNA is typically composed of tandem 5- to 26-bp repeats that are sufficient for telomere function and that serve as binding sites for telomeric proteins (32). The ribonucleoprotein enzyme telomerase adds telomeric repeats to chromosome ends and prevents the gradual shortening that would otherwise occur. Telomerase synthesizes new telomeric repeats onto chromosome ends by using part of its RNA subunit as a template (13, 14, 31). Cells without telomerase encounter growth and viability problems once telomeres begin to become too short to properly function. In most human cells, telomerase activity is greatly reduced or absent and the ensuing telomere shortening functions to inhibit the formation of cancer by limiting the number of divisions that cells can undergo (4, 7, 16, 30).
Recognition of a telomeric end by telomerase in vivo is complex and requires a number of different interactions between components of telomerase and components of the telomere (32). Specialized proteins that bind the 3′ single-stranded overhangs of telomeres, including the yeast Cdc13 protein, can interact directly with telomerase (9, 28). A critical aspect of telomerase's interaction with the telomeres comes through base pairing between the telomeric overhang and the template region of the telomerase RNA. In all known telomerases, the template region of the RNA subunit contains a region of telomere homology that is longer than the unit telomeric repeat. This presence of short sequence identities at the 3′ and 5′ borders of the template allow a newly synthesized telomeric repeat to translocate back to the 3′ end of the template prior to a second round of telomeric repeat synthesis (38).
Kluyveromyces lactis is an ascomycetous yeast species that is a valuable model organism for studying telomeres and telomerase. Unlike the better-studied yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, K. lactis has telomeres composed of repeats of uniform size (25 bp) and sequence (24). This indicates that the translocation step during a round of DNA synthesis by the telomerase enzyme normally occurs between precise positions at the two ends of the telomerase RNA template region. Point mutations at any of multiple positions within either of the two 5-nucleotide (nt)-long direct repeats that border the telomerase RNA template result in telomeric repeats of abnormal size (35). These appear to result from disruption of the normal base-pairing interactions between template and telomeric DNA during the translocation step.
Here we present genetic data that argue strongly that three additional nucleotides 3′ of the template and outside the region of continuous homology with the telomeric repeat are involved in the base pairing between telomeric DNA and the telomerase RNA template in K. lactis. Sequence data suggest that similar extended base-pairing regions are widespread in other yeast species.
MATERIALS AND METHODS
Strains.
All strains used in this study are derivatives of the Kluyveromyces lactis haploid strain 7B520 (ura3-1 his2-2 trp1) (37). The ter1 mutations were made in the plasmid pTER-BX-UA (26) with the QuikChange mutagenesis kit as described by the manufacturer (Stratagene, La Jolla, CA). Typically, oligonucleotides of 25 to 35 nt were used. The TpD mutation was constructed in two steps, one to add bases to one side of the template and the other to delete bases from the other side by using oligonucleotides of 59 and 45 bp, respectively. Yeast mutants were constructed in strain Bcl-6-2, a derivative of the 7B520 strain that had the TER1 gene recently replaced (within ∼100 cell divisions) with TER1-7C(Bcl) by the loop-in, loop-out protocol (27). The TER1-7C(Bcl) allele is phenotypically silent but creates BclI restriction sites in the TER1 gene and in any terminal telomeric repeats formed by the mutant telomerase. The presence of TER1-7C(Bcl) allowed us to use BclI digestion to determine whether any newly made plasmid loop-out derivative contained the newly introduced ter1 mutation or whether it retained the original TER1-7C(Bcl) allele (34). The use of recently constructed TER1-7C(Bcl) cells to start the new template mutagenesis ensured that the final mutants would contain relatively few Bcl telomeric repeats. Once mutant strains were passaged for approximately five streaks, they would be expected to have turned over all or nearly all of any Bcl repeats that might initially have been present. Previously characterized ter1 template mutants were built in the 7B520 strain (25, 26).
K. lactis transformation.
K. lactis cells were transformed via electroporation. Typically, a 500-ml yeast culture was grown to an optical density at 600 nm of 0.5 to 1.5 in YPD (yeast extract-peptone-dextrose) medium. Cells were then pelleted and resuspended in 80 ml cold sterile water. Ten milliliters of 10× TE (Tris-HCl, EDTA, pH 7.5) followed by 10 ml of 1 M lithium acetate (pH 7.5) was then added, and the cell suspension was shaken gently at 30°C for 45 min. A 2.5-ml amount of 1 M dithiothreitol was then added, and shaking was continued for an additional 15 min. Cells were then twice pelleted and washed in ice-cold sterile water. A final wash was then done with 25 ml ice-cold 1 M sorbitol, and cells were resuspended in 0.5 ml cold 1 M sorbitol. Forty microliters of these cells was incubated on ice for 2 to 5 min with plasmid DNA prior to electroporation at 1,500 to 1,700 V in an Eppendorf electroporator (model 2510) using 2-mm cuvettes. An 0.5-ml amount of 1 M ice-cold sorbitol was added to the cell mixture, which was then placed on synthetic defined dropout medium lacking uracil to select for transformants. Plasmid loop-outs were selected on synthetic complete plates containing 500 μg/ml 5-fluoroorotic acid.
To be fully confident that any phenotypes observed in yeast mutants were due to the introduction of the intended mutation, the experimental design included redundancy at two steps. For each mutation discussed in this work, two independent plasmids containing the desired mutation (confirmed by sequencing) were generated. From each of these, two independently derived K. lactis strains containing the mutation were then isolated from each plasmid. Thus, four yeast strains containing each mutation were isolated and used for serial passaging on YPD plates. This was carried out by restreaking to single colonies at 30°C every 3 to 4 days. Each streak is estimated to represent 20 to 25 cell divisions. The telomere phenotypes of the four strains made of a single mutant were typically very similar.
Southern blotting and hybridizations.
Yeast genomic DNA was prepared from 1.5 ml overnight liquid YPD cultures grown at 30°C with the final resuspension being in 30 μl TE. Genomic DNAs digested with restriction enzymes were generally separated on 0.8% agarose gels run at 30 V for 17 h. DNA digested with restriction enzyme BsrBI was run on 2.5% 3:1 agarose (Cambrex Bio Science Rockland, Inc.) gels run at 65 V for 19 h. All gels were blotted onto Hybond N+ membranes (Amersham Biosciences, Piscataway, NJ) after being stained with ethidium bromide. Hybridizations were carried out in 500 mM Na2HPO4 and 7% sodium dodecyl sulfate at 45 to 55°C as previously described (6) with [γ-32P]ATP-labeled oligonucleotide Klac1-25 (5′-ACGGATTTGATTAGGTATGTGGTGT-3′) used as a probe. Washes were done with 100 mM Na2HPO4-2% sodium dodecyl sulfate. TER1 sequence probes were labeled using [α-32P]dATP and the Prime-it II random primer labeling kit (Stratagene, La Jolla, CA). These probes were used at 65°C to determine which TER1 allele remained in yeast plasmid loop-out clones that had been digested with BclI and EcoRI.
RESULTS
Mutations at positions −2 to −4 of the K. lactis telomerase RNA template region affect telomere length in a manner consistent with base pairing with the telomeric repeat.
The telomerase RNA of the yeast Kluyveromyces lactis contains a 30-nt segment of perfect telomere homology that serves as a template for the synthesis of the uniform 25-bp telomeric repeats in this organism (26). The 5-nt repeats on the borders of this template region function during the translocation step of DNA synthesis by the telomerase enzyme (35). During translocation, the telomeric end formed by copying the 5′-terminal repeat is unwound from that repeat and base pairs with the 3′-terminal repeat. Underwood et al. recently hypothesized that an additional 3 nt just to the 3′ side of the template might also function in base pairing during translocation (35). This was based on the sequence match of this region to the corresponding positions of the telomeric DNA as well as on telomere length alterations caused by two point mutations in the region.
To more fully investigate the biological role of this telomerase template-adjacent region, additional base substitutions were constructed and tested for their effect on telomere length. As for all mutants described in this work, the mutant TER1 genes replaced the wild-type allele in haploid K. lactis cells through plasmid loop-in/loop-out mutagenesis. Because telomerase RNA template mutations alter both the RNA and its telomeric DNA repeat product and because the latter can be paired to the template at two different positions (before and after translocation), we shall refer to the mutations by names that include the position changed, the altered base present, and whether the most relevant molecule affected is the RNA or its DNA product. Thus, ter1-(−2AR) allele is an adenine substitution at the −2 position (relative to the 30-bp template) that is present in the telomerase RNA and ter1-28TD is a thymine substitution at position 28 of the telomeric DNA repeat as drawn in Fig. 1A. The numbering of the template matches the nomenclature used previously (34, 35).
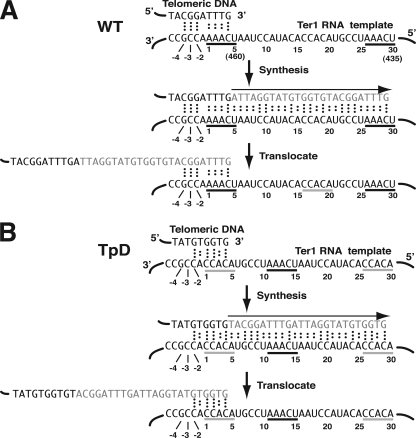
FIG. 1.
Putative base-pairing interactions between the telomere and the telomerase RNA template region of K. lactis wild type (WT) and the TpD template permutation mutant. (A) Diagram of telomeric repeat synthesis using a wild-type TER1 template. The top panel shows potential base-pairing interactions between a single-stranded end of telomeric DNA with the region immediately 3′ to the template of the wild-type Ter1 RNA. Dots between bases indicate hydrogen bonds of base pairs. Numbers indicate nomenclature used for base positions in this work. Numbers in parentheses indicate base positions numbered from the 5′ end of the Ter1 RNA. The middle panel shows the synthesis step when a 25-nt telomeric repeat (shown in gray) is copied. The bottom panel shows the same sequences after translocation of the newly synthesized DNA such that the new 3′ DNA end is base paired back at the 3′ end of the template. Sequences underlined in black indicate the 5-nt direct repeats of the template region. The sequence underlined in gray indicates the 5-nt sequence engineered to be the direct repeats of the template region in the TER1-TpD permutation mutant. (B) Diagram of telomeric repeat synthesis using a TER1-TpD template. The TER1-TpD mutant has a 30-nt template region that is circularly permuted relative to the normal template. The sequences underlined in gray are the novel 5-nt repeats, and the sequence underlined in black shows the position of the sequence that comprises the 5-nt repeats (bases 1 to 5 and 26 to 30) in the wild-type template. Aside from different start and stop points for DNA synthesis, the TpD template is expected to produce wild-type telomeric repeats. The region from −1 to −4 next to the template is identical in wild-type Ter1 and Ter1-TpD, but as shown, that region is expected to interact with different parts of the telomeric repeat in the two cell types.
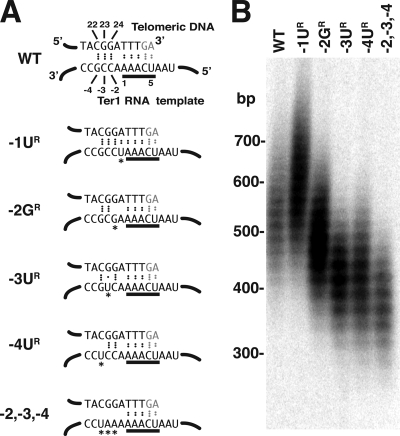
The mutations generated in the template-adjacent region and their predicted effects on the base pairing with a translocated telomeric repeat are shown in Fig. 2A, and the effects of the mutations on telomere length are shown in Fig. 2B. The BsrBI restriction digest used to examine telomeric DNA by Southern blotting cleaves exactly 3 bp internally from the start of the first base pair of telomere homology in 10 of the 12 K. lactis telomeres. This allowed accurate sizing of the telomeres on high-percentage agarose gels. We found that base substitutions at each of three new positions (−2, −3, and −4), postulated to be involved in base pairing to the telomere, result in telomeres that are substantially shorter than those of wild-type control cells (Fig. 2B). While wild-type telomeres averaged ∼530 bp in length, those of the −2, −3, and −4 mutants varied from 60 to 110 bp shorter. With each of these mutants, telomeres remained at the same shortened length for as long as they were passaged (a minimum of 10 streaks; equivalent to 200 to 250 cell divisions) (data not shown). This stability of altered telomere length phenotypes was characteristic of all mutants in this study unless otherwise noted.
FIG. 2.
Effects of minus-region (positions −1 to −4) mutations in the Ter1 template on telomere length. (A) Predicted base pairing between the telomeric DNA end (top strand) and the Ter1 template (bottom strand) in wild type (WT) and five minus-region mutants (each with a wild-type permutation of the template). Only the 3′ side of the template is shown for simplicity. Dots between bases indicate hydrogen bonds. Mutant bases are marked with asterisks. The bases underlined in black are the 3′ 5-nt repeat of the template. (B) Southern blot of BsrBI-digested genome DNA from wild-type (WT) and ter1 mutant strains probed with a telomeric repeat sequence. Sequences visible represent the 10 (of 12) telomeres where the BsrBI site is located close (3 bp) to the start of telomeric sequence. Positions of size markers are indicated.
In contrast to the mutations at positions −2, −3, and −4, a mutation at the −1 position that was predicted to replace a mismatched base with a base able to pair with the telomeric DNA repeat, and thus increase the total base-pairing potential of the −1 to −4 region, showed an increase in telomere length of 100 to 150 bp compared to that of the wild type. A mutation that altered all three of the bases at positions −2 to −4 showed telomeres only slightly shorter than those of the short-telomere phenotypes produced by the single point mutations tested. This suggests that the ter1-(−3UR) and ter1-(−4UR) mutations by themselves may largely disrupt the function encoded by the −2 to −4 region. These results add support to the idea that the −1 to −4 region plays a role in telomerase function and are consistent with the idea that this role involves base pairing between that part of the RNA and 22 to 24 positions of the telomeric repeat.
Another notable result from running the BsrBI-digested telomeric fragments on high-percentage agarose gels was that the telomeres from both wild-type and mutant cells were resolved into a ladder of bands rather than simply a heterogeneous smear as is typically observed in most Southern blots of telomeric fragments from K. lactis or from most other species. The ladders appeared to have step sizes of 25 bp, consistent with being multiples of the K. lactis telomeric repeats. While past cloning of K. lactis telomeres (24, 33) had suggested that they terminate rather randomly within the 25-bp unit, our results here suggest instead that the terminal positions of the telomere with respect to the 25-bp repeat are decidedly biased toward one or more positions or regions of the repeat.
Minus-region mutations have opposite effects on telomere length in a permuted template mutant.
To genetically assess a base-pairing interaction in an RNA molecule, it is desirable to generate compensatory base changes, pairs of mutations that individually disrupt the predicted pairing but together restore it. This approach is problematic with the telomerase RNA mutations being studied because the needed compensatory mutations would need to be at positions 22 to 24 of the DNA of the telomeric repeat. This region of the telomeric repeat lies within the binding site for the Rap1 protein, a key factor in telomeric length regulation in yeast (20, 23). Thus, it is likely that such “compensatory” mutations would affect telomere length by more than one mechanism. In an attempt to overcome this problem, we constructed a TER1 gene with a circularly permuted template. This mutation, TER1-TpD, has the 30-bp region of the template permuted by shifting it 10 bp in one direction (Fig. 1B and 3A). This creates novel 5-bp terminal repeats but leaves the minus region and all the rest of the Ter1 RNA identical to the wild type.
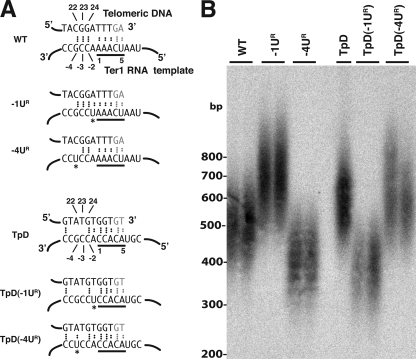
FIG. 3.
ter1 minus-region mutations have different effects on telomere length depending on the permutation of the template. (A) Predicted base pairing between the telomeric DNA end and the Ter1 template in the −1UR and −4UR mutations in the wild type and the TpD template permutation. Dots between bases indicate hydrogen bonds. Asterisks indicate mutant bases. Bases underlined in black are the 3′ 5-nt repeat of the template. (B) Southern blot of BsrBI-digested genome DNA from the same strains probed with a telomeric repeat sequence. Positions of size markers are indicated. WT, wild type.
As shown in Fig. 3A, the ter1-(−1UR) and ter1-(−4UR) mutations are predicted to have opposite effects on the base-pairing potential of the minus region with the telomere of the wild-type and TpD permutations. When the ter1-(−1UR) and ter1-(−4UR) mutations were combined with TpD template permutation, we observed that they had opposite effects on telomere length compared to the ter1-(−1UR) and ter1-(−4UR) mutations present with the wild-type template permutation (Fig. 3B). Whereas the ter1-(−1UR) mutation substantially increases the length of telomeres in the wild-type template permutation, it decreases telomere length by ∼200 bp in the TpD permutation. Conversely, the ter1-(−4UR) mutation decreases telomere length in the wild-type template permutation but has little effect on telomere length in the TpD permutation. These results provide support for the idea that the minus-region mutations affect telomere length through their effect on the base-pairing potential with the telomeric DNA.
Minus-region mutations partially suppress ter1 mutations that alter positions 23 and 24 of the telomeric repeat.
Mutations at positions 22 to 24 of the TER1 template were shown previously to produce substantially shorter than normal telomeres (34). This occurred in spite of the fact that these mutations altered the binding site of the protein Rap1, known to function as a negative regulator of telomere length in both S. cerevisiae and K. lactis (20, 22, 23). Mutations at positions 16 to 20 of the template, also in the Rap1 binding site, led to telomeres that rapidly elongated, the phenotype expected for disrupted binding of Rap1 (26, 34). It was suggested that the short telomeres produced by mutations at positions 22 to 24 of the Ter1 template were caused by disruption of a second, positive function that overlapped the Rap1 binding site. As these positions correspond exactly to the telomeric region expected to base pair with the −2 to −4 region of the Ter1 RNA, it seemed likely that disrupted base pairing involving this region contributed to the short-telomere phenotypes of the ter1 mutants with alterations at positions 22 to 24.
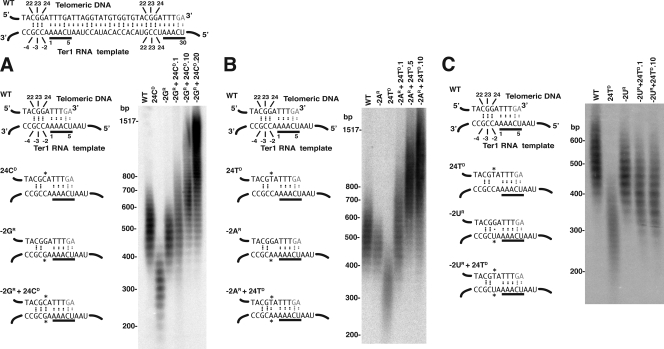
To address the above possibility, several base substitutions in the TER1 template at positions 22 to 24 were combined with the base changes at positions −2 to −4 predicted to restore base pairing. Combining the −2GR mutation with the ter1-24CD mutation was found to produce telomeres much longer than those produced by the ter1-24CD mutation alone (Fig. 4A). Indeed, the combined mutant acquired a gradual telomere elongation phenotype that is similar to that seen with mutants with mutations that affect the left side of the Rap1 binding site (template positions 16 to 20). Very similar results were seen with the −2AR and ter1-24TD(SnaB) pair. While both individual mutations produced shorter-than-normal telomeres, the double mutant produced a gradual elongation phenotype (Fig. 4B). In a control experiment, a −2UR mutation was combined in cis with the 24TD(SnaB) TER1 mutation (Fig. 4C). In this case, the double mutant had telomeres that were intermediate in length between those of the two individual single mutants and remained at that size even after 10 streaks of growth. We conclude that base changes at position 24 of the template could be strongly suppressed by mutations at the −2 position that were predicted to restore base-pairing interactions.
FIG. 4.
The short-telomere phenotypes caused by mutations at position 24 of the Ter1 template are suppressed by mutations at −2 that are predicted to restore base pairing with the telomeric repeat. (A) The 24CD and −2GR mutations. The left panel shows the predicted base pairing between the telomeric DNA end and the Ter1 template in the 24CD and −2GR mutations alone or in combination. On top is shown the complete template sequence and its predicted pairing to telomeric sequences for the wild type. The lower diagrams show only the left part of this sequence for the mutants indicated. Dots between bases indicate hydrogen bonds. Asterisks indicate mutant bases. Bases underlined in black are the 3′ 5-nt repeat of the template. The right panel shows a Southern blot of BsrBI-digested genome DNA from the same strains probed with a telomeric repeat sequence. The double mutant is shown 1, 10, and 20 streaks after its creation. (B) The 24TD and −2AR mutations. Predicted base-pairing interactions and a Southern blot are shown, as in panel A. The double mutant is shown 1, 5, and 10 streaks after its creation. (C) The 24TD and −2UR mutations. Predicted base-pairing interactions and a Southern blot are shown, as in panel A. The double mutant is shown 1 and 10 streaks after its creation. WT, wild type.
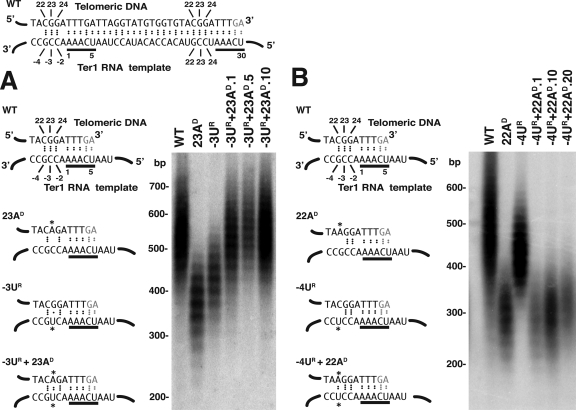
In other experiments, we tested whether the short telomeres of the ter1-22AD and ter1-23AD mutants could be suppressed by being combined in cis with minus-region mutations expected to restore base pairing. As shown in Fig. 5A, the −3UR mutation was found to suppress the very short telomere phenotype of ter1-(23AD) despite the fact that, by itself, it produced a short-telomere phenotype as well. Although the ter1-(−3UR)/(23AD) double mutant did not produce a gradual telomere elongation phenotype, it did produce telomeres that stabilized rapidly at a length longer than those in either single mutant. In contrast, the ter1-(−4UR)(22AD) mutant produced telomeres that were similar in length or only slightly longer than the ter1-(22AD) single mutant (Fig. 5B). The reason for this failure to observe suppression is unclear. Despite this particular result, our results provide further support for the model that disrupted base pairing with at least the −2 and −3 positions is involved in producing the short-telomere phenotypes of the ter1 mutants that alter positions 23 and 24 of the telomeric repeat.
FIG. 5.
Combining the ter1-23A and ter1-22A short-telomere mutants with minus-region mutations predicted to restore base pairing with the telomeric repeat. (A) The 23AD and −3UR mutations. The left panel shows the predicted base pairing between the telomeric DNA end and the Ter1 template in the 23AD and −3UR mutations alone or in combination. On top is shown the complete template sequence and its predicted pairing to telomeric sequences for the wild type. The lower diagrams show only the left part of this sequence for the mutants indicated. Dots between bases indicate hydrogen bonds. Asterisks indicate mutant bases. Bases underlined in black are the 3′ 5-nt repeat of the template. The right panel shows a Southern blot of BsrBI-digested genome DNA from the same strains probed with a telomeric repeat sequence. The double mutant is shown 1, 5, and 10 streaks after its creation. (B) The 22AD and −4UR mutations. Predicted base-pairing interactions and a Southern blot are shown, as in panel A. The double mutant is shown 1, 10, and 20 streaks after its creation. WT, wild type.
Multiple yeast species appear to have an extended template/telomere base-pairing region similar to that of K. lactis.
We next examined known telomerase RNA genes as well as newly identified putative telomerase RNA genes (15; L. Guo, D. Zhang, Y. Wu, R. Malmberg, M. McEachern, and L. Cai, unpublished data) from a variety of yeast species for the possible presence of an extended base-pairing potential between the 3′ side of the RNA template and the telomeric DNA, as exists in K. lactis. As shown in Fig. 6, most species examined were found to have template terminal repeats of 7 to 10 nt in size. In three species, Saccharomyces kluyveri, Ashbya gossypii, and Debaryomyces hansenii, the repeats were perfect matches of 8 to 10 nt. Eight other species, including five of six Kluyveromyces species as well as Candida albicans, Candida tropicalis, and Candida glabrata, had repeats that differed from one another by a single nucleotide near their middles. Kluyveromyces nonfermentans and Kluyveromyces aestuarii had repeats that differed from one another by a single extra nucleotide in the 5′ repeat. The templates of Candida guilliermondii and Pichia stipitis could be described as being bordered either by perfect 7- to 8-nt repeats or by imperfect 9- to 10-nt repeats. S. cerevisiae, as well as some other closely related Saccharomyces species, has a template that can be drawn having a few slightly different pairs of 7- to 9-nt terminal repeats. Only Saccharomyces castellii, with 4-nt terminal repeats, and S. pombe (data not shown) could not be reasonably drawn to have extended repeats. We conclude that the enlarged telomere base-pairing domain at the 3′ end of the telomerase RNA template is a feature common to many yeast telomerase RNAs. Notably, S. cerevisiae, S. castellii, and S. pombe are species that have telomeric repeats that are highly heterogeneous in size and sequence. Finally, we would point out that our results slightly complicate the common usage of the term template to mean the region of a telomerase RNA with perfect telomere homology.
FIG. 6.
Long imperfect direct repeats border the template region of telomerase RNAs from multiple yeast species. Shown are template regions of telomerase RNAs from a variety of yeast species. Terminal repeats of the template are shown underlined. Nonidentical bases within the repeats are indicated by gaps while missing bases (relative to the other repeat) are indicated by carets. S. cerevisiae has three possible pairs of direct repeats, as indicated. The second pair in S. cerevisiae as well as the pair of direct repeats in C. guilliermondii is shown in two independent lines in the figure because of their overlap. The yeast species are shown next to an approximate phylogenetic tree based on previous studies (2, 10, 18, 21). The templates are aligned according to the region encoding the expected Rap1 binding site of the telomeric repeat (shaded gray). In some species, the Rap1 binding site is encoded by two discontinuous parts of the template, which are copied into consecutive Rap1 binding sites in tandem telomeric repeats.
DISCUSSION
Our results here suggest that many yeast species including K. lactis use a 7- to 10-nt domain at the 3′ end of the telomerase RNA template for base pairing to telomeric DNA. The full extent of this base-pairing domain had been overlooked previously because of the lack of perfect sequence homology between this domain and the telomeric 3′ overhang. A variety of experimental data from K. lactis support the idea that positions −2 to −4 of the Ter1 template RNA region form base-pairing interactions with telomeric DNA. Perhaps the most compelling are the opposite effects of the ter1-(−1UR) and ter1-(−4UR) mutations on the wild-type and permuted TpD templates. Such effects are difficult to account for by alternative hypotheses but can readily be explained by the alternate positions of the telomeric sequences expected to base pair with the −2 to −4 region in the two different permutations of the template. The ter1-(−1UR) mutation, in particular, produces large and opposite telomere length phenotypes depending on the permutation of the template. With the wild-type permutation, the ter1-(−1UR) mutation is predicted to improve base pairing and produces longer telomeres, while in the TpD permutation, the ter1-(−1UR) mutation is predicted to disrupt base pairing and produces shorter telomeres (Fig. 3). The effect of the ter1-(−4UR) mutation is less dramatic in both permutations of the template but is again consistent with the base-pairing hypothesis. In the permuted TpD template, the ter1-(−4UR) mutation restores base pairing between position −4 of the telomerase RNA and position 22 of the telomeric repeat. However, unlike the situation with the wild-type permutation, this base pair is separated by a mismatch at the −3/23 position (Fig. 3A). This may explain why the ter1-(−4UR) mutation has a negligible effect on telomere length on the TpD permutation. Consistent with this, simultaneously altering the −3, −4, and −5 positions of the TpD permutation to base pair with the telomeric repeat led to longer telomeres than were present in cells carrying the TpD telomerase without these mutations (data not shown).
The TpD permutation, by itself, produces a slight telomere lengthening relative to the wild-type permutation despite the apparent presence of one fewer potential base-pairing interaction at the 3′ region of the template (Fig. 3A and B). Although a permuted telomerase RNA template might be able to alter telomere length through a number of mechanisms, it can be noted that the predicted base pairs at the 3′ region of the TpD template (specifically positions 1 and 2) have more of the stronger GC base pairs than does the wild-type interaction at the same positions. These might serve to compensate for the smaller number of base pairs able to form with the TpD template.
In vitro work with the S. cerevisiae telomerase has indicated that the length of hybrid between the telomerase RNA and a telomeric DNA primer remains constant at around 7 nt in both primer binding and DNA synthesis (11). It is quite likely that the same or similar size limitations to the RNA-DNA hybrid occur in other yeasts such as K. lactis. This would predict that the full 9-nt region (positions −4 to 5) at the 3′ end of the K. lactis template cannot be simultaneously base paired with telomeric DNA. It should be noted, however, that the K. lactis telomerase does not typically copy as far as position 30 (the last position of telomere homology) in the template in vitro or in vivo (12, 35). In vitro, it most frequently stops after copying position 28 in the template. This would lead to a 7-nt region from positions −4 to 3 of the 3′ side of template being available to base pair with telomeric DNA.
Conversely, without the base pairing from the −2 to −4 region, there would be only three T residues (from positions 26 to 28) at the end of a newly synthesized telomeric repeat to allow base pairing with the 3′ region of the telomerase RNA template. By themselves, these could potentially align with template A residues at positions 1 to 3 in more than one register and lead to repeats of slightly different sizes being synthesized. The −2 to −4 region therefore likely contributes to the sequence and length uniformity of K. lactis telomeric repeats. Notably, yeast species that lack the extended template base-pairing potential, such as S. castellii and S. pombe, or that have alternate base-pairing positions, such as S. cerevisiae, produce telomeric repeats that are irregular in sequence.
In theory, use of the −2 to −4 region of K. lactis Ter1 in base pairing with telomeric DNA might lead to a specific type of telomeric repeat sequence dimorphism. If reverse transcription of the −1 position occurred during repeat synthesis by telomerase, this would lead to the incorporation of a T residue at position 1 of the telomeric repeat instead of the normal A residue. As the variant repeat expected from this has not been observed in telomeres from K. lactis that have been sequenced to date (24, 33, 35), we conclude that reverse transcription of the −1 position is rare or absent.
It is conceivable that an enlarged template/telomere base-pairing region could also be an adaptation related to the synthesis of the large telomeric repeats that are present in many yeast species such K. lactis. Large telomeric repeat size will necessitate telomerase translocation steps that involve movements of greater lengths of RNA than would occur with telomerases that synthesized the shorter 5- to 8-bp telomeric repeats present in most organisms. Reestablishment of base-pairing interactions between the telomerase RNA and the telomeric DNA upon the translocation step of the enzyme might therefore be predicted to benefit from a somewhat larger and strengthened base-pairing domain.
The presence of imperfect homology between the 3′ domain of the telomerase RNA template and telomeric DNA of multiple yeast species including K. lactis could suggest that this mismatch may play some useful function in some species. However, given that a mutation at the −1 position in K. lactis that improves base pairing also leads to an increase in telomere length, any postulated role of mismatched sequence is unlikely to be related to increasing telomerase's overall ability to extend a telomere. Perhaps a more likely possibility is that during selection for an extended base-pairing region, imperfect pairing, at least away from the 3′ telomeric end, was not detrimental to function and therefore simply not selected against.
The precise mechanism by which increased telomere/template base pairing leads to longer telomeres is not clear but is likely due to an increase in the ability of telomerase to extend telomeres. The simplest possibility would be that improved base pairing directly promotes telomerase RNA's binding to at least a subset of telomeric ends. Those ends terminating with bases copied from the end of the template (positions 28 to 30) would have the shortest potential stretches of base pairing to the 3′ region of the template and would be most likely to be influenced by the base-pairing potential of the −2 to −4 region of the template. Such ends would of course be produced each time that telomerase synthesizes DNA by copying the full template. An additional or alternative possibility is that the paired minus-region/telomere interacts with the catalytic protein of telomerase activity and alters its activity.
Some evidence from in vitro telomerase assays done with both K. lactis (12) and S. cerevisiae (5) suggests that telomeric primer mutations that weakened telomere/template base pairing led to greater telomerase activity, the opposite of what our results might suggest. However, in these cases, the annealed telomere/template region was both larger and some distance away from the direct repeats of the template, the regions of relevance to our work. Thus, we believe it remains likely that K. lactis telomeres terminating at nucleotides copied from the end of the template (positions 28 to 30) will be more efficiently elongated when supplemental base pairing provided by the −2 to −4 region is available.
Our observations here help in the interpretation of K. lactis mutants that have telomeric repeats with base changes at positions 22 to 24, a region of the repeat that encodes part of the binding site for the negative regulator of telomere length, Rap1. While TER1 mutants with mutations affecting other parts of the Rap1 binding site typically produce telomere elongation, TER1 mutants with mutations at positions 22 to 24 produce telomeres that remain substantially shorter than normal (34). The ter1-24T(SnaB) mutation (which creates a SnaBI restriction site) has been the most extensively studied of this group. When tandem 24T(SnaB) repeats are inserted basally at an otherwise wild-type telomere, that telomere becomes abnormally long (1). This indicates that 24T(SnaB) repeats are defective at negatively regulating telomere length as would be expected for a Rap1 binding defect. It was therefore concluded that the ter1-24T(SnaB) mutant, which uses a mutant telomerase to add 24T(SnaB) repeats onto the ends of all telomeres, must have one or more additional defects that inhibit telomere elongation and act to block the telomere elongation that would otherwise occur from the Rap1 binding defect. Our results here show that the short-telomere phenotypes of mutations at positions 23 and 24 of the Ter1 template, including ter1-24T(SnaB), can be converted into a gradual telomere elongation phenotype by mutations at positions −2 and −3 that are predicted to restore base-pairing potential between the mutant telomeric repeat and the Ter1 RNA. Thus, a template base-pairing defect appears to be an important contributor to the short-telomere phenotypes of ter1 template mutations from positions 23 and 24. The exact extent of the suppression in each particular mutation combination is difficult to ascertain because, unlike the permuted templates, the mutant templates with mutations at positions 22 to 24 lead to mutant telomeric repeats that, by themselves, perturb telomere function.
An important point regarding the interpretation of mutations affecting positions 22 to 24 is that each of the mutants with alterations at those positions had substantially shorter telomeres than did mutants with alterations only at positions −4 to −2 (Fig. 4 and 5). If disrupting the base pairing between the telomerase RNA and the telomeric DNA was the only defect present, mutating the 22 to 24 region should have produced the same degree of shortening as that seen by mutating the −2 to −4 region. Our results therefore argue that positions 22 to 24 of the telomeric repeat probably perturb yet another function that contributes positively to telomere maintenance. One good possibility for this is an effect on the binding of the single-strand telomere binding protein Cdc13, the binding site of which in S. cerevisiae telomeric repeats overlaps closely with the sequence of the Rap1 binding site (8). This putative third function encoded by positions 22 to 24 of the template could underlie the failure of the −4UR mutation to suppress the short telomeres of the ter1-22AD mutant. If the 22AD mutation produces its short-telomere phenotype primarily via disruption of the second positive function rather than by disruption of telomere/template base pairing, than the −4UR mutation might not be expected to suppress it. Additionally, if the 22AD mutation did not appreciably disrupt Rap1 binding, than no suppression of it would be expected to include telomeres elongated to larger than normal sizes as occurs with the −2AR/24TD(SnaB), −2GR/24CD, and −3UR/23AD mutation pairs.
One unexpected observation from our studies was the apparent partial suppression of the 24TD mutation by the −2UR mutation, which did not restore base pairing (Fig. 4C). This suppression is much less dramatic than that produced by the −2AR mutation (which does restore base pairing) but nonetheless appears real. Its basis remains unclear. One possibility is that the T-U mismatch produced in ter1-(−2UR)(24TD) is somehow less detrimental than the T-C mismatch produced in ter1-(24TD). The 24T telomeric repeats, with their compensatory positive and negative defects for telomere length regulation, might also be especially sensitive to perturbations in telomerase function. A second possibility is that the −2 to −4 region might have a second function in addition to base pairing with telomeric sequence that is differentially affected in the −2UR and −2AR mutations.
In examining the consequences of TER1 mutations for telomere length, we made the important observation that K. lactis telomeres run on high-resolution agarose gels can be resolved into a ladder of diffuse but very distinct bands. As the telomeres that we resolved on these gels all have the same sequence starting from the BsrBI site adjacent to each telomere, the variation in size of the telomeric signal in these ladders of bands observed comes from (i) different numbers of telomeric repeats and (ii) a nonrandom terminal structure with regard to the 25-bp telomeric repeat unit. Previous cloning of small numbers of K. lactis telomeres (24, 33, 35) had suggested that their natural end points were scattered throughout the 25-bp unit of the telomeric repeat and might therefore be relatively or completely random. Our results here indicate instead that K. lactis telomeres must have preferred terminal end points or regions of end points on one or both strands, as is the case with ciliate and mammalian telomeres (17, 19, 29). The basis for the difference from cloning results is not clear, although it might in part be due to some cloning results coming from mutants with perturbed telomere function. The exact nature of the K. lactis telomere terminal structure, such as the size and sequence of 3′ overhangs, remains to be determined. The various examples of Ter1 mutants that had the wild-type template permutation but often very different telomere lengths appeared to all have the bands of the telomere ladder in the same register. This is consistent with those mutants all having the same terminal sequence structure as that of the telomeres of wild-type cells. The TpD template permutation, in contrast, produced a less distinct banding pattern. Interestingly, detailed studies of it and other Ter1 template permutations have shown that altered template permutations do alter the terminal telomeric sequences of K. lactis telomeres (Z. Wang, R. Zinzen, and M. McEachern, unpublished data).
Acknowledgments
We thank Robert Zinzen for assistance with construction of a precursor to the TpD template permutation mutant.
This work was supported by grants from the National Institutes of Health (GM 61645 and GM 072080).
Footnotes
Published ahead of print on 17 August 2009.
REFERENCES
- 1.Bechard, L. H., B. D. Butuner, G. J. Peterson, W. McRae, Z. Topcu, and M. J. McEachern. 2009. Mutant telomeric repeats in yeast can disrupt the negative regulation of recombination-mediated telomere maintenance and create an alternative lengthening of telomeres-like phenotype. Mol. Cell. Biol. 29:626-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergeron, J., and G. Drouin. 2008. The evolution of 5S ribosomal RNA genes linked to the rDNA units of fungal species. Curr. Genet. 54:123-131. [DOI] [PubMed] [Google Scholar]
- 3.Bertuch, A. A., and V. Lundblad. 2006. The maintenance and masking of chromosome termini. Curr. Opin. Cell Biol. 18:247-253. [DOI] [PubMed] [Google Scholar]
- 4.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. Chiu, M. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 5.Bosoy, D., and N. F. Lue. 2004. Yeast telomerase is capable of limited repeat addition processivity. Nucleic Acids Res. 32:93-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, Y., S. S. Chan, and S. Chang. 2008. Telomere dysfunction and tumour suppression: the senescence connection. Nat. Rev. Cancer 8:450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eldridge, A. M., W. A. Halsey, and D. S. Wuttke. 2006. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry 45:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 10.Fitzpatrick, D. A., M. E. Logue, J. E. Stajich, and G. Butler. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forstemann, K., and J. Lingner. 2005. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep. 6:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulton, T. B., and E. H. Blackburn. 1998. Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol. 18:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 14.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 15.Gunisova, S., E. Elboher, J. Nosek, V. Gorkovoy, Y. Brown, J. F. Lucier, N. Laterreur, R. J. Wellinger, Y. Tzfati, and L. Tomaska. 2009. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15:546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hathcock, K. S., Y. J. Chiang, and R. J. Hodes. 2005. In vivo regulation of telomerase activity and telomere length. Immunol. Rev. 205:104-113. [DOI] [PubMed] [Google Scholar]
- 17.Jacob, N. K., R. Skopp, and C. M. Price. 2001. G-overhang dynamics at Tetrahymena telomeres. EMBO J. 20:4299-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James, T. Y., F. Kauff, C. L. Schoch, P. B. Matheny, V. Hofstetter, C. J. Cox, G. Celio, C. Gueidan, E. Fraker, J. Miadlikowska, H. T. Lumbsch, A. Rauhut, V. Reeb, A. E. Arnold, A. Amtoft, J. E. Stajich, K. Hosaka, G. H. Sung, D. Johnson, B. O'Rourke, M. Crockett, M. Binder, J. M. Curtis, J. C. Slot, Z. Wang, A. W. Wilson, A. Schussler, J. E. Longcore, K. O'Donnell, S. Mozley-Standridge, D. Porter, P. M. Letcher, M. J. Powell, J. W. Taylor, M. M. White, G. W. Griffith, D. R. Davies, R. A. Humber, J. B. Morton, J. Sugiyama, A. Y. Rossman, J. D. Rogers, D. H. Pfister, D. Hewitt, K. Hansen, S. Hambleton, R. A. Shoemaker, J. Kohlmeyer, B. Volkmann-Kohlmeyer, R. A. Spotts, M. Serdani, P. W. Crous, K. W. Hughes, K. Matsuura, E. Langer, G. Langer, W. A. Untereiner, R. Lucking, B. Budel, D. M. Geiser, A. Aptroot, P. Diederich, I. Schmitt, M. Schultz, R. Yahr, D. S. Hibbett, F. Lutzoni, D. J. McLaughlin, J. W. Spatafora, and R. Vilgalys. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818-822. [DOI] [PubMed] [Google Scholar]
- 19.Klobutcher, L. A., M. T. Swanton, P. Donini, and D. M. Prescott. 1981. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl. Acad. Sci. USA 78:3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krauskopf, A., and E. H. Blackburn. 1996. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383:354-357. [DOI] [PubMed] [Google Scholar]
- 21.Kurtzman, C. P. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 4:233-245. [DOI] [PubMed] [Google Scholar]
- 22.Lustig, A. J., S. Kurtz, and D. Shore. 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250:549-553. [DOI] [PubMed] [Google Scholar]
- 23.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 24.McEachern, M. J., and E. H. Blackburn. 1994. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc. Natl. Acad. Sci. USA 91:3453-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822-1834. [DOI] [PubMed] [Google Scholar]
- 26.McEachern, M. J., and E. H. Blackburn. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376:403-409. [DOI] [PubMed] [Google Scholar]
- 27.McEachern, M. J., D. H. Underwood, and E. H. Blackburn. 2002. Dynamics of telomeric DNA turnover in yeast. Genetics 160:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 29.Sfeir, A. J., W. Chai, J. W. Shay, and W. E. Wright. 2005. Telomere-end processing the terminal nucleotides of human chromosomes. Mol. Cell 18:131-138. [DOI] [PubMed] [Google Scholar]
- 30.Shawi, M., and C. Autexier. 2008. Telomerase, senescence and ageing. Mech. Ageing Dev. 129:3-10. [DOI] [PubMed] [Google Scholar]
- 31.Shippen-Lentz, D., and E. H. Blackburn. 1990. Functional evidence for an RNA template in telomerase. Science 247:546-552. [DOI] [PubMed] [Google Scholar]
- 32.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 33.Tzfati, Y., T. B. Fulton, J. Roy, and E. H. Blackburn. 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288:863-867. [DOI] [PubMed] [Google Scholar]
- 34.Underwood, D. H., C. Carroll, and M. J. McEachern. 2004. Genetic dissection of the Kluyveromyces lactis telomere and evidence for telomere capping defects in TER1 mutants with long telomeres. Eukaryot. Cell 3:369-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Underwood, D. H., R. P. Zinzen, and M. J. McEachern. 2004. Template requirements for telomerase translocation in Kluyveromyces lactis. Mol. Cell. Biol. 24:912-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verdun, R. E., and J. Karlseder. 2007. Replication and protection of telomeres. Nature 447:924-931. [DOI] [PubMed] [Google Scholar]
- 37.Wray, L. V., M. M. Witte, R. C. Dickson, and M. I. Riley. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, G. L., J. D. Bradley, L. D. Attardi, and E. H. Blackburn. 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344:126-132. [DOI] [PubMed] [Google Scholar]