Abstract
The PTS1-dependent peroxisomal matrix protein import is facilitated by the receptor protein Pex5 and can be divided into cargo recognition in the cytosol, membrane docking of the cargo-receptor complex, cargo release, and recycling of the receptor. The final step is controlled by the ubiquitination status of Pex5. While polyubiquitinated Pex5 is degraded by the proteasome, monoubiquitinated Pex5 is destined for a new round of the receptor cycle. Recently, the ubiquitin-conjugating enzymes involved in Pex5 ubiquitination were identified as Ubc4 and Pex4 (Ubc10), whereas the identity of the corresponding protein-ubiquitin ligases remained unknown. Here we report on the identification of the protein-ubiquitin ligases that are responsible for the ubiquitination of the peroxisomal protein import receptor Pex5. It is demonstrated that each of the three RING peroxins Pex2, Pex10, and Pex12 exhibits ubiquitin-protein isopeptide ligase activity. Our results show that Pex2 mediates the Ubc4-dependent polyubiquitination whereas Pex12 facilitates the Pex4-dependent monoubiquitination of Pex5.
The maintenance of peroxisome function depends on the formation of the peroxisomal membrane and the subsequent import of both membrane and matrix proteins. Without exception, peroxisomal matrix proteins are nucleus encoded, synthesized on free ribosomes, and subsequently imported in a posttranslational manner (40). The peroxisomal import apparatus can facilitate the transport of folded and oligomeric proteins over the peroxisomal membrane, with the basic principle of this translocation event still being unknown. Based on the concept of cycling receptors (9, 31), the receptor cycle is divided into four steps. In the first step, the cargo proteins are recognized in the cytosol by their cognate receptor protein Pex5 or Pex7. In general, this initial step depends on either one of the two well-characterized PTSs (peroxisomal targeting signals), PTS1 and PTS2, which are recognized and bound by the corresponding receptor proteins Pex5 and Pex7, respectively. In the second step, the cargo-loaded receptors dock with distinct proteins accessible at the surface of the peroxisomal membrane, namely, Pex13 and Pex14. These two proteins together with Pex17 are established components of the docking complex. A second complex of the peroxisomal protein import machinery acts downstream of the docking event and consists of the three peroxins Pex2, Pex10, and Pex12. A common feature of these proteins is a C-terminal RING (really interesting new gene) finger domain. The RING finger subcomplex and the docking subcomplex are both linked in a Pex8-dependent manner to form a larger assembly, the importomer (1). In the third step of the receptor cycle, the cargo is delivered to the peroxisomal matrix, and finally, the receptor is released from the peroxisomal membrane in an ATP-dependent manner and thus made available for proteasomal degradation or another round of import (for a review, see reference 27).
With respect to the PTS1 receptor Pex5, recent reports demonstrated that this final ATP-dependent step in the receptor cycle is catalyzed by the AAA (ATPases associated with various cellular activities) peroxins Pex1 and Pex6 (33, 37). The signal for the export process is the attachment of a monoubiquitin moiety or, alternatively, the anchoring of a polyubiquitin chain (5, 35). This protein modification is in general facilitated by a three-step enzyme cascade (20). The ubiquitin (Ub)-activating enzyme (E1) activates the Ub and transfers it to the Ub conjugation enzyme (E2). In a final step, a protein-Ub ligase (E3) binds both E2 and substrate and thereby facilitates the conjugation of the Ub moiety onto the substrate protein. Saccharomyces cerevisiae harbors genes coding for one E1 enzyme, 11 E2 enzymes, and approximately 80 to 100 putative E3 enzymes (18, 29). It was demonstrated that the polyubiquitination of Pex5 primarily depends on the E2 protein Ubc4, which upon deletion can be partly replaced by Ubc5 or Ubc1 (22, 25, 36). Polyubiquitination of Pex5 is not a prerequisite for its function in peroxisomal protein import but might be a crucial step of a quality control system for the disposal of dysfunctional Pex5 (10, 22, 25, 36). Pex5 monoubiquitination is facilitated by the E2 protein Pex4 (Ubc10) in yeast or the Pex4-like UbcH5a/b/c in humans (14, 35, 47). The modification of Pex5 by a single Ub primes the receptor for its export back to the cytosol, where the Ub supposedly is removed prior to the initiation of a new receptor cycle (5, 14, 35). Although the functional relevance and the cognate E2 protein required for the different Ub modifications of Pex5 were identified, the factor(s) determining the substrate specificity, the protein-Ub ligase(s), remained unknown.
Here we report on the discovery of the function of Pex2 and Pex12 as E3 proteins required for ubiquitination of the import receptor Pex5. These RING peroxins, defects of which cause the lethal peroxisome biogenesis disorders in humans, exhibit Ub-protein isopeptide ligase activity with Pex5 as the molecular target. Pex2 is shown to mediate the Ubc4-dependent polyubiquitination whereas Pex12 facilitates the Pex4-dependent monoubiquitination of Pex5.
MATERIALS AND METHODS
Yeast strains and culture conditions.
The Saccharomyces cerevisiae strain UTL-7A (MATa ura3-52 trp1 leu2-3,112) was used as the wild-type strain for the generation of several isogenic deletion strains by the short flanking homology method as described previously (15). The resulting deletion strains were pex4Δ (46); pex5Δ (13); pex2Δ, pex10Δ, and pex12Δ (17); and pex2Δ/pex4Δ, pex10Δ/pex4Δ, and pex12Δ/pex4Δ (36) strains. Strains in which the genomic copies of genes express proteins fused to protein A (ProtA), tobacco etch virus (TEV)-ProtA, or hemagglutinin (HA) tags were generated according to the method in reference 23. The strains constructed comprise Pex4-HA, Pex2(1-221)-ProtA, Pex2(1-221)-ProtA pex4Δ, Pex2-TEV-ProtA Pex4-HA, Pex2-TEV-ProtA pex4Δ pex5Δ, Pex10(1-238)-ProtA, Pex10(1-238)-ProtA pex4Δ, Pex12(1-333)-ProtA, and Pex12(1-333)-ProtA pex4Δ strains. Primers used for this purpose are listed in Table 1. The Pex2-ProtA, Pex2-ProtA pex4Δ, Pex10-ProtA, Pex10-ProtA pex4Δ, Pex12-ProtA, and Pex12-ProtA pex4Δ strains were described earlier in reference 1. Yeast media have been described previously (11).
TABLE 1.
Oligonucleotides used in this study
| Purpose and name | Protein or other element(s) (strand) | Plasmid or DNA | Sequence |
|---|---|---|---|
| Integration | |||
| PEX2 | ProtA (sense) | pProtA/HU | 5′ GATGCCTGTGGATCCTCTGGGAGACTGACCGCCTCACCAGTGTACGGTGAAGCTCAAAAACTTATT 3′ |
| ProtA (antisense) | pProtA/HU | 5′ ATAGTATACACATATATAGAGATACAAGCGAGGGAACGGGGCCCATTTTACTTATAATACAGTTTTTTAG 3′ | |
| PEX10 | ProtA (sense) | pProtA/HU | 5′ GCAGACAACACTGTCAACCACAGGAAATTCTGGTCCTGCGGCAAGGTGAAGCTCAAAAACTTATT 3′ |
| ProtA (antisense) | pProtA/HU | 5′ GGCCTGTGGACAATGCTAAAAGAGTAGTCAAATTATTGATTATTTACTTATAATACAGTTTTTTAG 3′ | |
| PEX12 | ProtA (sense) | pProtA/HU | 5′ ACATACAACAAACACACGAATAAATGGGAAGTTGTGACAGGTATTAGGAAGCTACTAATCGGTGAAGCTCAAAAACTTATT 3′ |
| ProtA (antisense) | pProtA/HU | 5′ TGCTTGTAACACAACTGCAAAGAGGTGGAACAAGTGACATGAATTACCTGCTGGATAACATTTACTTATAATACAGTTTTTTAG 3′ | |
| PEX2 1 to 221 | ProtA (sense) | pProtA/HU | 5′ AAAAAACCACCAAGGTCTCGTTCTACGACTACTTATAAGACAGTTGGTGAAGCTCAAAAACTTATT 3′ |
| ProtA (antisense) | pProtA/HU | 5′ AGCGATCTGGTATGGGTTGGTTGGAAAACCGCCGCAACGGGGGCAATTTTACTTATAATACAGTTTTTTAG 3′ | |
| PEX10 1 to 238 | ProtA (sense) | pProtA/HU | 5′ TCTACGCTCGGGTCCTGGATTTATGGGAGAAAAAGGACCAATGATGGTGAAGCTCAAAAACTTATT 3′ |
| ProtA (antisense) | pProtA/HU | 5′ ATGTTCTGAACGCTCCTGTAGACCAACAGACGACCTAGTAATTGAATTTTACTTATAATACAGTTTTTTAG 3′ | |
| PEX12 1 to 333 | ProtA (sense) | pProtA/HU | 5′ TCACATAGTGACAAAACGGAAGATAAAGAGGGAGTTTCTGAGGCCGGTGAAGCTCAAAAACTTATT3′ |
| ProtA (antisense) | pProtA/HU | 5′ TTCCAATACACAGGGGTTTTGTACAGTTTTTTCGCAAACGGGACAATTTTACTTATAATACAGTTTTTTAG 3′ | |
| PEX2 | TEV-ProtA (sense) | pYM8 | 5′ GATGCCTGTGGATCCTCTGGGAGACTGACCGCCTCACCAGTGTACCGTACGCTGCAGGTCGAC 3′ |
| ProtA (antisense) | pYM8 | 5′ ATAGTATACACATATATAGAGATACAAGCGAGGGAACGGGGCCCTATCGATGAATTCGAGCTCG 3′ | |
| PEX4 | HA (sense) | pYM3 | 5′ AAAGTACTTCCTAGCAGAAAGAGAGCGGATC 3′ |
| HA (antisense) | pYM3 | 5′ CCCATTGTTTGCCATTCGAACACATCCATCC 3′ | |
| Cloning | |||
| UBC7 | ORFa plus EcoRI (sense) | Genomic DNA | 5′ GAATTCATGTCGAAAACCGCTCAG 3′ |
| ORF plus NotI (antisense) | Genomic DNA | 5′ GCGGCCGCTCAGAATCCTAATGATTT 3′ |
ORF, open reading frame.
Plasmids and cloning strategies.
Glutathione S-transferase (GST) fusions were constructed by cloning DNA fragments of the RING finger domains of PEX2 (amino acids [aa] 215 to 271), PEX10 (aa 238 to 337), and PEX12 (aa 293 to 399) in the bacterial expression vector pGEX-4T2 (GE Healthcare, Freiburg, Germany) as described previously (24). In order to generate fusion proteins of the RING domains and the S peptide, PEX2 (aa 215 to 271) and PEX10 (aa 238 to 337) were excised by EcoRI/XhoI, while PEX12 (aa 293 to 399) was excised by EcoRI/NotI. The obtained fragments were cloned into the pET-42b vector (Novagen, Madison, WI), resulting in pET 42b + PEX2 (aa 215 to 271), pET 42b + PEX10 (aa 238 to 337), and pET 42b + PEX12 (aa 293 to 399). The open reading frame of UBC7 was amplified from genomic DNA (Table 1) and was cloned into pGEX-4T1 (EcoRI; NotI), resulting in pGEX 4T1 + UBC7. The generation of pGEX 4T2 + PEX4 (35), pET 15b + UBC4 (26), and pET 9d + PEX5 (21) has been described previously.
Recombinant proteins.
Genes coding for recombinant proteins were expressed in Escherichia coli BL21(DE3). Cells were harvested, diluted in phosphate-buffered saline buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.3) containing protease inhibitors (8 μM antipain, 0.3 μM aprotinin, 1 μM bestatin, 10 μM chymostatin, 5 μM leupeptin, 1.5 μM pepstatin, 1 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride [Boehringer, Mannheim, Germany]), and broken using a French press. The 100,000 × g supernatant containing the soluble fusion proteins was loaded on affinity columns. Samples derived from cells expressing GST-Pex2(RING), GST-S peptide-Pex2(RING), GST-Pex10(RING), GST-S peptide-Pex10(RING), GST-Pex12(RING), GST-S peptide-Pex12(RING), GST-Pex4, and GST-Ubc7 were loaded on glutathione-Sepharose 4B (Pharmacia, Freiburg, Germany), washed and eluted from the column with 10 mM glutathione, or cleaved from the fusion tag with 8 U thrombin as described previously (35). The method for the purification of His fusion proteins like His-Ubc4 and His-Pex5 has been described previously (21). In the case of His-Ubc4, the fusion tag was cleaved off with 8 U thrombin.
Recombinant yeast E1, Ub, methylated ubiquitin, and His-Ub were purchased from Sigma (Munich, Germany). S protein was purchased from Biozyme (San Diego, CA).
In vivo ubiquitination assays.
Oleate-induced yeast cells were harvested, washed twice, and resuspended in lysis buffer (0.2 M HEPES, 1 M potassium acetate, and 50 mM magnesium acetate, pH 7.5) and protease inhibitors (8 μM antipain, 0.3 μM aprotinin, 1 μM bestatin, 10 μM chymostatin, 5 μM leupeptin, 1.5 μM pepstatin, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 5 mM NaF [Boehringer, Mannheim, Germany]). For the preparation of polyubiquitinated (polyUb) Pex5 from wild-type cells, lysis buffer was supplied with 100 μM MG132 (Sigma, St. Louis, MO), while 20 mM N-ethylmaleimide (NEM; Sigma, St. Louis, MO) was added to accumulate the monoubiquitinated form of Pex5. Additionally, 0.1 mM TPEN [N,N,N′,N′-tetrakis-(2-pyrididylmethyl)ethylenediamine] or 0.1 mM ZnCl2 was added to the samples where indicated. After addition of 3 g of glass beads (0.5 mm), breakage was achieved by vortex mixing (10 60-s bursts with breaks of at least 60 s on ice). Samples were transferred to Corex tubes and were centrifuged at 1,500 × g (Eppendorf rotor A-4-81) for 10 min. Supernatants were normalized for protein and volume, and membranes were sedimented by centrifugation at 100,000 × g, followed by trichloroacetic acid (TCA) precipitation and sample preparation (36).
Immunoprecipitation of denatured proteins was carried out using acetone-washed and dried sediments of TCA-precipitated samples. The sediment was resuspended in 100 μl of urea cracking buffer (50 mM Tris-Cl, pH 7.5, 6 M urea, 1% sodium dodecyl sulfate [SDS]) and incubated for 10 min at 65°C. Subsequently, 1 ml of Tween 20-IP buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5% Tween 20, 0.1 mM EDTA) and 10 μl of 100 mg/ml bovine serum albumin were added. After sedimentation of nondissolved material, Pex5 antiserum was added, and the mixture was incubated under continuous swirling for 4 h at 4°C. Subsequently, 75 μl of preswollen ProtA-Sepharose beads (GE Healthcare, Uppsala, Sweden) was added, and the mixture was further incubated for 1 h at 4°C. The immunoprecipitated material was subsequently subjected to sedimentation, washed twice with Tween 20-IP buffer, once with Tween 20-urea buffer (100 mM Tris-Cl, pH 7.5, 200 mM NaCl, 2 M urea, 0.5% Tween 20), and once with Tris-buffered saline buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl). Finally, the beads were boiled in 50 μl of IP sample buffer (62.5 mM Tris, 3% SDS, 5% β-mercaptoethanol, 10% glycerol, 0.05% bromphenol blue, pH 8.0) and prepared for immunoblot analysis.
In vitro ubiquitination assays.
Autoubiquitination assays of the E2 enzymes Pex4, Ubc4, and Ubc7 were carried out as described previously (35). Autoubiquitination reaction mixtures of the RING finger domains of Pex2, Pex10, and Pex12 contained 0.6 μg of each GST-RING protein or GST, 0.1 μg of yeast E1, 0.8 μg of E2 enzyme as indicated, 5 μg of Ub species as indicated, and 10 μl of buffer containing 2 mM ATP, 50 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 2.5 μM ZnCl2, and 0.1 mM dithiothreitol. After incubation at 30°C for 90 min with gentle shaking, the reaction was stopped by addition of SDS sample buffer and heating at 95°C for 5 min.
Reactions for the ubiquitination of the S protein were carried out in a similar way, with the difference that S peptide-RING finger fusions were used instead of the GST-RING fusions and that the reaction mixture contained additionally 1.8 μg of S protein.
Reaction mixtures for the in vitro ubiquitination of recombinant His-Pex5 contained 0.5 μg of GST-RING fusion proteins and 0.5 μg of His-Pex5.
Miscellaneous procedures.
Immunopurification of native protein complexes from yeast cells by using immunoglobulin G (IgG)-Sepharose was described in reference 1. Immunoreactive complexes were visualized using anti-rabbit or anti-mouse IgG-coupled horseradish peroxidase in combination with the ECL system from Amersham Pharmacia Biotech (Uppsala, Sweden). Polyclonal rabbit antibodies were raised against Pex2 (this study), Pex5 (3), Pex10 (1), Pex12 (2), Pex13 (13), Pex14 (3), Pex15 (37), Ubc4 (8), and Ub (Sigma, Munich, Germany). Monoclonal mouse antibodies were raised against HA (12CA5; Roche, Penzberg, Germany), GST (Sigma, Munich, Germany), porin (1), and Ub (clone FK2; Biomol, Hamburg, Germany).
RESULTS
Ubiquitination of Pex5 depends on the presence of zinc ions and requires the presence of the RING peroxins.
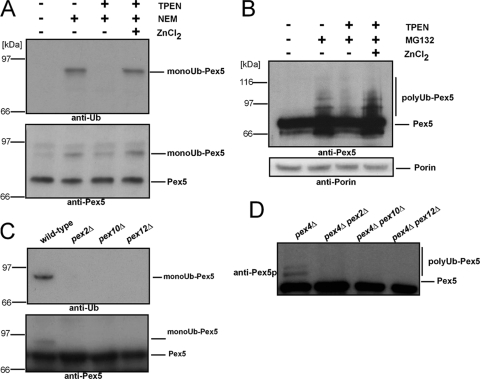
Previous results obtained from yeast and human cells demonstrated that the PTS1 receptor Pex5 is either mono- or polyubiquitinated (5, 22, 25, 36). This protein modification is facilitated by Pex4 or Ubc4 as E2 enzymes for Pex5 mono- and polyubiquitination, respectively (14, 22, 35, 36, 47). As Pex5 ubiquitination takes place exclusively at the peroxisomal membrane, the required protein-Ub ligases (E3s) were considered to be peroxisomal enzymes. E3s have the ability to bind both to an E2 and to the substrate via a RING finger, U box, or HECT (homologous to E6-associated protein C terminus) “catalytic” domain. In addition, certain E3s of the RING type are part of complexes where substrate recognition is assigned to a separate subunit (29). To specify which class of E3s is involved in the ubiquitination of the PTS1 receptor, we monitored the mono- and polyubiquitination of Pex5 in the presence and absence of TPEN. This reagent acts as a specific chelator for zinc ions and thus should inhibit the activity of RING finger-containing protein-Ub ligases, whereas HECT and U-box-like E3 remain unaffected with respect to their activity. When wild-type Pex5 was isolated via immunoprecipitation, ubiquitinated Pex5 species were visible only when NEM was present (Fig. 1A, lane 2). Prior addition of TPEN prevented the formation of monoubiquitinated Pex5 even after NEM was added (Fig. 1A, lane 3). The effect could be compensated for by the addition of larger amounts of zinc (Fig. 1A, lane 4). Similar results were obtained when polyUb Pex5 was monitored. Inhibition of the proteasome by MG132 led to the accumulation of polyUb Pex5 (Fig. 1B, lane 2). This receptor modification was not present when TPEN was applied to the sample during preparation. Addition of zinc quenched the function of TPEN, and polyUb Pex5 was again detectable (Fig. 1B, lanes 3 and 4). The MG132-independent porin served as a loading control.
FIG. 1.
Ubiquitination of Pex5 depends on the presence of zinc ions and the RING finger peroxins. (A) Pex5 was isolated by immunoprecipitation with anti-Pex5 antibodies from whole-cell membranes of oleic acid-induced wild-type cells in the presence or absence of TPEN, zinc chloride, and NEM to inhibit deubiquitination enzymes. (B) Wild-type cells were broken by glass bead treatment in the presence or absence of TPEN, zinc chloride, and MG132 to inhibit proteasomal degradation. Proteins were subsequently precipitated by TCA, and samples were subjected to immunoblot analysis. Detection of the mitochondrial porin served as a loading control. (C) Pex5 was isolated from whole-cell membranes of the indicated strains in the presence of NEM and subjected to immunoblot analysis. Deletion of either one of the RING peroxins resulted in a loss of Pex5 ubiquitination. (D) Whole-cell TCA lysates of indicated strains were prepared and analyzed by immunoblot analysis. Whereas deletion of PEX4 stabilized the polyubiquitinated form of Pex5, no modification was detectable when in addition one of the RING peroxins was deleted, indicating that each of the ring finger peroxins is required for Pex5 polyubiquitination. Detection of Pex5 and ubiquitinated Pex5 was performed with antibodies against Pex5 or anti-Ub antibodies as indicated.
The results so far indicated that protein-Ub ligases of the RING finger type are involved in this process, and the best candidates were the RING peroxins Pex2, Pex10, and Pex12, which form a complex at the peroxisomal membrane and are required for peroxisomal matrix protein import. Accordingly, we addressed the question of whether the Pex4-dependent monoubiquitination of Pex5 requires the presence of the RING peroxins. Wild-type cells or cells deficient in PEX2, PEX10, or PEX12 were grown on oleic acid-containing medium to induce proliferation of peroxisomal membranes. Cells were harvested and lysed in the presence of NEM to prevent deubiquitination (25). Pex5 was isolated by immunoprecipitation and analyzed by immunoblot analysis. As shown in Fig. 1C, deficiency in either one of the RING peroxins led to the complete inhibition of Pex5 monoubiquitination. Next we analyzed whether the RING peroxins are necessary for Pex5 polyubiquitination. Pex5 is polyUb in strains lacking either a component of the AAA or the Pex4 complex (22, 36). Accordingly, polyUb Pex5 species are visible in whole-cell TCA lysates of a pex4Δ strain (Fig. 1D, lane 1). In contrast, no polyubiquitination was observed when PEX4 was deleted together with PEX2, PEX10, or PEX12 (Fig. 1D). Taken together, the data indicate that the three RING peroxins (Pex2, Pex10, and Pex12) are indirectly or directly involved in the mono- and polyubiquitination event of the PTS1 receptor Pex5.
The peroxisomal RING complex contains Pex4 and Ubc4 and exhibits ubiquitin ligase activity.
The importomer at the peroxisomal membrane is described as a protein complex of the peroxisomal protein import machinery consisting of the docking complex (Pex13, Pex14, and Pex17), which is connected to the complex of the RING finger peroxins in a Pex8-dependent manner (1). Further experiments demonstrated that also the AAA complex (Pex1 and Pex6) and its membrane anchor protein Pex15 are part of the peroxisomal importomer (39). If the RING finger peroxins functioned as protein-Ub ligases, we assumed that also the corresponding E2 proteins might be components of the importomer. To prove this assumption, we constructed strains expressing RING peroxin Pex2 fused to two IgG binding domains derived from Staphylococcus aureus ProtA, with a cleavage site for the TEV protease inserted between Pex2 and the ProtA tag (23). In addition, the E2 protein Pex4 was genomically tagged with HA as a tool for its detection. The corresponding wild-type strain was initially tested for growth on plates containing oleic acid as the sole carbon source, which will support cell growth only if peroxisomal β-oxidation is functional. The corresponding strain did grow at the same rate as that of the nontagged wild type, indicating that Pex2-TEV-ProtA and Pex4-HA are functional (data not shown).
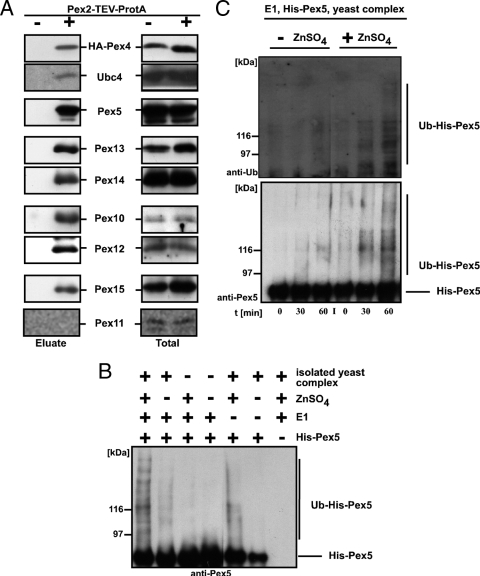
Protein complexes containing Pex2-TEV-ProtA were isolated from digitonin-solubilized membranes. Aliquots of solubilized material and eluates were subjected to immunoblot analysis. A wild-type strain without expression of Pex2-TEV-ProtA served as a control for unspecific binding of proteins to the Sepharose matrix. As shown in Fig. 2A and in agreement with published data, the Pex2 complex contained the other two peroxisomal RING peroxins, Pex10 and Pex12; the docking complex components Pex13 and Pex14; and the PTS1 receptor Pex5 and the AAA membrane anchor protein Pex15 (1, 39). In addition, Pex4 and Ubc4 coeluted with Pex2 as bait, indicating that these two E2 proteins required for either mono- or polyubiquitination of Pex5 also are components of the peroxisomal importomer. The specificity of the isolation was verified by the absence of the dominant peroxisomal membrane protein Pex11, which is not part of the importomer (1).
FIG. 2.
The RING complex exhibits Ub ligase activity. (A) The membrane-bound protein import complex was isolated from digitonin-solubilized membranes of wild-type cells expressing Pex2-TEV-ProtA as well as HA-tagged Pex4 via IgG-Sepharose and subsequent TEV protease cleavage. As a control, wild-type cells expressing no ProtA fusion protein were treated equally. The corresponding digitonin-solubilized membranes (totals) are shown in the right column. Indicated proteins of the TEV protease eluates and totals were detected by immunoblot analysis. (B and C) Recombinant Pex5 was incubated in the presence or absence of recombinant E1, zinc ions, and the Pex2 complex isolated from pex5Δ membranes. Reactions were stopped by the addition of SDS sample buffer after 45 min (B) or indicated time points (C), and modified Pex5 species were detected by immunoblot analysis. Ubiquitinated Pex5 was clearly detectable only when all constituents were present. Lack of either zinc or E1 led to significantly smaller amounts of modified Pex5.
Our observation of an association of the E2 protein Pex4 and Ubc4 with the putative E3 ligase Pex2, Pex10, and Pex12 tempted us to investigate whether this complex is sufficient to ubiquitinate recombinant Pex5. The Pex2 complex from a pex5Δ pex4Δ strain was prepared and, together with recombinant E1 protein and Pex5, subjected to in vitro ubiquitination. High-molecular-weight Pex5 species were detectable when the E1 and the RING complex were present in the assay (Fig. 2B, lane 1), and the amount of this Pex5 species was significantly decreased in the absence of zinc sulfate (Fig. 2B, lane 2). As expected for an enzyme-catalyzed reaction, the zinc-dependent ubiquitination occurred in a time-dependent manner (Fig. 2C). The formation of modified Pex5 species strictly depended on the presence of the RING complex (Fig. 2B). When the E1 protein was missing, a residual zinc-dependent modification of Pex5 was visible, which might have been due to a copurification of residual E1 or activated Ub which was bound to the isolated yeast complex. However, the results of this experiment show that the isolated yeast complex harbors the enzyme activities required for ubiquitination of Pex5.
The RING peroxins Pex2, Pex10, and Pex12 exhibit E3 ligase activity.
A characteristic feature of protein-Ub ligases is their ability to catalyze autoubiquitination. Ligase activity and the E2-binding site are in general located within the RING domain (30, 49). As all attempts failed to obtain recombinant full-length RING peroxins, we used truncated versions of these proteins. Coding sequences of Pex2 aa 215 to 271 (Pex2RING), Pex10 aa 238 to 337 (Pex10RING), and Pex12 aa 293 to 399 (Pex12RING) were fused to GST (24), and the recombinant fusion proteins were isolated by affinity chromatography. Recombinant Pex4 and Ubc4 were obtained as described previously (26, 35), and GST-Ubc7 served as a negative control. The functionality of all E2-enzymes was tested beforehand by in vitro autoubiquitination (data not shown).
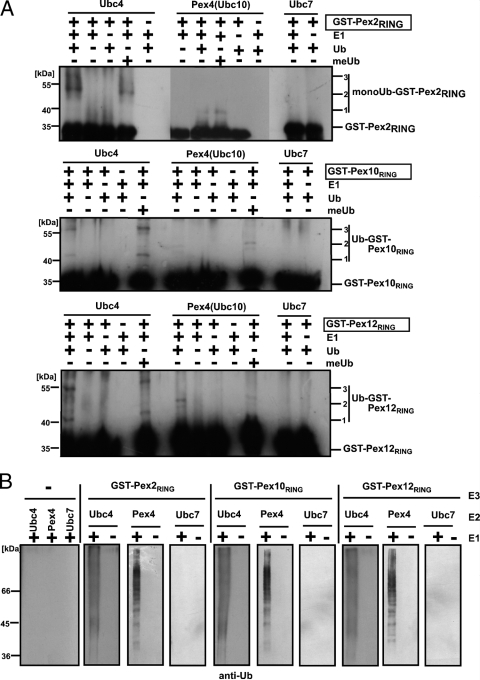
To test for E3 activity, the isolated RING domains of each RING peroxin were subjected to an in vitro autoubiquitination assay as outlined in Fig. 3. Assays were performed in the presence or absence of Ub-activating enzyme and in combination with Ubc4, Pex4, or Ubc7 as E2 protein. In the presence of Ub and E1 protein, all three RING finger domains led to a Ubc4-dependent formation of higher-molecular-weight bands, indicative of an autoubiquitination (Fig. 3A, lanes 1 to 5, and 3B). Omission of either the E1 or Ub precluded the formation of modified RING domains. When methylated Ub was used, autoubiquitination was also observed and the resulting pattern was indistinguishable from the one obtained with wild-type Ub (Fig. 3A). As the methylated Ub species is unable to form chains and thus would block polyubiquitination, the appearance of the same modification pattern suggests that the RING finger domains of Pex2, Pex10, and Pex12 in the presence of Ubc4 as E2 most likely undergo multiple monoubiquitinations at different lysine residues.
FIG. 3.
Autoubiquitination of the recombinant RING domains of Pex2, Pex10, and Pex12. Isolated GST fusions of the RING finger domains of the three RING peroxins, Pex2 (aa 215 to 271), Pex10 (aa 238 to 337), and Pex12 (aa 293 to 399), were incubated with the indicated E2 enzymes in the presence or absence of E1, Ub, and methylated Ub (meUb) (A) or with Ub in the presence or absence of E1 (B). Autoubiquitination of the proteins was monitored by immunoblot analysis with antibodies against GST (A) or Ub (B). Ubc4 and Pex4 but not Ubc7 facilitated the modifications of the three RING domains.
Pex4 represents the second E2 protein which is involved in ubiquitination events essential for peroxisomal matrix protein import (14, 35). Similarly to Ubc4, the Pex4-catalyzed autoubiquitination reactions resulted in E1- and Ub-dependent modifications with multiple monoubiquitin moieties at all three RING finger domains (Fig. 3A, lanes 6 to 10, and B). However, the high-molecular-weight bands detected differ in intensity and size from those obtained in the reactions with Ubc4. This result indicates that the RING domains could be modified in different ways, depending on the E2 protein. The E2 protein Ubc7, which has not been implicated in the peroxisomal ubiquitination cascade (14, 22, 25, 35, 36), served as a control for the specificity of the in vitro autoubiquitination. Thus, the RING finger domains of Pex2, Pex10, and Pex12 all exhibit protein-Ub ligase activity with specificity for the E2 protein Pex4 and Ubc4.
RING peroxin-mediated transfer of Ub to target proteins.
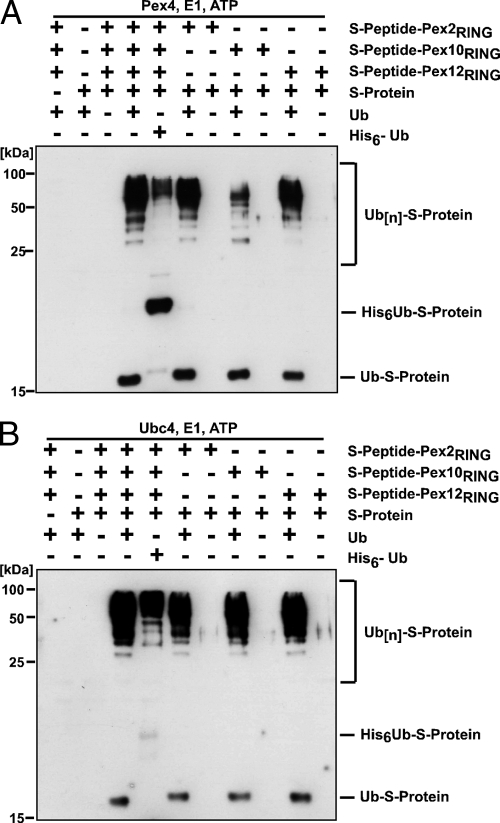
A variety of RING motifs will bind to E2s and catalyze self-addition of Ub when incorporated into fusion proteins (30). However, the principal in vivo function of an E3 is to catalyze the transfer of Ub from a charged E2 to a target protein. Using a test substrate, we demonstrated that the three RING finger peroxins posses such a transfer activity. The assay is based on the high affinity between the 10-kDa S protein and the 15-aa S peptide (38) and has been successfully applied to prove the enzyme activity of putative E3 ligases (4, 43). Therefore, we fused the S peptide to the GST-tagged RING domains of Pex2, Pex10, and Pex12, producing RING fusions with a high-affinity binding site for the S protein. The idea is that the S protein is targeted to the putative protein-Ub ligase via the S peptide. The close proximity of the S protein and the protein-Ub ligase would then allow ubiquitination of the model substrate. The Ub-activating enzyme combined with either Ubc4 or Pex4 as E2 protein and the RING fusions were incubated in the presence or absence of S protein and either Ub or His-tagged Ub. All three RING fusion proteins efficiently catalyzed the transfer of Ub to the S protein in vitro, which is shown by the appearance of Ub-immunoreactive bands at Mr values corresponding to those of monoubiquitinated and polyUb S protein (Fig. 4). These bands shifted to a higher molecular weight when, instead of Ub, a His-tagged Ub was applied, thereby proving the Ub nature of these bands (Fig. 4A and B, lane 4). No ubiquitination of the S protein was observed when either the RING fusion or Ub was omitted. Interestingly, the reaction could be facilitated by Pex4 (Fig. 4A) and Ubc4 (Fig. 4B) but not by Ubc7 (data not shown), indicative of an E2-selective catalytic mechanism of the RING peroxins. Our findings show that the three RING peroxins Pex2, Pex10, and Pex12 are capable of mediating the ubiquitination of the S protein, which proves their protein-Ub ligase activity.
FIG. 4.
RING peroxin-dependent ubiquitination of a synthetic model substrate. Indicated combinations of the recombinant model substrate GST-S peptide-RING finger fusion proteins, the S protein, and either Ub or Ub fused to a His tag (His6-Ub) were incubated with Pex4 (A) or Ubc4 (B) and analyzed by immunoblotting with antibodies against Ub. The appearance of ubiquitinated S protein depends on the presence of any of the three RING domains and Ub.
Pex2 and Pex12 function as protein-Ub ligases in Pex5 ubiquitination.
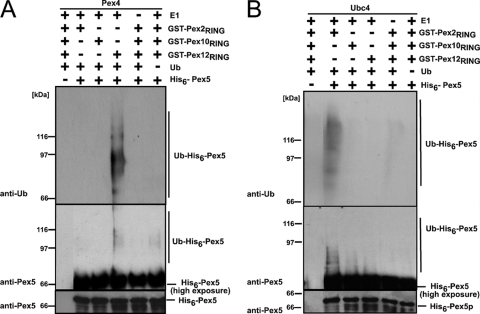
Depending on the E2 protein, Pex5 is modified by Ub in two different ways. Monoubiquitination of Pex5 depends on Pex4, whereas Ubc4 is responsible for Pex5 polyubiquitination (14, 22, 35, 36, 47). Our findings that all three peroxisomal RING finger proteins exhibit protein-Ub ligase activity in the presence of either Pex4 or Ubc4 raised the question of whether they all are capable of ubiquitinating Pex5. Pex4-dependent ubiquitination of the His-tagged PTS1 receptor was observed when the RING domain of Pex12 was present in the assay (Fig. 5A). Neither Pex2 nor Pex10 was capable of facilitating the Pex4-dependent ubiquitination of Pex5. However, Pex5 ubiquitination occurred in the concomitant presence of Pex2 and Ubc4, which was detectable both with Ub and with Pex5 antibody (Fig. 5B). These data indicate that Pex12 seems to act jointly with Pex4 while Pex2 seems to cooperate with Ubc4 in the Pex5-specific ubiquitination. The E2- and E3-dependent ubiquitination of Pex5 (Fig. 5) proved to be much more pronounced than the autoubiquitination of the RING domains (Fig. 3), which required a longer exposure for detection.
FIG. 5.
RING peroxin-dependent ubiquitination of the PTS1 receptor Pex5. Purified GST fusions of the RING finger domains were incubated with recombinant His-Pex5 and either Pex4 (A) or Ubc4 (B) as E2 enzyme. Pex5 ubiquitination was monitored by immunoblot analysis with Pex5- and Ub-specific antibodies. Slower-migrating His-Pex5 forms representing the ubiquitinated receptor were detected when Pex4 was combined with Pex12-RING and when Ubc4 was combined with Pex2-RING. These data demonstrate that these two RING peroxins harbor Pex5-specific protein-Ub ligase activity. The data also indicate that Pex4 and Pex12 as well as Ubc4 and Pex2 form cooperating pairs in the Pex5-specific ubiquitination.
Selective RING finger-dependent ubiquitination of Pex5 in vivo.
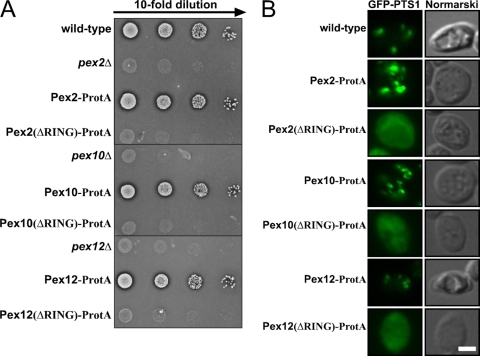
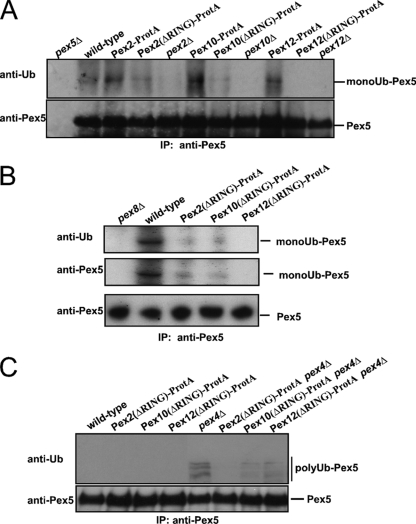
To test for the physiological relevance of the observed protein-Ub ligase activity of the RING peroxins, we monitored Pex5 ubiquitination in different mutant strains. Previous experiments demonstrated that the different RING finger proteins are rapidly degraded if one of them is missing, indicating that they stabilize each other (1, 16, 50), which seems to account for the loss of mono- and polyubiquitination upon deletion of either of the proteins (Fig. 1C and D). To allow testing for the contribution of each of the RING finger peroxins to the different modifications of the PTS1 receptor, we had to inactivate the proteins without destabilization of the RING complex. To this end, we replaced the RING domains of either of the RING peroxins with ProtA, which did not result in destabilization of the proteins (data not shown) and thus allowed the analysis of the function of the RING domains individually. The functional analysis of these strains revealed that each of the three RING domains is essential for peroxisome biogenesis, as the strains expressing the RING-truncated versions exhibit a growth defect on oleic acid as the single carbon source (Fig. 6A) and a defect in the peroxisomal import of PTS1-green fluorescent protein (GFP) (Fig. 6B). In order to investigate the contribution of each RING domain to the ubiquitination of PTS1 receptor, oleic acid-induced wild-type cells were lysed in the presence of NEM and Pex5 was isolated by immunoprecipitation. Monoubiquitinated Pex5 species were detected in wild-type cells as well as in strains expressing either of the full-length RING peroxins fused to ProtA (Fig. 7A, lanes 2, 3 6, and 9). Deletion of PEX2, PEX10, and PEX12 interfered with the Pex5 modification completely as already described above (Fig. 1A and Fig. 7A, lanes 5, 8, and 11). Interestingly, Pex5 was still modified, albeit less efficiently, when the RING domain of Pex2 or Pex10 was replaced with protein A (Fig. 7A, lanes 4 and 7, and B). In contrast, the deletion of the RING domain of Pex12 resulted in the absence of detectable monoubiquitinated Pex5 (Fig. 7A, lane 10, and B). From this, we conclude that under physiological conditions Pex12 is responsible for the monoubiquitination of Pex5.
FIG. 6.
Phenotypic analysis of RING truncation mutants. (A) The growth behavior of indicated S. cerevisiae wild-type and mutant strains on oleic acid as the sole carbon source was investigated using serial dilutions. These were spotted and incubated for 7 days at 30°C. (B) Strains expressing GFP-PTS1 were examined for GFP fluorescence by bright-field microscopy as indicated. Bar = 10 μm. Analysis of live cells for GFP fluorescence was performed with a Zeiss Axioplan microscope and AxioVision 4.1 software (Zeiss, Jena, Germany). The RING domains of each of the three RING peroxins were found to be essential for the import of GFP-PTS1 into peroxisomes.
FIG. 7.
Pex12-RING is required for Pex5 monoubiquitination while Pex2-RING is essential for Pex5 polyubiquitination. The influence of genomic truncations on the monoubiquitination (A and B) and polyubiquitination (C) of Pex5 was analyzed in vivo. Pex5 was isolated by immunoprecipitation from total cell membranes prepared from the indicated oleic acid-induced wild-type strains, null mutants, or strains harboring the genomic truncation of the RING peroxins. The pex8Δ strain served as a negative control. Samples were analyzed by immunoblot analysis with antibodies against Pex5 and Ub. (A and B) To monitor monoubiquitination, NEM was applied to the cells prior to breakage. Genomic truncation of the Pex12-RING prevents the receptor monoubiquitination whereas this modification is still present upon truncation of either the RING domain of Pex2 or that of Pex10. This result indicates that Pex12 is responsible for the Pex5 monoubiquitination. (C) Polyubiquitination occurs in mutants affected in late stages of the peroxisomal protein import pathway represented here by the pex4Δ strain. Truncation of the RING domain of Pex2 but not of Pex10 or Pex12 prevents the receptor polyubiquitination in a pex4Δ background. This result demonstrates that the RING domain of Pex2 is required for Pex5 polyubiquitination.
Polyubiquitination of Pex5 is supposed to be part of a quality control pathway and usually is not observed under wild-type conditions but takes place in mutants affected in late steps of the peroxisomal protein import process, like the pex4Δ strain. Thus, we analyzed the function of the genomically truncated RING peroxins in a pex4Δ background which accumulates polyUb Pex5 at the peroxisomal membrane (22, 36). Accordingly, polyUb Pex5 species were observed in samples derived from a pex4Δ strain (Fig. 7C, lane 5), whereas this modification was absent in the wild-type control (Fig. 7C, lane 1). We now analyzed the consequence of the genomic truncation of either of the RING domains of the three RING peroxins on Pex5 polyubiquitination. Polyubiquitination of Pex5 still took place in cells with truncations of Pex10 and Pex12, although the pattern was less intense. However, no polyubiquitination of Pex5 occurred in cells harboring the genomically truncated Pex2, indicating that the RING domain of Pex2 is required for the modification. Thus, we conclude that Pex2 is the E3 enzyme responsible for the in vivo polyubiquitination of the PTS1 receptor.
DISCUSSION
In order to elucidate whether the RING peroxins Pex2, Pex10, and Pex12 function as E3-Ub ligases, we used several approaches. (i) We show that the RING complex is sufficient for the zinc-dependent ubiquitination of Pex5, thereby demonstrating that at least one of the complex constituents exhibits protein-Ub ligase activity (Fig. 2B). (ii) We demonstrate that all three RING peroxins exhibit a Pex4- or Ubc4-dependent autoubiquitination and transubiquitination activity (Fig. 3 and 4). Both approaches clearly demonstrated that Pex2, Pex10, and Pex12 are protein-Ub ligases. Pex4 and Ubc4 could not be replaced by Ubc7, indicating that all three RING finger domains exhibit a selectivity for the E2 enzymes which is typical for RING finger E3 ligases. In general, this specificity is mediated by the RING domain itself, which is known to bind the E2 proteins (7, 51). (iii) Our results also show that Pex5 can be ubiquitinated by Pex2 and Pex12 in vitro in an assay mixture which contained only recombinant proteins. It turns out that Pex4 forms a functional pair with Pex12 whereas Ubc4 cooperates with Pex2 in the ubiquitination of the PTS1 receptor. Higher-molecular-weight bands within a wide range which might represent Ub modifications at multiple sites were observed (Fig. 5). The finding that the modification pattern differs between the in vivo and in vitro situations is a common observation for this type of ubiquitination assay (32). As Pex5 is embedded into the peroxisomal membrane, where it becomes a part of a multiprotein complex in vivo, lysine residues which normally are hidden may become accessible and thus become possible ubiquitination targets in vitro. Moreover, the fact that only the RING domains and not the entire RING finger proteins were used in our assay may have reduced the specificity of the protein-Ub ligases with respect to their target amino acid residues. (iv) Finally, the identification of the protein-Ub ligase activity of the RING proteins and the specific cooperation of the E2/E3 pairs were corroborated by the in vivo analysis of mutants harboring genomic replacements of the RING domain with ProtA. This approach inactivated each RING peroxin separately without affecting the interdependent stabilization of the proteins within the RING complex. We could demonstrate that the substitution of the RING domain of Pex2 results in a complete loss of the Ubc4-dependent polyubiquitination, whereas truncation of the Pex12-RING domain inhibited the Pex4-dependent monoubiquitination of the PTS1 receptor (Fig. 7).
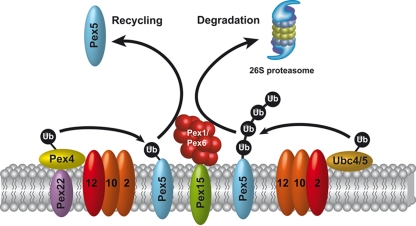
Taken together, the results place Pex2 as the responsible protein-Ub ligase for the Ubc4-dependent polyubiquitination into the proteolytic quality control pathway that leads to proteasomal degradation of Pex5, whereas Pex12 functions as the E3 responsible for the Pex4-dependent monoubiquitination and thus represents a central part of the receptor cycle (Fig. 8).
FIG. 8.
The ubiquitination cascade of the peroxisomal protein import machinery. Pex5, the import receptor for peroxisomal matrix proteins harboring a type I signal sequence (PTS1), is monoubiquitinated and polyUb. Polyubiquitination of Pex5 requires the Ub-conjugating enzyme Ubc4 and the RING peroxin Pex2 as protein-Ub ligase (E3). polyUb Pex5 is supposed to be recognized by the AAA peroxins Pex1 and Pex6 and directed to proteasomal degradation as a part of a quality control system. Monoubiquitination of the receptor is catalyzed by the peroxisomal E2 enzyme Pex4 and Pex12 as the responsible protein-Ub ligase (E3). Monoubiquitination of Pex5 can be regarded as an export signal supposedly recognized by the AAA peroxins, which displace the receptor from the membrane to the cytosol, where it is then available for another round of import.
The importance of Pex12 for peroxisomal matrix protein import is indicated by the fact that approximately 50% of Zellweger syndrome patients affected in the RING-peroxin complex suffer from mutations in the RING finger domain of PEX12 (42). As monoubiquitination is a prerequisite for the recycling of yeast and human Pex5 (5, 14, 35), our finding that the specificity to this Pex4-dependent modification is provided by Pex12 may serve as a mechanistic explanation for the accumulation of Pex5 in the Pex12(S320F) mutant (6).
Pex2 was the first protein which could be connected to Zellweger syndrome (41, 45), and Pex2 also ranks among the first proteins that were recognized as harboring a RING domain (12, 34). As Pex2 is essential for peroxisomal biogenesis, but the polyubiquitination of Pex5 is not (35, 48), the dependency of peroxisomal matrix protein import on Pex2 may indicate additional functions of the protein. One possibility is a contribution of Pex2 to the structural integrity of the RING complex which may be due to a function in the stabilization of Pex10, which is unstable in pex2Δ strains (1, 16). As Pex10 also exhibits intrinsic E3 activity but is not directly involved in the receptor ubiquitination both in vivo and in vitro, the existence of a putative target for this ligase remains to be investigated. A recent report postulates that S. cerevisiae Pex10 is required for polyubiquitination of Pex5, which is based on the observation that the mutation L288A of Pex10 has an effect on the polyubiquitination of Pex5 in vivo (47). Our in vivo results indicate that while substitution of the RING domain of Pex10 reduces the polyubiquitination of Pex5, Pex2 is essential for the polyubiquitination of the receptor (Fig. 7). Moreover, the in vitro data indicate that Pex2 but not Pex10 can directly ubiquitinate Pex5 in an Ubc4-dependent manner (Fig. 5). The fact that beside Pex5 the PTS2 coreceptors Pex18 of S. cerevisiae and Pex20 of P. pastoris are also known to be modified by Ub (28) indicates that there is more than one target protein for the peroxisomal ubiquitination machinery. Future experiments should address the question of whether Pex10 is involved in these processes.
The identification of Pex2, Pex10, and Pex12 as protein-Ub ligases of the peroxisomal protein import machinery completes the peroxisomal ubiquitination cascade acting on the peroxisomal import receptor Pex5. The RING peroxins may account for the overall zinc requirement of the matrix protein import into peroxisomes and glyoxysomes (19, 44). Furthermore, the reported results give insight into the nature of the peroxisomal protein import complex, the importomer. With the exception of the docking complex, all other subcomplexes of the importomer seem to be mechanistically connected to receptor ubiquitination (RING complex and Pex4 complex) or the Ub-mediated receptor release from the membrane (AAA complex). Deficiency in each of these components results in an import defect for peroxisomal matrix proteins, indicating that ubiquitination is an essential step in the peroxisomal protein import pathway. Whether ubiquitination of the receptor, besides being a precondition for receptor recycling, also contributes to other receptor-related functions like cargo release or the still-unresolved mechanism of transport of folded proteins across the peroxisomal membrane remains to be investigated.
Acknowledgments
We are grateful to Uta Ricken and Sigrid Wüthrich for technical assistance and to Wolfgang Schliebs for the reading of the manuscript. We thank Bart Kus and Kiran Madura for kindly providing the His-Ubc4 plasmid and Ubc4 antibody.
Harald W. Platta was supported by the Sophia & Fritz Heinemann Stiftung. This work was supported by the Deutsche Forschungsgemeinschaft (SFB642) and by the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print on 17 August 2009.
REFERENCES
- 1.Agne, B., N. M. Meindl, K. Niederhoff, H. Einwächter, P. Rehling, A. Sickmann, H. E. Meyer, W. Girzalsky, and W. H. Kunau. 2003. Pex8p. An intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell 11:635-646. [DOI] [PubMed] [Google Scholar]
- 2.Albertini, M., W. Girzalsky, M. Veenhuis, and W.-H. Kunau. 2001. Pex12p of Saccharomyces cerevisiae is a component of a multi-protein complex essential for peroxisomal matrix protein import. Eur. J. Cell Biol. 80:257-270. [DOI] [PubMed] [Google Scholar]
- 3.Albertini, M., P. Rehling, R. Erdmann, W. Girzalsky, J. A. K. W. Kiel, M. Veenhuis, and W.-H. Kunau. 1997. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89:83-92. [DOI] [PubMed] [Google Scholar]
- 4.Bays, N. W., R. G. Gardner, L. P. Seelig, C. A. Joazeiro, and R. Y. Hampton. 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3:24-29. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho, A. F., M. P. Pinto, C. P. Grou, I. S. Alencastre, M. Fransen, C. Sá-Miranda, and J. E. Azevedo. 2007. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282:31267-31272. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C. C., D. S. Warren, K. A. Sacksteder, and S. J. Gould. 1999. PEX12 interacts with PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J. Cell Biol. 147:761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, D. E., P. S. Brzovic, and R. E. Klevit. 2007. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14:941-948. [DOI] [PubMed] [Google Scholar]
- 8.Chuang, S. M., and K. Madura. 2005. Saccharomyces cerevisiae Ub-conjugating enzyme Ubc4 binds the proteasome in the presence of translationally damaged proteins. Genetics 171:1477-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodt, G., and S. J. Gould. 1996. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdmann, R., and W. Schliebs. 2005. Peroxisomal matrix protein import: the transient pore model. Nat. Rev. Mol. Cell Biol. 6:738-742. [DOI] [PubMed] [Google Scholar]
- 11.Erdmann, R., F. F. Wiebel, A. Flessau, J. Rytka, A. Beyer, K. U. Fröhlich, and W.-H. Kunau. 1991. PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell 64:499-510. [DOI] [PubMed] [Google Scholar]
- 12.Freemont, P. S., I. M. Hanson, and J. Trowsdale. 1991. A novel cysteine-rich sequence motif. Cell 64:483-484. [DOI] [PubMed] [Google Scholar]
- 13.Girzalsky, W., P. Rehling, K. Stein, J. Kipper, L. Blank, W.-H. Kunau, and R. Erdmann. 1999. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2 dependent protein import into peroxisomes. J. Cell Biol. 144:1151-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grou, C. P., A. F. Carvalho, M. P. Pinto, S. Wiese, H. Piechura, H. E. Meyer, B. Warscheid, C. Sa-Miranda, and J. E. Azevedo. 2008. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J. Biol. Chem. 283:14190-14197. [DOI] [PubMed] [Google Scholar]
- 15.Güldener, U., S. Heck, T. Fiedler, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazra, P. P., I. Suriapranata, W. B. Snyder, and S. Subramani. 2002. Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic 3:560-574. [DOI] [PubMed] [Google Scholar]
- 17.Hettema, E. H., W. Girzalsky, M. van Den Berg, R. Erdmann, and B. Distel. 2000. Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19:223-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicke, L., H. L. Schubert, and C. P. Hill. 2005. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6:610-621. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, T. L., and L. J. Olsen. 2003. Import of the peroxisomal targeting signal type 2 protein 3-ketoacyl-coenzyme A thiolase into glyoxysomes. Plant Physiol. 133:1991-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerscher, O., R. Felberbaum, and M. Hochstrasser. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159-180. [DOI] [PubMed] [Google Scholar]
- 21.Kerssen, D., E. Hambruch, W. Klaas, H. W. Platta, B. de Kruijff, R. Erdmann, W. H. Kunau, and W. Schliebs. 2006. Membrane association of the cycling peroxisome import receptor Pex5p. J. Biol. Chem. 281:27003-27015. [DOI] [PubMed] [Google Scholar]
- 22.Kiel, J. A., K. Emmrich, H. E. Meyer, and W. H. Kunau. 2005. Ubiquitination of the PTS1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J. Biol. Chem. 280:1921-1930. [DOI] [PubMed] [Google Scholar]
- 23.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 24.Koellensperger, G., S. Daubert, R. Erdmann, S. Hann, and H. Rottensteiner. 2007. Characterisation of zinc-binding domains of peroxisomal RING finger proteins using size exclusion chromatography/inductively coupled plasma-mass spectrometry. Biol. Chem. 388:1209-1214. [DOI] [PubMed] [Google Scholar]
- 25.Kragt, A., T. M. Voorn-Brouwer, M. Van den Berg, and B. Distel. 2005. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J. Biol. Chem. 280:7867-7874. [DOI] [PubMed] [Google Scholar]
- 26.Kus, B. M., C. E. Caldon, R. Andorn-Broza, and A. M. Edwards. 2004. Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins 54:455-467. [DOI] [PubMed] [Google Scholar]
- 27.Léon, S., J. M. Goodman, and S. Subramani. 2006. Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim. Biophys. Acta 1763:1552-1564. [DOI] [PubMed] [Google Scholar]
- 28.Leon, S., L. Zhang, W. H. McDonald, J. R. Yates, J. M. Cregg, and S. Subramani. 2006. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J. Cell Biol. 172:67-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, W., M. H. Bengtson, A. Ulbrich, A. Matsuda, V. A. Reddy, A. Orth, S. K. Chanda, S. Batalov, and C. A. Joazeiro. 2008. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One 3:e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marzioch, M., R. Erdmann, M. Veenhuis, and W.-H. Kunau. 1994. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 13:4908-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyase, S., S. Tateishi, K. Watanabe, K. Tomita, K. Suzuki, H. Inoue, and M. Yamaizumi. 2005. Differential regulation of Rad18 through Rad6-dependent mono- and polyubiquitination. J. Biol. Chem. 280:515-524. [DOI] [PubMed] [Google Scholar]
- 33.Miyata, N., and Y. Fujiki. 2005. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 25:10822-10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patarca, R., and M. A. Fletcher. 1992. Ring finger in the peroxisome assembly factor-1. FEBS Lett. 312:1-2. [DOI] [PubMed] [Google Scholar]
- 35.Platta, H. W., F. El Magraoui, D. Schlee, S. Grunau, W. Girzalsky, and R. Erdmann. 2007. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177:197-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platta, H. W., W. Girzalsky, and R. Erdmann. 2004. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem. J. 384:37-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platta, H. W., S. Grunau, K. Rosenkranz, W. Girzalsky, and R. Erdmann. 2005. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7:817-822. [DOI] [PubMed] [Google Scholar]
- 38.Richards, F. M. 1958. On the enzymic activity of subtilisin-modified ribonuclease. Proc. Natl. Acad. Sci. USA 44:162-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenkranz, K., I. Birschmann, S. Grunau, W. Girzalsky, W.-H. Kunau, and R. Erdmann. 2006. Functional association of the AAA-complex and the peroxisomal importomer. FEBS J. 273:3804-3815. [DOI] [PubMed] [Google Scholar]
- 40.Schrader, M., and H. D. Fahimi. 2008. The peroxisome: still a mysterious organelle. Histochem. Cell Biol. 129:421-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimozawa, N., T. Tsukamoto, Y. Suzuki, T. Orii, Y. Shirayoshi, T. Mori, and Y. Fujiki. 1992. A human gene responsible for Zellweger syndrome that affects peroxisome assembly. Science 255:1132-1134. [DOI] [PubMed] [Google Scholar]
- 42.Steinberg, S. J., G. Dodt, G. V. Raymond, N. E. Braverman, A. B. Moser, and H. W. Moser. 2006. Peroxisome biogenesis disorders. Biochim. Biophys. Acta 1763:1733-1748. [DOI] [PubMed] [Google Scholar]
- 43.Swanson, R., M. Locher, and M. Hochstrasser. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15:2660-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terlecky, S. R., J. E. Legakis, S. E. Hueni, and S. Subramani. 2001. Quantitative analysis of peroxisomal protein import in vitro. Exp. Cell Res. 263:98-106. [DOI] [PubMed] [Google Scholar]
- 45.Tsukamoto, T., S. Miura, and Y. Fujiki. 1991. Restoration by a 35K membrane protein of peroxisome assembly in a peroxisome-deficient mammalian cell mutant. Nature 350:77-81. [DOI] [PubMed] [Google Scholar]
- 46.Wiebel, F. F., and W.-H. Kunau. 1992. The PAS2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature 359:73-76. [DOI] [PubMed] [Google Scholar]
- 47.Williams, C., M. van den Berg, E. Geers, and B. Distel. 2008. Pex10p functions as an E(3) ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem. Biophys. Res. Commun. 374:620-624. [DOI] [PubMed] [Google Scholar]
- 48.Williams, C., M. van den Berg, R. R. Sprenger, and B. Distel. 2007. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282:22534-22543. [DOI] [PubMed] [Google Scholar]
- 49.Xie, Y., and A. Varshavsky. 1999. The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 18:6832-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, L., S. Leon, and S. Subramani. 2006. Two independent pathways traffic the intraperoxisomal peroxin PpPex8p into peroxisomes: mechanism and evolutionary implications. Mol. Biol. Cell 17:690-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng, N., P. Wang, P. D. Jeffrey, and N. P. Pavletich. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102:533-539. [DOI] [PubMed] [Google Scholar]