Abstract
Interleukin-6 (IL-6), involved in cancer-related inflammation, acts as an autocrine and paracrine growth factor, which promotes angiogenesis, metastasis, and subversion of immunity, and changes the response to hormones and to chemotherapeutics. We explored transcription mechanisms involved in differential IL-6 gene expression in breast cancer cells with different metastatic properties. In weakly metastatic MCF7 cells, histone H3 K9 methylation, HP1 binding, and weak recruitment of AP-1 Fra-1/c-Jun, NF-κB p65 transcription factors, and coactivators is indicative of low chromatin accessibility and gene transcription at the IL-6 gene promoter. In highly metastatic MDA-MB231 cells, strong DNase, MNase, and restriction enzyme accessibility, as well potent constitutive transcription of the IL-6 gene promoter, coincide with increased H3 S10 K14 phosphoacetylation and promoter enrichment of AP-1 Fra-1/c-Jun and NF-κB p65 transcription factors and MSK1, CBP/p300, Brg1, and Ezh2 cofactors. Complementation, silencing, and kinase inhibitor experiments further demonstrate involvement of AP-1 Fra-1/c-Jun and NF-κB p65/RelB members, but not of the alpha estrogen receptor in promoting chromatin accessibility and transcription across the IL-6 gene promoter in metastatic breast cancer cells. Finally, the natural withanolide Withaferin A was found to repress IL-6 gene transcription in metastatic breast cancer cells upon dual inhibition of NF-κB and AP-1 Fra-1 transcription factors and silencing of IL-6 promoter chromatin accessibility.
Today, various links have been established between chronic inflammation and cancer, converging on the transcription factor NF-κB (48, 53). Controlled expression of NF-κB-driven cytokine genes is an essential component of the immune response and crucial for homeostasis. However, upon switching from inducible to constitutive NF-κB activity, cellular growth becomes deregulated. As observed in many cancers, cells become resistant to apoptosis or demonstrate a propensity to metastasize due to constitutive expression of chemokines, cytokines, and growth factors (21, 50). In addition, hyperactivated activator protein 1 (AP-1) transcription factors are closely involved in cancer invasive gene expression programs, including cytokine gene expression (10, 23, 59).
Besides its health-beneficial role in inflammation, the cytokine interleukin-6 (IL-6) also acts as a paracrine/autocrine growth factor of many tumor cells and confers cancer therapy resistance, which correlates with worse prognosis in breast cancer patients (12, 57, 80, 82, 89). Transcription of the highly conserved IL-6 gene promoter requires binding of AP-1, cyclic AMP-responsive element-binding protein (CREB), and NF-κB transcription factors in response to various stimuli (76, 79), with strict stereospecific requirements for optimal cofactor recruitment, promoting a promoter enhanceosome model with multiple transcription factor-cofactor interactions, in which NF-κB is the primary trigger for IL-6 gene induction in response to tumor necrosis factor (TNF) (76, 79). TNF-induced IL-6 gene expression also requires activation of the mitogen- and stress-activated protein kinase-1 (MSK1) to elicit selective chromatin relaxation at the IL-6 gene promoter, upon phosphorylation of NF-κB p65 S276 and histone H3 S10, followed by further CBP-dependent acetylation of the IL-6 enhanceosome/chromatin environment (85). Nowadays, it is clear that proper expression of each NF-κB-dependent gene requires dynamic cross-talk of promoter enhanceosomes with the local chromatin environment (1, 6, 56, 62, 78). However, it still remains elusive whether transcription factor activation is a prerequisite for chromatin remodeling or, reciprocally, whether local chromatin settings dictate transcription factor binding activities to trigger selective cytokine gene expression.
Within this report, we explored whether chromatin-based mechanisms are responsible for differential IL-6 gene expression in weakly or highly invasive breast cancer cell types. Cancer is nowadays also recognized as en epigenetic disease, since various environmental exposures, chronic inflammation, and aberrant growth factor pathways can deregulate histone/DNA modifications and chromatin remodeling factors (7, 25, 45, 78). Unequal nuclease sensitivities between tumor and normal cell genomes already demonstrated different chromatin organization in benign and malignant cells (52, 63). We have applied chromatin immunoprecipitation (ChIP), chromatin accessibility assays (DNase, micrococcal nuclease [MNase], and restriction enzyme accessibility), and functional approaches (small interfering RNA [siRNA] and inhibitor studies) to further unravel the relationship between transcription factor activation and changes in chromatin structure in relation to IL-6 gene expression levels, in various breast cancer cell types representative of weak or strong metastatic breast tumor samples.
MATERIALS AND METHODS
Cell lines and reagents.
The human breast cancer cell lines MCF7, MCF7-Fra1, MDA-MB231, MDA-MB468, ZR75, and T47D were grown as previously described (10, 88). Recombinant TNF, produced in Escherichia coli and purified to at least 99% homogeneity in our laboratory, has been described elsewhere (77). Kinase inhibitors were purchased from Calbiochem (IKK2i IV), Chromadex (Withaferin A), Promega (U0126), or Alexis (p38 inhibitor SB2053810 and Jun N-terminal protein kinase [JNK] inhibitor SPC600125).
RNA isolation and real-time qPCR analysis.
For induction experiments, breast cancer cells were seeded in six-well dishes and treated with or without 2,500 IU/ml TNF as indicated in the figure legends. Total RNA was extracted by the acid guanidinium thiocyanate-phenol-chloroform method using the Trizol reagent (Invitrogen, Merelbeke, Belgium). Reverse transcription (RT) was performed on 500 ng of total RNA in a 30-μl total volume. After 1/5 dilution, quantitative real-time PCR (qPCR) was performed on 5 μl of mixture for each condition using Invitrogen Sybr green platinum Supermix-UDG on a iCycler apparatus (Bio-Rad, Eke, Belgium). A serial dilution of a cDNA mix standard was used to determine the efficiency of the PCR. All amplifications were performed in duplicate or triplicate, and data were analyzed using Genex software (Bio-Rad, Eke, Belgium), taking primer set efficiency into account. The following qPCR primers were used: for IL-6, sense primer 5′GACAGCCACTCACCTCTTCA3′ and antisense primer 5′AGTGCCTCTTTGCTGCTTTC34, and for β-actin, sense primer 5′GGATGCAGAAGGAGATCACTG3′ and antisense primer 5′CGATCCACACGGAGTACTTG3′. For classic semiquantitative RT-PCR (Fig. 1B), we used the following primers: for IL-6, sense primer 5′GGAGTACCATAGCTACCTGG3′ and antisense primer 5′GACCACAGTGAGGAATGTCC3′, and for the IL-6 cDNA amplicon of 331 bp and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), sense primer 5′GTCCATGCCATCACTGCCA3′ and antisense primer 5′GTGGGAGTTGCTGTTGAAG3′, with a GAPDH amplicon of 342 bp. PCR conditions for RT-PCR were as follows: 94°C for 120 s; 30 cycles of 94°C for 30 s, 57°C for 30 s, and 68°C for 45 s; and final elongation at 68°C for 8 min. PCR products were resolved on a 1.2% agarose gel.
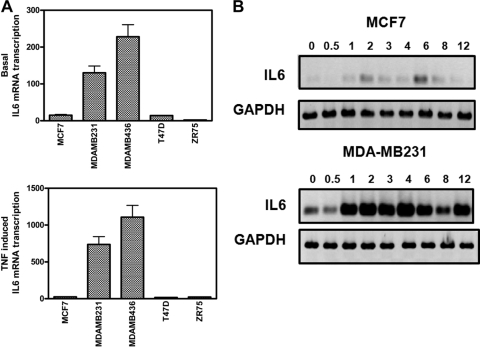
FIG. 1.
Weakly and highly invasive breast cancer cell lines demonstrate weak and strong IL-6 gene expression, respectively. (A) MCF7, MDA-MB231, MDA-MB468, T47D, and ZR75 cells were left untreated (upper panel) or were treated with 2,500 IU/ml TNF (lower panel) for 5 h. Total RNA was isolated, and relative IL-6 mRNA transcription levels were determined by qPCR. (B) Total RNA was extracted from MCF7 and MDA-MB231 cells, left untreated or treated with 2,500 IU/ml TNF for various time points (as indicated on the figure). IL-6 mRNA levels from the two breast carcinoma cells were compared and normalized using housekeeping GAPDH mRNA levels by semiquantitative RT-PCR. Results are representative of two independent experiments.
ELISA.
IL-6 secretion in cell supernatants collected at various time points following TNF induction, was measured by sandwich enzyme-linked immunosorbent assay (ELISA) according to the manufacturer instructions.
Western blot analysis and antibodies.
Western blot analysis was performed as previously described (85). Antibodies directed against Fra-1 (H50), c-Jun (H79), CREB (C21)X, C/EBPβ (C19)X, RelB (C19), Sp1 (PEP2), and p53 (D-01) were obtained from Santa-Cruz Biotechnology, Inc. Antibodies against p65 and phospho-specific antibodies against S73 c-Jun and S536 p65 were from Cell Signaling Technology (Beverly, MA). β-Actin antibodies were obtained from Sigma.
Nuclease digestions, restriction enzyme accessibility, and IEL.
The indirect end-labeling (IEL) technique was performed as previously described (81, 83, 84) with minor modifications. Briefly, cell nuclei were purified from exponentially growing cells. Nuclease digestion (with either DNase I, MNase, or restriction enzymes) was performed in situ on intact nuclei in the presence of the specific digestion buffer for each nuclease: for DNase I, digestion buffer A (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.3 M sucrose); for MNase, buffer A supplemented with 10 mM CaCl2; and for restriction enzymes, the recommended buffer from New England Biolabs (Beverly, MA) supplemented with 100 mg/ml bovine serum albumin and 0.1 mM phenylmethylsulfonyl fluoride. Sodium butyrate was added to all buffers at a final concentration of 1 mM. Nuclei were digested for 10 min on ice for DNase I, 20 min at 22°C for MNase, and 30 min at the recommended temperature for restriction enzymes. Reactions were stopped by addition of 2× proteinase K buffer (100 mM Tris, pH 7.5, 200 mM NaCl, 2 mM EDTA, 1% sodium dodecyl sulfate). Following proteinase K and RNase treatment, genomic DNA underwent phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform extractions and was resuspended in sterile water after ethanol precipitation. Purified DNA (15 to 30 μg) was incubated overnight at 37°C with an excess of PstI. Samples were then analyzed by electrophoresis on a 1.5% agarose gel. Commercial length markers that are highly enriched for repeat sequences were used as markers, where 1 μl was added to 15 or 30 μg genomic DNA and electrophoresed (100-bp marker; New England Biolabs). Following electrophoresis, Southern blotting was carried out: samples were hybridized on a Hybond N+ membrane with a [32P]dCTP-radiolabeled probe spanning nt +1757 to nt +2453 from the IL-6 gene (corresponding to the HindIII-PstI fragment), and the membrane was washed and exposed to X-ray films for autoradiography or to Phospho-Imager screens. Alternative to the HindIII-PstI hybridization probe, a SmaI-EcoRI probe was designed, as schematically depicted in Fig. 4A. IEL hybridization results were analyzed and quantified by free ImageJ sofware (http://rsb.info.nih.gov/ij/), and hypersensitive DNase or MNase positions along the IL-6 gene locus have been calculated according to length marker standard curves.
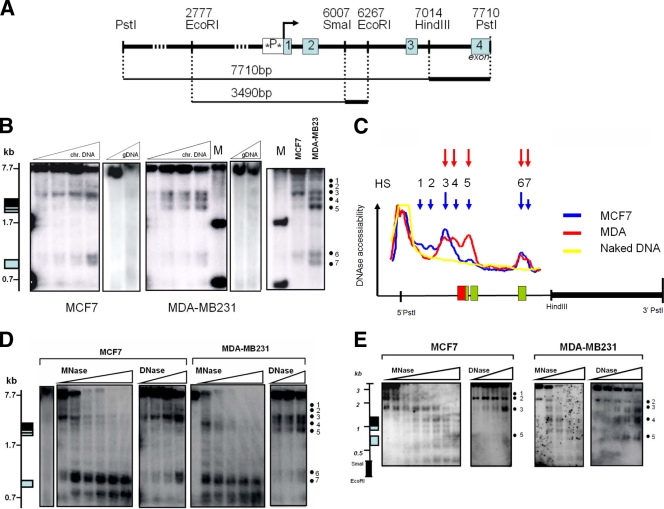
FIG. 4.
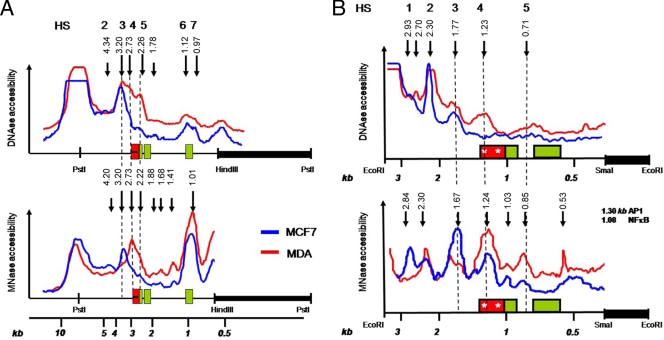
Weakly and highly invasive breast cancer cell types reveal weak and strong chromatin accessibilities for DNase and MNase, respectively, across the IL-6 gene promoter. (A) Schematic representation of both hybridization probes applied in DNase and MNase restriction IEL assays with indication of the relative fragment lengths, starting from either the 3′ PstI (upper line) or 3′ EcoRI (lower line) restriction site end along the IL-6 gene locus (Ensemble Gene ID ENSG00000136244). The IL-6 promoter box is colored black, whereas exons are shaded gray. Stars in the promoter box indicate the AP-1-CREB-C/EBP region and the NF-κB transcription motif, localized at 2.68 and 2.53 kb, respectively, from the 3′ PstI restriction end (1.23 and 1.08 kb, respectively, from the 3′ EcoRI restriction end). (B) DNase accessibility pattern of MCF7 and MDA-MB231 IL-6 gene locus chromatin (chr.) DNA, revealed by IEL with the HindIII-PstI probe. gDNA, genomic DNA; M, molecular size markers. (C) Densitometric profile of DHS as revealed by IEL and analyzed by ImageJ software. The IL-6 promoter box is colored red, whereas exons are shaded green. (D) Comparison of DNase versus MNase accessibility patterns (with increasing enzyme concentrations from left to right) of MCF7 and MDA-MB231 IL-6 gene locus chromatin DNA, revealed by IEL with the HindIII-PstI probe. (E) Comparison of DNase versus MNase accessibility patterns (with increasing enzyme concentrations from left to right) of MCF7 and MDA-MB231 IL-6 gene locus chromatin DNA, revealed by IEL with the SmaI-EcoRI probe.
CHART qPCR.
Determination of chromatin accessibility by real-time PCR (CHART PCR) using the NheI restriction enzyme was performed as previously described (30). Normalization of NheI-cut DNA samples was performed against an upstream DNA region lacking NheI restriction sites. The relative percentage of NheI-accessible chromatin DNA was calculated by subtracting the relative amount of amplified uncut IL-6 promoter DNA (i.e., normalized for the starting amount of DNA by qPCR amplification of an NheI-insensitive DNA region) from 100% starting material (as accessible plus unaccessible chromatin fractions added up to 100%). The following qPCR primer sets were used: set 1 (containing the NheI restriction site), forward primer 5′ CGTGCATGACTTCAGCTTTAC 3′ and reverse primer 5′ TGCAGCTTAGGTCGTCATTG 3′; and set 2 (no NheI restriction site), forward primer 5′ GAAGGCTGTGGAATTCTGAC 3′ and reverse primer 5′ CAGCTCGGCTATATCGGTTC 3′.
ChIP assay.
ChIP analysis was performed as described previously (85). ChIP antibodies used were against Fra1 (H50), c-Jun (H79), NF-κB p50 (NLS), Brg1 (H88), p300 (C20), CBP (C20), IKK1 (Santa Cruz Biotechnology, Inc.), HP1, Ezh3, histone H3, H3 K9me2 (Abcam), NF-κB p65 (Biomol), MSK1 (Sigma), H3 S10 P, H3 K9 Ac, K14 Ac, HDAC1 (Upstate), and RNA polymerase II (Pol II) C-terminal domain (CTD) (Babco). qPCR of coimmunoprecipitated genomic DNA fragments was performed on a Bio-Rad iCycler with the following promoter-specific primers: sense primer 5′-CGTGCATGACTTCAGCTTTAC-3′ and reverse primer 5′-TGCAGCTTAGGTCGTCATTG-3′. ChIP results are presented as ratio of specific to aspecific-antibody-bound immunoprecipitate DNA to input fraction. ChIP experiments were repeated between two and six times. The standard error of the mean of the different ChIP experiments varies between 15 and 30% between experiments.
siRNA transfection experiments.
siRNA against p65 was designed by Cell Signaling. siRNAs against Fra-1, c-Jun, RelB, and ERα were synthesized by MWG-Biotech and target the following sequences: Fra-1, 5′-AACACCATGAGTGGCAGTCAG-3′; c-Jun, 5′-TCCTGAAACAGAGCATGACCCTGAA-3′; RelB, 5′-AAGAACCATCAGGAAGTAGAC-3′; and estrogen receptor alpha (ERα), 5′-AAGATCCTGAAACAGAGCATG-3′. Duplexes (3.6 nM final concentration) were transiently transfected in cells using Lipofectamine RNAiMAX reagent (Invitrogen) by reverse transfection according to the manufacturer's protocol.
RESULTS
Weakly and highly invasive breast cancer cell lines demonstrate weak and strong IL-6 gene expression, respectively.
In line with gene expression profiles demonstrating increased IL-6 expression levels at an advanced metastatic breast cancer stage in patient samples (12, 80, 82), we started with the quantification of IL-6 gene expression levels in different breast cancer cell lines with a weakly invasive (i.e., MCF7, T47D, and ZR75) or highly invasive phenotype (i.e., MDA-MB231 and MDA-MB436). To compare IL-6 transcription levels, all cell lines were left untreated or were treated for 5 h with TNF, after which mRNA was collected for qPCR analysis of IL-6 mRNA levels. As can be observed from Fig. 1A, a significant difference in basal IL-6 transcription levels (upper panel) can be observed in different weakly and highly invasive breast cancer cell types. Furthermore, although a 5-h TNF induction can significantly increase IL-6 transcription levels in all cancer cell types, TNF stimulation in weakly invasive cell types never reaches basal or inducible IL-6 transcription levels similar to those present in the highly invasive cell types (Fig. 1A, lower panel). Along the same line, a time course of IL-6 gene transcription levels in weakly or highly metastatic MCF7 or MDA-MB231 cells, respectively, demonstrates inducible IL-6 gene expression in both cell types, although with strongly different magnitudes (Fig. 1B).
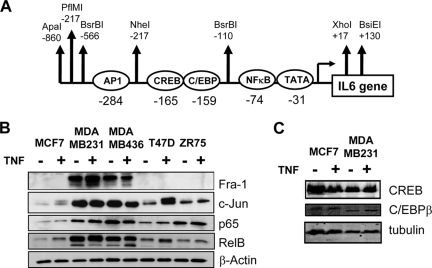
Since overall IL-6 promoter regulation and gene expression strongly depend on coordinated interplay of AP-1 and, NF-κB transcription factor activities (76) (Fig. 2A), we next compared expression levels of the major transcription factor members in the different breast cancer cell types by Western analysis. From Fig. 2B, it appears that highly invasive breast cancer cell lines are strongly enriched for AP-1 Fra-1 and c-Jun and NF-κB p65 and RelB members. In contrast, CREB and C/EBPβ expression levels are not significantly different (Fig. 2C).
FIG. 2.
Weakly and highly invasive breast cancer cell lines demonstrate overexpression of NF-κB and AP-1 transcription factor families. (A) Schematic representation of the highly conserved IL-6 promoter region with indication of the various embedded transcription factor and restriction enzyme motifs. (B) On total cell lysates of MCF7, MDA-MB231, MDA-MB468, T47D, and ZR75 cells, left untreated or treated for 30 min with TNF, we performed Western analysis against Fra-1, c-Jun, NF-κB p65, RelB, and β-actin, as indicated. (C) On total cell lysates of MCF7 or MDA-MB231 cells, left untreated or treated for 30 min with TNF, we performed Western analysis against CREB, C/EBPβ, and tubulin.
Weakly and highly invasive breast cancer cell lines demonstrate heterochromatic and euchromatic chromatin architectures at the IL-6 gene promoter, respectively.
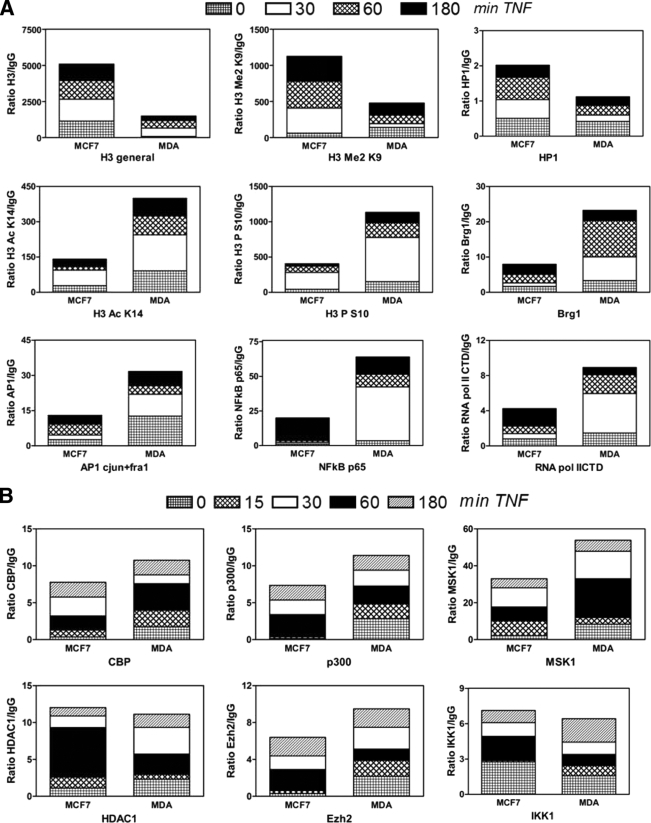
In fibroblasts and breast cancer cells, we previously showed that increased IL-6 transcription levels mirror increased histone H3 phosphoacetylation at the IL-6 promoter enhanceosome upon interplay of the CBP acetylase and MSK kinase cofactors with AP-1, NF-κB, and CREB transcription factors (77, 78, 85). In this respect, we performed ChIP analysis to compare endogenous enhanceosome compositions in MCF7 and MDA-MB231 cells, representative of a weakly or highly invasive breast cancer cell phenotype, respectively. We determined the presence of specific histone marks indicative for gene activation/silencing and evaluated promoter recruitment of various cofactors and transcription factors during 3 h of basal and TNF-inducible transcription in both cell types. Some interesting conclusions can be drawn from Fig. 3A. With respect to histone marks, the proximal IL-6 promoter in MDA-MB231 cells is enriched for gene activation histone marks, such as phospho H3 S10 and acetyl H3 K14. In contrast, enrichment of dimethylated H3 K9 and heterochromatic protein HP1 in MCF7 cells is rather indicative of a silenced IL-6 promoter configuration. Along the same line, MCF7 cells reveal a higher H3 density in the proximal IL-6 promoter than MDA-MB231 cells, in further support of a silenced compact chromatin configuration.
FIG. 3.
Weakly and highly invasive breast cancer cell lines demonstrate heterochromatic and euchromatic chromatin architectures at the IL-6 gene promoter, respectively. MCF7 and MDA-MB231 cells were left untreated or were treated with TNF (2,500 IU/ml) for various time points (as indicated on the figure). ChIP was performed against various histone modifications and transcription factors (A) or cofactors (B). Antibody-bound IL-6 promoter DNA was quantified by qPCR, and ChIP results are represented as relative amount of bound antibody-specific immunoprecipitated DNA to immunoglobulin G (IgG)-aspecific immunoprecipated DNA, normalized for relative amount of input material. ChIP results of the various time points are presented as stacked bar graphs and are means of two ChIP experiments. The standard error of the mean varies 15 to 30% between two experiments.
Upon further looking at levels of transcription factor recruitment, we found increased amounts of NF-κB p65 and AP-1 members c-Jun and Fra-1 at the proximal IL-6 promoter in MDA-MB231 cells, compared to MCF7 cells, which fits with the elevated expression of these factors in MDA-MB231 cells (Fig. 3B). In contrast, both breast cancer cell lines recruit similar amounts of NF-κB p50 (data not shown). Other transcription factor families were not evaluated by ChIP, since CREB, C/EBPβ, Sp1, and RBP-Jκ did not reveal major differences in expression levels when both cell types were compared (Fig. 2C) (data not shown) and similar DNA-binding profiles have already been demonstrated by in vivo footprinting assays (6). Of further interest, excessive IL-6 gene transcription levels in MDA-MB231 cells also correspond with stronger recruitment of RNA Pol II and the chromatin-remodeling factor Brg1, the kinase MSK1, the acetylases CBP and p300, and the histone methyltransferase Ezh2, whereas levels of HDAC1 and IKK1 recruitment are comparable in both cell types. In line with our results, Ezh2 has been characterized as a coactivator in estrogen/wnt receptor target genes in metastatic cancer (67). Altogether our ChIP results indicate a more transcription-permissive euchromatin-like IL-6 gene promoter in MDA-MB231 cells than in MCF7 cells, in full correspondence with the different magnitudes in IL-6 gene transcription.
Weakly and highly invasive breast cancer cell types reveal a weak and strong chromatin accessibility, respectively, across the IL-6 gene promoter.
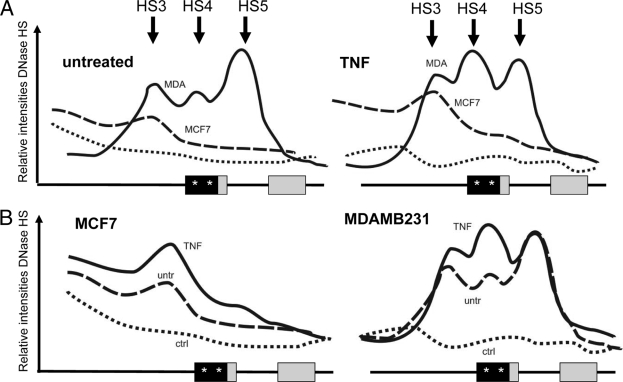
As chromatin structure represents the ultimate integration site of both environmental and differentiative inputs which determines proper gene expression, we next compared the chromatin structures at the IL-6 gene promoter in both cell lines by measuring relative endonuclease (i.e., DNase, MNase, and restriction enzymes) accessibility of chromosomal IL-6 gene DNA. Two DNA hybridization probes were designed downstream of the transcription start site, as indicated in Fig. 4A, and IEL with a PstI-HindIII probe followed by Southern hybridization was applied to map the DNase I hypersensitive sites (HS) in a 7.7-kb region of the endogenous IL-6 gene locus surrounding the promoter region. Altogether, about seven constitutive HS were mapped, among which five were detected in the MCF7 cells (HS1 to -3, -6, and -7) and another combination of five HS in the MDA-MB231 cells (HS3 to -7). Of special note, both cell types show strongly enhanced DNase accessibility near exon3, i.e., HS6 and -7 (Fig. 4B and C). Interestingly, the strongest difference in DNase accessibility was observed at the conserved proximal promoter, in the vicinity of the AP-1-CREB/C-EBPβ enhancer and the NF-κB-binding motif (HS4), i.e., close to the transcription start site, and in exons 1 and 2 (HS5) of the IL-6 gene (Fig. 4B and C). Furthermore, when evaluating TNF-induced DNase accessibility in both cell lines, no major changes were observed in HS positions as compared to the basal condition. However, TNF quantitatively increases accessibility of HS3 and HS4 in MDA-MB231 cells, or HS 3 but not HS4 in MCF7 cells (Fig. 5). Of special note, lack of HS4 in MCF7 cells suggests a locked or silenced chromatin configuration at the proximal IL-6 promoter which prevents strong IL-6 gene expression, in line with the ChIP results discussed above. Reciprocally, high accessibility of HS4 and -5 is clearly consistent with abundant expression of the IL-6 gene in MDA-MB231 cells as compared to MCF7 cells. Altogether, these data suggest that in MDA-MB231 cells, the proximal promoter and transcription start site region of the IL-6 gene may be either temporarily or permanently accessible: i.e., devoid of nucleosomes or may contain a more relaxed nucleosomal structure compared to MCF7 cells. In this respect, we used additional nuclease protocols with MNase and restriction enzymes in conjunction with DNase I to enable a more quantitative assessment of the presence or absence of nuclesomes along a given DNA in the total cell population. MNase preferentially digests nucleosome-free DNA, including linker regions between nucleosomes, and digests essentially the entire chromatin into nucleosome core particles, as either mono-, di-, or trinucleosomes. By IEL, the presence of a nucleosome or nucleosomes is mapped to a fixed restriction site in the DNA. Upon mapping MNase-sensitive sites at the IL-6 gene in MCF7 and MDA-MB231 cells by means of IEL and Southern hybridization, we clearly detected a laddering pattern, consistent with the presence of positioned nucleosomes along the 7.7-kb IL-6 gene locus (Fig. 4D). Furthermore, the presence of these nuclease-sensitive sites was only observed in vivo, thus confirming that they were a consequence of chromatin organization and were not due to secondary sequence-directed cleavage of the nucleases (Fig. 4D). Of special interest, the region with the strongest MNase accessibility at the proximal promoter nicely corresponds with the increased DNase I HS4 in MDA-MB231 cells, as indicated by the dashed lines (Fig. 6). In MCF7 cells, MNase and DNase accessibility is more concentrated upstream of the proximal promoter in the vicinity of HS3 (Fig. 4D). Similarly to the DNase I results, these MNase sites only revealed moderate quantitative but not qualitative changes by TNF induction (data not shown). Taken together, these findings suggest that MCF7 and MDA-MB231 cells have a silenced and accessible nucleosome accessibility, respectively, at the AP-1-CREB/C-EBPβ-NF-κB enhancer region in the proximal IL-6 promoter, of which the latter accessibility can be further increased in the presence of TNF (Fig. 5).
FIG. 5.
Densitometric profiles of DNase accessibility patterns of chromatin DNA isolated from untreated (untr) or TNF-treated (180 min) MCF7 and MDA-MB231 cells, revealed by IEL with the HindIII-PstI probe. Curves are represented in a treatment-specific (A) or cell-type-specific (B) combination. ctrl, control. Stars in the proximal promoter box indicate AP-1 and NF-κB transcription factor motifs.
FIG. 6.
(A) Densitometric profile of DNase- and MNase-sensitive sites as revealed by restriction IEL in Fig. 4D and analyzed by ImageJ software. Stars in the proximal promoter box indicate AP-1 and NF-κB transcription motifs. (B) Densitometric profile of DHS and MNase HS as revealed by IEL in Fig. 4E and analyzed by ImageJ software. Stars in the proximal promoter box indicate AP-1 and NF-κB transcription factor motifs.
To further confirm our results with a second DNA probe, we mapped DNase- and MNase-sensitive sites along a 3.5-kb IL-6 gene locus fragment with an SmaI-EcoRI probe downstream of the transcription start site. Although the obtained hybridization signal intensities were generally weaker than with the HindIII-PstI probe (due to smaller probe size with lower specific radioactivity), the smaller DNA fragment sizes obtained in the IEL experiment allowed a higher-resolution mapping in the promoter region and revealed some additional hypersensitive sites. Similarly as with the HindIII-PstI probe, MDA-MB231 cells reveal three major HS surrounding the proximal promoter (marked as HS3 to -5), whereas we only detected 1 major HS (HS3) in MCF7 cells (Fig. 4E and 6). Furthermore, preferred MNase accessibility can again be observed in the vicinity of HS4 in MDA-MB231 and HS3 in MCF7 cells. In Fig. 7, the major DNase- and MNase-sensitive regions, revealed by both probes, are summarized and indicated relative to the IL-6 promoter transcription start site.
FIG. 7.
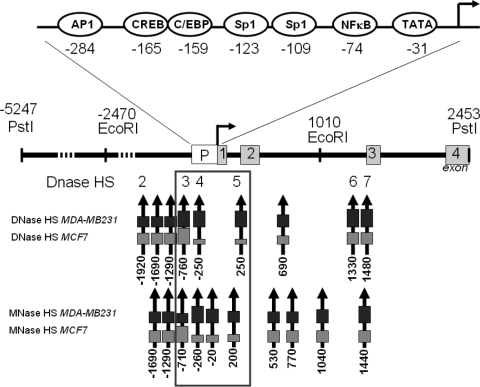
Schematic overview of the various DNase- and MNase-sensitive sites along the IL-6 gene promoter, relative to the major transcription start site (as demonstrated in Fig. 4 to 6). Arrows indicate HS positions, and box size reflects relative accessibility. The three HS localized near the proximal IL-6 promoter are the major focus of this report and are outlined by a box. The numbers correspond to the positions relative to the transcription start site, arbitrarily set as 1.
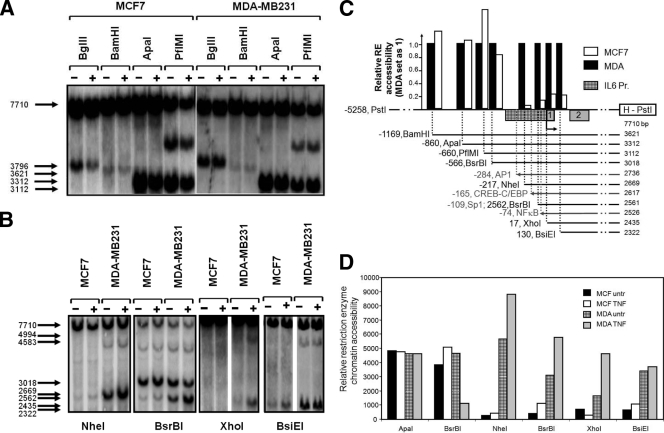
Finally, complementary to DNase and MNase accessibility assays, digestion of chromatin with restriction endonucleases has been very informative too for fine mapping chromatin accessibility in the IL-6 promoter region. By combination of chromatin restriction enzyme digestion with IEL and Southern hybridization with the HindIII-PstI probe, the relative accessibility of specific restriction sites in the promoter region of interest can be easily determined upon evaluation of the cleavage efficiency. Isolated nuclei from both cancer cell lines, both untreated and TNF induced, were partially digested with eight different restriction enzymes cutting within the IL-6 promoter region spanning nt −1200 to +100, thus encompassing the accessible region in the vicinity of HS3 to -5. Upstream of the AP-1-CREB/C-EBPβ enhancer region, comparable constitutive restriction accessibility by BglII, BamHI, ApaI, or PflMI could be detected at the IL-6 promoter in vivo in MCF7 as well as in MDA-MB231 cells (Fig. 8A). In contrast, at the AP-1-CREB/C-EBPβ enhancer and transcription start region surrounding the NF-κB motif, in vivo digestion by NheI, BsrBI, XhoI, or BsiEI enzymes reveals a strongly different accessibility in MCF7 as compared to MDA-MB231 cells (Fig. 8B, or schematically depicted in panel C). This specific difference in accessibility in both regions is especially clear in case of the restriction enzyme BsrBI, which cuts the IL-6 promoter twice: once with similar efficacy upstream of the AP-1-CREB/C-EBPβ enhancer region and once with a clearly different efficacy within the AP-1-CREB/C-EBPβ-NF-κB region. Furthermore, sequence analysis of genomic DNA purified from MCF7 and MDA-MB231 cells demonstrates the absence of polymorphisms, which could account for differential restriction enzyme accessibility of NheI, BsrBI, XhoI, and BsiEI (data not shown). Furthermore, the accessibility by NheI, BsrBI, and XhoI sites surrounding the NF-κB motif can significantly be increased in the presence of TNF (Fig. 8B, or represented in a bar graph in panel D), in line with the increased DNase and MNase accessibility across the HS4 promoter region and elevated IL-6 gene expression upon TNF treatment. Taken together, DNase, MNase, and restriction accessibility assays similarly indicate that the proximal enhancer region of the IL-6 gene may be either temporarily or permanently devoid of nucleosomes or contain a more accessible nucleosomal structure in MDA-MB231 cells than in MCF7 cells. As can be deduced from the MNase accessibility plot (Fig. 6), maximal internucleosome accessibility peaks around 1,670 bp and 1,240 bp upstream of the EcoRI probe end label: i.e., upstream and within the AP-1-CREB/C-EBPβ-NF-κB promoter region of the IL-6 gene. The ability of the MDA-MB231, but not MCF7, cells to constitutively transcribe increased levels of IL-6 nicely corresponds with the observation that the chromatin near the IL-6 proximal promoter enhancer region is highly accessible in the highly metastatic MDA-MB231 cells, but not in the weakly invasive MCF7 cells.
FIG. 8.
Weakly and highly invasive breast cancer cell types reveal weak and strong chromatin accessibilities, respectively, for restriction enzymes across the IL-6 gene promoter. (A and B) Chromatin DNA, isolated from MCF7 and MDA-MB231 cells which were left untreated (untr) or were treated with TNF for 3 h and exposed to different restriction enzymes (as indicated in the figure). Accessible IL-6 gene chromatin fragments which are digested are revealed by IEL with the HindIII-PstI probe. (C and D) Bar graphs representing densitometric analysis of the restriction enzyme accessibilities, detected by IEL in panels A and B and quantified by ImageJ software. A schematic representation of the restriction site motifs and transcription factor binding sites along the IL-6 promoter (Pr.) sequence is shown in panel C. The IL-6 promoter box is hatched, whereas exons are shaded gray. Negative numbers in panel C indicate relative nucleotide positions of the restriction site to the transcription start site. Numbers below HindIII-PstI probe indicate the fragment lengths detected by the HindIII-PstI probe obtained for the different restriction enzymes.
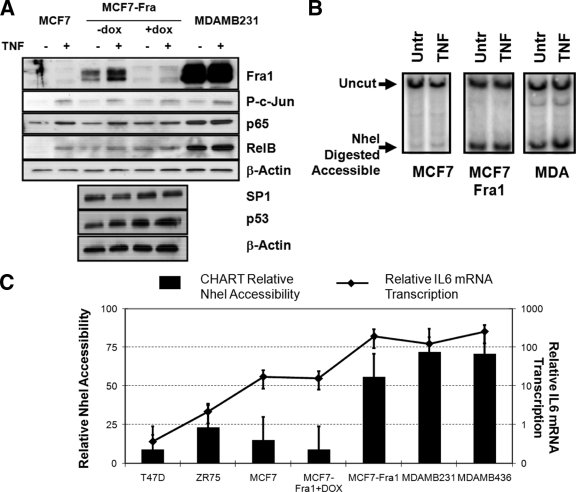
Increased AP-1 Fra-1 transcription factor levels in transgenic MCF7-Fra-1 cells or in highly invasive breast cancer cell types increase IL-6 promoter chromatin accessibility and gene expression.
Next, we wanted to evaluate whether the silenced IL-6 promoter chromatin in MCF7 cells could be switched to a more euchromatic configuration of MDA-MB231 cells by manipulating differentially expressed transcription factor levels. Since AP-1 Fra-1 was found to be the major expressed transcription factor enriched at the IL-6 gene promoter in unstimulated MDA-MB231 cells (Fig. 2 and 3) and was previously identified as a key player in breast cancer invasive programs and IL-6 gene expression (10, 28), we set out to analyze NheI restriction accessibility at the IL-6 gene promoter in Fra-1-overexpressing MCF7 cells in the absence or presence of doxycycline-inducible silencing (10, 28) (Fig. 9A and B). In parallel, we compared NheI accessibility in various other breast cancer cell lines with different expression levels of AP-1 Fra-1 (Fig. 9C). From Fig. 9B, it is clear that Fra-1 overexpression significantly increases NheI accessibility across the IL-6 promoter region in MCF7 cells, as revealed by IEL hybridization. Similar results were obtained by CHART qPCR (Fig. 9C). Here, doxycycline-induced Fra-1 silencing in Fra-1-overexpressing MCF7 cells again reduces NheI accessibility at the IL-6 promoter, in full agreement with decreased IL-6 gene expression levels (Fig. 9C). Moreover, upon comparing NheI accessibility in the different breast cancer cell lines, characterized for AP-1 Fra-1 levels in Fig. 2B, a strong correlation can again be observed between relative Fra-1 protein expression, NheI accessibility, and IL-6 gene expression, three parameters which together increase from left (weakly invasive cell type) to right (highly invasive cell type) (Fig. 9C). This suggests that AP-1 Fra-1 is indeed an important player in establishing accessible chromatin at the enhancer region and is able to catalyze chromatin opening in a stochastic concentration-dependent fashion.
FIG. 9.
Increased Fra1 expression levels increase chromatin accessibility and gene expression across the IL-6 gene promoter. MCF7, MCF7-Fra1, doxycycline (dox)-cultured MCF-Fra-1, MDA-MB231, MDA-MB468, T47D, and ZR75 cells were left untreated (Untr) or were treated for 3 h with TNF (2,500 IU/ml) as indicated in panels A to C. (A) Total cell extracts were analyzed by Western analysis for the presence of AP-1 Fra-1, (P)-c-Jun, NF-κB p65 and RelB, Sp1, p53, and β-actin expression as indicated. (B) Chromatin DNA, isolated from different cell types left untreated or treated with doxycycline for 24 h and/or TNF for 3 h as indicated, was exposed to NheI restriction enzyme. NheI-accessible chromatin fragments which are digested were revealed by restriction IEL with the HindIII-PstI probe for the samples indicated. (C) Chromatin DNA, isolated from various cell types left untreated or treated with doxycycline for 24 h as indicated, was exposed to NheI restriction enzyme. NheI-unaccessible chromatin fragments which remain undigested were quantified by CHART qPCR and subtracted from CHART qPCR amounts obtained with intact undigested chromatin DNA to yield the percent NheI chromatin accessibility (as accessible plus unaccessible fractions add up to 100%). Corresponding IL-6 mRNA transcription levels of the various cell types left untreated or treated with doxycycline for 24 h were quantified by qPCR and are indicated as the line graph in panel C (right axis).
Constitutive NF-κB and AP-1 transcription factor activity increases chromatin accessibility across the IL-6 gene promoter, which promotes malignant IL-6 gene expression in highly invasive breast cancer cells.
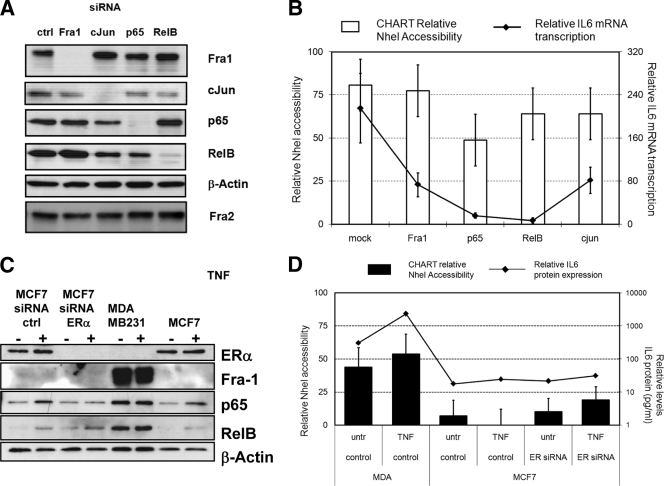
In a reciprocal approach, we wanted to evaluate whether open IL-6 promoter chromatin in MDA-MB231 cells could again be reversed to a silenced configuration upon elimination of NF-κB and AP-1 transcription factors which are highly expressed and/or enriched at the IL-6 gene promoter in MDA-MB231 cells.
In the first series of experiments, AP-1 Fra-1, c-Jun, NF-κB p65, or RelB transcription factors were individually silenced by transient siRNA transfection of MDA-MB231 cells, which were subsequently analyzed for NheI accessibility across the IL-6 gene promoter. Western blot analysis of the different proteins demonstrates specific and highly efficient silencing of each transcription factor in the current experimental setups, whereas β-actin and Fra-2 protein levels were not affected under any condition (Fig. 10A). Upon analysis of IL-6 mRNA gene expression, silencing of NF-κB members p65 and RelB potently decreases constitutive IL-6 gene transcription, whereas the effects of Fra-1 and c-Jun silencing are rather moderate (Fig. 10B). Similar results were obtained upon stimulation of TNF (data not shown). Surprisingly, only silencing of NF-κB p65 is able to significantly reduce NheI accessibility, whereas chromatin silencing effects of Fra-1, c-Jun, and RelB silencing are rather weak or redundant in MDA-MB231 cells (Fig. 10B). As NF-κB and AP-1 activities are also controlled by ER activity (10, 88), we next evaluated whether the differential ER statuses of MCF7 (ER+) and MDA-MB231 (ER−) might codetermine IL-6 promoter accessibility and gene expression. However, despite specific and highly efficient ERα silencing in MCF7 cells (Fig. 10C), no significant changes in IL-6 promoter accessibility and gene expression could be observed (Fig. 10D). Altogether, these findings indicate that the highly accessible IL-6 promoter chromatin configuration in MDA-MB231 cells is not the consequence of a single transcription factor recruited and may be regulated in a stochastic (nuclear concentration dependent) fashion by multiple factors and/or signals.
FIG. 10.
Silencing of individual NF-κB, AP1, or ER transcription factor expression weakly changes chromatin accessibility across the IL-6 gene promoter. (A) MDA-MB231 cells were transiently transfected with specific siRNAs against Fra-1, c-Jun, p65, and RelB. Total cell extracts were prepared 60 h posttransfection for Western analysis of Fra-1, Fra-2, c-Jun, p65, RelB, and β-actin. ctrl, control. (B) Relative IL-6 promoter chromatin accessibility by the NheI restriction enzyme was measured by CHART PCR in MDA-MB231 cells transfected with mock siRNA or siRNA against Fra-1, c-Jun, p65, and RelB. Relative NheI accessibility, presented as bar graphs, is calculated by subtracting the percentage of NheI-unaccessible IL-6 promoter DNA (i.e., amplified amount of uncut IL-6 promoter DNA normalized for the starting amount of DNA by qPCR amplification of an NheI-insensitive promoter region) from 100% (as accessible plus unaccessible fractions add up to 100%). Corresponding IL-6 mRNA transcription levels were quantified by qPCR and are indicated as a line graph in panel B (right axis). (C) MCF7 cells were transiently transfected with specific siRNA against ERα. Total cell extracts were prepared 60 h posttransfection for Western analysis of ERα, Fra1, p65, RelB, and β-actin. (D) Relative IL-6 promoter chromatin accessibility by the NheI restriction enzyme was measured by CHART qPCR in MCF7 cells transfected with mock siRNA or ERα-specific siRNA as compared to MDA-MB231 cells. Relative NheI CHART accessibility is presented as bar graphs. Cell supernatants were collected for quantification of secreted IL-6 protein levels by IL-6 ELISA. Corresponding IL-6 protein expression levels are indicated as line graph in panel D (right axis). untr, untreated.
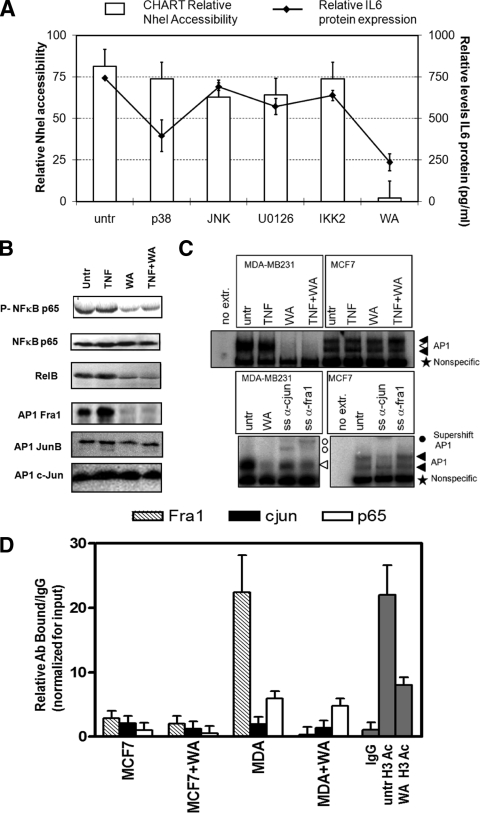
Furthermore, in the second series of experiments, we verified whether synthetic (SB205380, U0126, SPC60012, and IKK2i IV) or natural (Withaferin A) (41) kinase inhibitors of IKK2, p38, extracellular signal-related kinase (ERK), or JNK mitogen-activated protein kinase (MAPK), which target NF-κB and AP-1 family members, were able to silence IL-6 gene transcription in MDA-MB231 cells concomitantly with chromatin silencing (Fig. 11A). Interestingly, none of the pharmacological inhibitors was found to have a strong impact on chromatin accessibility and/or IL-6 expression levels in these cancer cells. In contrast, the natural withanolide Withaferin A significantly reduced NheI accessibility concomitantly with a (strong) reduction in IL-6 gene expression (Fig. 11A). Furthermore, upon further evaluation of Withaferin A-dependent effects on NF-κB family members, we observed that Withaferin A reduces NF-κB p65 Ser536 phosphorylation and RelB protein levels. With respect to AP-1 factors, Withaferin A completely eliminates Fra-1 protein levels (Fig. 11B) and subsequent AP-1/DNA binding in MDA-MB231 cells (Fig. 11C). Finally, we also demonstrate that Withaferin A strongly decreases histone H3 acetylation (K9 and K14), Fra-1 recruitment and, to a lesser extent, c-Jun and p65 recruitment at the IL-6 gene promoter (Fig. 11D). In line with previous reports on cross-coupling of AP-1 and NF-κB members in gene expression (43, 46, 69, 72), our results further indicate that combined inhibition of NF-κB and AP-1 family members, rather than elimination of individual transcription factors, is required for full silencing of chromatin accessibility across the IL-6 gene promoter and for abolishing IL-6 gene expression in MDA-MB231 cells (Fig. 10 and 11).
FIG. 11.
Combined inhibition of NF-κB or AP1 transcription factors by Withaferin A strongly silences chromatin accessibility and gene expression across the IL-6 gene promoter. (A) MDA-MB231 cells were treated for 3 h with IKK2 inhibitor IV (1 μM), p38 inhibitor SB205380 (10 μM), ERK inhibitor U0126 (10 μM), JNK inhibitor SP600125 (10 μM), or Withaferin A (WA; 2.8 μg/ml) as indicated. Corresponding IL-6 promoter chromatin accessibility by the NheI restriction enzyme was measured by CHART PCR in MCF7 cells or in MDA-MB231 cells, left alone (untreated [untr]) or treated for 3 h with the specific inhibitors indicated. Cell supernatants were collected for quantification of secreted IL-6 protein levels by IL-6-ELISA. Relative IL-6 protein expression levels are indicated as line graph in panel A (right axis). (B) Total cell extracts were prepared from MDA-MB231 cells treated for 3 h with Withaferin A (WA; 2.8 μg/ml) alone or cotreated for the last hour with TNF. Western analysis was performed for NF-κB p65, phospho-Ser536-p65, RelB, Fra-1, JunB, and c-Jun. (C) MCF7 and MDA-MB231 cells were treated for 2 h with TNF alone or TNF following 2 h of pretreatment with Withaferin A (2.8 μg/ml). Nuclear lysates were prepared, and AP-1 electrophoretic mobility shift assays were performed essentially as described previously (76). For supershift (SS) analysis, anti-c-Jun and anti-Fra-1 antibodies were added to the extracts 15 min before addition of the probe. (D) ChIP was performed against Fra-1, c-Jun, and NF-κB p65, acetylated histone H3 (antibody pool for acetyl K9 and acetyl K14) in MDA-MB231 cells, left alone or treated for 3 h with Withaferin A (2.8 μg/ml). Immunoprecipitate-bound IL-6 promoter DNA was quantified by qPCR, and ChIP results are represented as relative amount of bound antibody-specific immunoprecipitated DNA/immunoglobulin G (IgG)-aspecific immunoprecipitated DNA, normalized for the relative amount of input material. ChIP results are presented as bar graphs and are means of two ChIP experiments.
DISCUSSION
We have further explored how differential IL-6 gene regulation is achieved in weakly invasive (MCF7) versus highly invasive (MDA-MB231) breast cancer cell types (Fig. 1 and 2) to improve our mechanistic understanding of malignant IL-6 gene expression during tumorigenesis and cancer metastasis (see the introduction).
By ChIP assays in MDA-MB231 cells, we found strongly elevated histone H3 S10 K14 phosphoacetylation and reduced H3 K9 dimethylation levels, concomitantly with favorable recruitment of the histone acetylases CBP and p300, the methylase Ezh2, the kinase MSK1, and the chromatin-remodeling factor Brg1 at the IL-6 gene promoter region (Fig. 3) during IL-6 gene transcription. In contrast, in MCF7 cells, sustained histone H3 K9 dimethylation may exclude a potent increase of histone H3 S10 K14 phosphoacetylation in the presence of heterochromatin protein HP1 and HDAC1, which may establish a predominant silenced proximal-promoter conformation. Alternatively, elevated levels of H3 histone phosphoacetylation at the IL-6 gene promoter in MDA-MB231 cells may prevent silencing by relieving HP1 chromatin tethering (55). Furthermore, ChIPs against total H3 levels present at the IL-6 gene promoter show that increased H3 phosphoacetylation levels in MDA-MB231 cells coincide with lower histone H3 density, which further confirms that the IL-6 promoter chromatin is more relaxed in MDA-MB231 cells than in MCF7 cells. In line with our results, recent genome-wide nucleosome studies have revealed that highly transcribed genes show partial loss of histones H3 and H4 due to the reduced nucleosome occupancy near their transcription start site (33, 39, 47, 60, 65, 75). Despite previous reports on the nuclear role of the H3 kinase IKK1 in cancer metastasis, we failed to demonstrate significant IKK1 enrichment at the IL-6 gene promoter in highly invasive MDA-MB231 cells (Fig. 3B) (51). Interestingly, besides its role as H3 kinase, the Drosophila MSK1 orthologue JIL1 was demonstrated to maintain euchromatic regions and to prevent higher-order heterochromatin spreading of dimethyl H3 K9 marks and HP1 recruitment (93). Furthermore, HP1 displacement by MSK1 was found to allow recruitment of Brg1 and RNA Pol II (86). Similarly, in MDA-MB231 cells, MSK1 may protect the proximal promoter from a silent chromatin conformation by promoting release of HP1 and suppressing H3 K9 methylation in favor of H3 phosphoacetylation (54, 55). Alternatively, decreased expression of HP1 may favor MSK recruitment upon metastasis formation (42). As such, IL-6 promoter activity may depend on an H3 methyl/phospho-acetyl switch chromatin structure, regulated in a stochastic and time-dependent manner by HDAC/HP1 or MSK1/CBP/Brg1/Ezh2 cofactor complexes (27, 49, 54). Of particular interest, histone activation and repression marks as well as cofactors and corepressors can simultaneously be detected at the IL-6 gene promoter in both cell types. However, the current ChIP approach does not allow to discriminate whether these gene activation/repression signatures originate from different cell pools in the asynchronous cell population or reflect differential expression of both cytokine alleles (4, 31, 34, 64). Alternatively, the IL-6 gene may demonstrate a bivalent chromatin signature (8, 61). At the transcription factor level, ChIP experiments demonstrate significant enrichment of NF-κB p65, AP-1 Fra-1, and c-Jun members at the IL-6 promoter, coinciding with elevated IL-6 gene expression, in MDA-MB231 cells, as compared to MCF7 cells. Of special note, interplay between NF-κB and AP-1 transcription factors in constitutive IL-6 gene expression has already been demonstrated in various aggressive cancers (10, 23, 29, 92, 94). Interestingly, c-Jun was found to control histone modifications, NF-κB recruitment, and RNA Pol II function to activate the ccl2 gene (91). During previous work, we have already demonstrated that CBP/MSK cofactors are essential for NF-κB-inducible IL-6 gene expression, whereas AP-1 and CREB-C/EBP rather act as general enhancer factors (76, 85). Since AP-1-dependent MSK1 recruitment has also been demonstrated at gene promoters in breast cancer cells, (24), AP-1 members may synergize with NF-κB for MSK recruitment at the IL-6 gene promoter, in line with our cofactor ChIP results (Fig. 3). Future studies need to address how MSK1 kinase activity regulates activity and/or recruitment of CBP/HDAC, Brg1/Bmi, and HP1/Ezh2 cofactors at the IL-6 gene promoter, as p65 phosphorylation-dependent cofactor exchange was found to be crucial in epigenetic control of NF-κB-dependent genes (16, 22, 62, 87).
Next we prepared experiments to compare chromatin accessibility in MCF7 and MDA-MB231 cells with a heterochromatic or euchromatic IL-6 promoter chromatin signature, respectively. Upon comparing DNase-hypersensitive sites (DHS), we mapped two minor and five major sites, along a 7.7-kb fragment at the IL-6 gene locus (Fig. 4 to 6). Interestingly, three highly accesssible DHS can be identified within MDA-MB231 cells: i.e., distal (DHS3; approximately −760 bp relative to the transcription start site) and proximal promoter (DHS4; approximately −250 bp) sequences, as well as an area close to exon 2 (DHS5; approximately +250 bp), which allow strong constitutive IL-6 gene expression (Fig. 7). In contrast, the accessibility in MCF7 cells is concentrated at the distal promoter sequences (DHS3), with reduced accessiblity in the vicinity of the proximal promoter and transcription start site, which results in low background levels of constitutive IL-6 gene expression. Furthermore, TNF treatment does not change the number of HS, but rather increases accessibility of HS3 and HS3-HS4 in MCF7 and MDA-MB231 cells, respectively. Additional MNase experiments further confirm that a maximal nucleosome accessibility for MCF7 is concentrated around HS3 and in MDA-MB231 cells near HS4 and HS5, suggesting a different nucleosome organization in both cell types. Similarly, upon verifying our IEL results obtained with a second DNA probe (SmaI-EcoRI), we could reconfirm the differential accessibility of the major DNase- and MNase-hypersensitive regions. The MNase results further indicate that both cell types have comparable phasing of nucleosomes and similar regions of (inter)nucleosome accessibility, although significant quantitative differences can be observed. This suggests subtle differences in (translational) nucleosome positioning and important effects on local nucleosome accessibility (for example by rotational positioning and/or nucleosome exchange or eviction). Altogether, DNase- and MNase-sensitive sites were mapped in both cell types by two different probes, which both revealed maximal accessibility near HS3 in MCF7 cells and maximal accessibility near HS3, HS4, and HS5 in MDA-MB231 in the IL-6 promoter region (Fig. 4 to 7). Of special note, maximal chromatin accessibility peaks either upstream of the IL-6 promoter in MCF7 cells or else within the proximal IL-6 promoter in MDA-MB231 cells (Fig. 5 to 7). This is further corroborated by restriction enzyme accessibility assays at the IL-6 promoter region in both breast cancer cells (Fig. 8). In line with the DNase and MNase results, we observe a comparable and constitutive accessibility upstream of the proximal promoter (near HS3) in both cell types. Furthermore, whereas a sharp decline in restriction accessibility is apparent at the proximal promoter harboring AP-1, CREB-C/EBP, and NF-κB transcription factor motifs (near HS4) in MCF7 cells, strong restriction enzyme accessibility is still present in MDA-MB231 cells. Our results therefore, demonstrate a pivotal role of chromatin accessibility at the proximal promoter to mediate constitutive and TNF-inducible IL-6 gene expression in metastatic breast cancer cells (Fig. 5, 6, and 8 and summarized in Fig. 7).
Interestingly, although we find changes in the number of DNase or MNase HS at the IL-6 promoter in response to TNF, a significantly increased accessibility (as determined by DNase, MNase, or restriction enzyme digestion) can be observed in the proximal promoter, which suggests that TNF-inducible factors (such as AP-1 and NF-κB) trigger small local changes in chromatin accessibility of the IL-6 promoter, in line with previous biochemical studies (Fig. 5) (3). In further support of this model, overexpression of AP-1 Fra-1 in MCF7 cells unambiguously increases the chromatin accessibility at the proximal enhancer region of the IL-6 promoter, concomitantly with a strong increase in IL-6 gene expression levels, whereas doxycycline-induced Fra-1 silencing could again reverse promoter accessibility and IL-6 gene expression (Fig. 9). Along the same line, comparison of different breast cancer cell lines with variable AP-1 Fra-1 expression levels revealed a strong positive correlation with chromatin accessibility, IL-6 gene expression, and metastatic properties (Fig. 9). In further agreement with our results, AP-1 binding to its responsive element incorporated into a nucleosome resulted in the complete disruption of nucleosomal structure and was sufficient to facilitate the subsequent binding of a second transcription factor (38, 58). In another cellular context, the ATF/AP-1 element of the IL-8 gene promoter was demonstrated to be crucial for cell-type-specific recruitment of a macro-H2A-containing nucleosome, which prohibits recruitment of NF-κB and coactivators and keeps the promoter in a strongly repressed state (1).
Surprisingly, the reverse experiment, in which NF-κB p65, RelB, and AP1 Fra-1 and c-Jun members were individually silenced in MDA-MB231 cells by siRNA transfection or inhibitor studies, showed that elimination of one factor or kinase has only weak effects on IL-6 promoter chromatin accessibility and IL-6 gene expression levels (Fig. 10) This suggests that epigenetic requirements, chromatin remodeling, and transcription are regulated at multiple interdependent levels (14, 15, 44). As such, highly accessible IL-6 promoter chromatin in MDA-MB231 cells may be regulated in a stochastic fashion by cooperative activities of different pathways (MAPK, IKK, Ras, p38, PI3K, etc.) converging on multiple NF-κB and AP1 members, which may explain the moderate chromatin effects in transient silencing or in inhibitor experiments against individual components (2, 19, 66). It is worth noting that MDA-MB231 cells highly overexpress many members of the AP-1 family, including Fra-1, c-Fos, c-Jun, Fra-2, and different NF-κB factors (10, 88). Furthermore, various reports illustrate important cross-talk of AP-1 and NF-κB members in various aggressive cancers (29, 92, 94). As such, whereas overexpression of Fra-1 alone in MCF7 cells may be sufficient to attract a remodeling complex involved in chromatin opening, elimination of a single factor (either transcription factor or kinase) in MDA-MB231 cells may not be sufficient to abrogate recruitment of this remodeling complex, since other hyperactivated AP-1 and NF-κB members could still take over. Of special note, the observation that for some silencing or inhibitor setups IL-6 gene expression is significantly decreased, whereas effects on chromatin accessibility are rather weak, is not necessarily inconsistent: recent reports indeed illustrate that strong transcriptional activators at enhancers are able to bypass epigenetic settings (44). In addition, transient transcription factor silencing may transiently decrease IL-6 transcription with minor chromatin silencing, whereas permanent transcription factor silencing could be required for heritable chromatin silencing of the IL-6 gene through mitosis.
In favor of interconnected IKK/MAPK pathways regulating NF-κB/AP-1 activity, recent studies demonstrate a crucial function of IKK1 and IKK2 kinases in AP-1 activation (besides NF-κB activation) which also depends on p38/JNK kinases (43, 69). Along the same line, our experiments with the natural withanolide Withaferin A (36, 41) reveal reduced NF-κB p65 Ser536 phosphorylation, and decreased RelB and AP-1 Fra-1 protein levels in MDA-MB231 cells (Fig. 11B). Furthermore, Withaferin A treatment strongly abolishes AP-1 Fra-1/DNA binding (Fig. 11C) and recruitment at the IL-6 gene promoter in MDA-MB231 cells (Fig. 11D). As such, combined inhibition of NF-κB p65, RelB, and AP-1 Fra-1 members by Withaferin A in MDA-MB231 cells (Fig. 11A) results in a strong silencing effect on IL-6 promoter chromatin accessibility coinciding with strongly reduced histone acetylation and IL-6 gene expression. The latter may explain the ability of withanolides from Withania somnifera (alternative name in Ayurvedic medicine is “Ashwagandha”) to inhibit cancer metastasis and support its polypharmaceutical use in integrative oncology (36, 70, 71, 90). Nevertheless, it cannot be excluded that additional Withaferin-sensitive transcription (co)factors, kinases, or epigenetic modulators may also contribute to regulation of IL-6 chromatin accessibility besides AP-1 p65 and Fra-1 (5, 30, 37).
Altogether, we demonstrate that AP-1 and NF-κB have a prominent role in modulating local chromatin accessibility of the proximal IL-6 promoter in metastatic breast cancer. Moreover, cooperation of AP-1 Fra-1 and NF-κB p65 activities is translated into unique cofactor requirements, chromatin accessibility, and transcription dynamics, which trigger malignant IL-6 gene expression in metastatic breast cancer cells. According to our model, chromatin accessibility of the IL-6 gene promoter follows a stochastic (concentration-dependent) cooperative process of multiple factors and chromatin complexes, rather than a simple binary switch mechanism. Similarly, tumorigenic expression of MCP1 in HPV18-positive cervical carcinoma cells strongly depends on the presence of specific DHS genomic regions regulated by c-Jun-Fra-1 heterodimers (26). Along the same line, cooperation between different signaling pathways and NF-κB, NFAT, and AP-1 transcription factors is essential for extensive nucleosome reorganization at the granulocyte-macrophage colony-stimulating factor promoter enhancer, resulting in cell-specific granulocyte-macrophage colony-stimulating factor expression (11, 14, 15, 35, 40). In view of the high kinetic complexity of time- and cell-specific NF-κB-dependent gene expression programs, stochastic chromatin regulation by signaling and transcription factor cooperativity may be indispensable for fine tuning gene transcription with high fidelity in the nucleoplasm (4, 13, 56). In this respect, cancer chemopreventive natural compounds, such as Withaferin A, genistein, curcumin, and green tea polyphenols, which interfere with multiple oncogenic pathways converging on the transcription factors NF-κB and AP-1 (9, 17, 18, 20, 32, 41, 68, 73, 74, 77), may soon join the arsenal of epigenetic drugs.
Acknowledgments
We thank K. Anasinska, V. Quivy, J. Claes, and P. Faes for expertise and technical support and all lab members for suggestions and critical comments.
This work was supported by the IAP5/12 program (Brussels, Belgium). W.V.B. is a postdoctoral fellow with the FWO-Vlaanderen. Work in the C.V.L. lab was supported by grants from the Belgian Fund for Scientific Research (FRS-FNRS, Belgium), the Televie-Program of the FRS-FNRS, the Action de Recherche Concertée du Ministère de la Communauté Francaise (Université Libre de Bruxelles, ARC program no. 04/09-309), the Programme d'Excellence “Cibles” of the Region Wallonne, and the Internationale Brachet Stiftung. C.V.L. is Directeur de Recherches of the FRS-FNRS.
Footnotes
Published ahead of print on 17 August 2009.
REFERENCES
- 1.Agelopoulos, M., and D. Thanos. 2006. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 25:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ancrile, B., K. H. Lim, and C. M. Counter. 2007. Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes Dev. 21:1714-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angelov, D., F. Lenouvel, F. Hans, C. W. Muller, P. Bouvet, J. Bednar, E. N. Moudrianakis, J. Cadet, and S. Dimitrov. 2004. The histone octamer is invisible when NF-kappaB binds to the nucleosome. J. Biol. Chem. 279:42374-42382. [DOI] [PubMed] [Google Scholar]
- 4.Apostolou, E., and D. Thanos. 2008. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134:85-96. [DOI] [PubMed] [Google Scholar]
- 5.Armenante, F., M. Merola, A. Furia, and M. Palmieri. 1999. Repression of the IL-6 gene is associated with hypermethylation. Biochem. Biophys. Res. Commun. 258:644-647. [DOI] [PubMed] [Google Scholar]
- 6.Armenante, F., M. Merola, A. Furia, M. Tovey, and M. Palmieri. 1999. Interleukin-6 repression is associated with a distinctive chromatin structure of the gene. Nucleic Acids Res. 27:4483-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backdahl, L., A. Bushell, and S. Beck. 2009. Inflammatory signalling as mediator of epigenetic modulation in tissue-specific chronic inflammation. Int. J. Biochem. Cell Biol. 41:176-184. [DOI] [PubMed] [Google Scholar]
- 8.Balch, C., K. P. Nephew, T. H. Huang, and S. A. Bapat. 2007. Epigenetic “bivalently marked” process of cancer stem cell-driven tumorigenesis. Bioessays 29:842-845. [DOI] [PubMed] [Google Scholar]
- 9.Belguise, K., S. Guo, S. Yang, A. E. Rogers, D. C. Seldin, D. H. Sherr, and G. E. Sonenshein. 2007. Green tea polyphenols reverse cooperation between c-Rel and CK2 that induces the aryl hydrocarbon receptor, slug, and an invasive phenotype. Cancer Res. 67:11742-11750. [DOI] [PubMed] [Google Scholar]
- 10.Belguise, K., N. Kersual, F. Galtier, and D. Chalbos. 2005. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene 24:1434-1444. [DOI] [PubMed] [Google Scholar]
- 11.Bert, A. G., B. V. Johnson, E. W. Baxter, and P. N. Cockerill. 2007. A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol. Cell. Biol. 27:2870-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertucci, F., P. Finetti, J. Rougemont, E. Charafe-Jauffret, N. Cervera, C. Tarpin, C. Nguyen, L. Xerri, R. Houlgatte, J. Jacquemier, P. Viens, and D. Birnbaum. 2005. Gene expression profiling identifies molecular subtypes of inflammatory breast cancer. Cancer Res. 65:2170-2178. [DOI] [PubMed] [Google Scholar]
- 13.Bosisio, D., I. Marazzi, A. Agresti, N. Shimizu, M. E. Bianchi, and G. Natoli. 2006. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 25:798-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brettingham-Moore, K. H., S. Rao, T. Juelich, M. F. Shannon, and A. F. Holloway. 2005. GM-CSF promoter chromatin remodelling and gene transcription display distinct signal and transcription factor requirements. Nucleic Acids Res. 33:225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brettingham-Moore, K. H., O. R. Sprod, X. Chen, P. Oakford, M. F. Shannon, and A. F. Holloway. 2008. Determinants of a transcriptionally competent environment at the GM-CSF promoter. Nucleic Acids Res. 36:2639-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cha, T. L., B. P. Zhou, W. Xia, Y. Wu, C. C. Yang, C. T. Chen, B. Ping, A. P. Otte, and M. C. Hung. 2005. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310:306-310. [DOI] [PubMed] [Google Scholar]
- 17.Dashwood, R. H., and E. Ho. 2007. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin. Cancer Biol. 17:363-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dashwood, R. H., M. C. Myzak, and E. Ho. 2006. Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis 27:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Santa, F., M. G. Totaro, E. Prosperini, S. Notarbartolo, G. Testa, and G. Natoli. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130:1083-1094. [DOI] [PubMed] [Google Scholar]
- 20.Dijsselbloem, N., S. Goriely, V. Albarani, S. Gerlo, S. Francoz, J. C. Marine, M. Goldman, G. Haegeman, and W. Vanden Berghe. 2007. A critical role for p53 in the control of NF-kappaB-dependent gene expression in TLR4-stimulated dendritic cells exposed to genistein. J. Immunol. 178:5048-5057. [DOI] [PubMed] [Google Scholar]
- 21.Dijsselbloem, N., W. Vanden Berghe, A. De Naeyer, and G. Haegeman. 2004. Soy isoflavone phyto-pharmaceuticals in interleukin-6 affections: multi-purpose nutraceuticals at the crossroad of hormone replacement, anti-cancer and anti-inflammatory therapy. Biochem. Pharmacol. 68:1171-1185. [DOI] [PubMed] [Google Scholar]
- 22.Dong, J., E. Jimi, H. Zhong, M. S. Hayden, and S. Ghosh. 2008. Repression of gene expression by unphosphorylated NF-kappaB p65 through epigenetic mechanisms. Genes Dev. 22:1159-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eferl, R., and E. F. Wagner. 2003. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3:859-868. [DOI] [PubMed] [Google Scholar]
- 24.Espino, P. S., L. Li, S. He, J. Yu, and J. R. Davie. 2006. Chromatin modification of the trefoil factor 1 gene in human breast cancer cells by the Ras/mitogen-activated protein kinase pathway. Cancer Res. 66:4610-4616. [DOI] [PubMed] [Google Scholar]
- 25.Esteller, M. 2007. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 8:286-298. [DOI] [PubMed] [Google Scholar]
- 26.Finzer, P., U. Soto, H. Delius, A. Patzelt, J. F. Coy, A. Poustka, H. zur Hausen, and F. Rosl. 2000. Differential transcriptional regulation of the monocyte-chemoattractant protein-1 (MCP-1) gene in tumorigenic and non-tumorigenic HPV 18 positive cells: the role of the chromatin structure and AP-1 composition. Oncogene 19:3235-3244. [DOI] [PubMed] [Google Scholar]
- 27.Fischle, W., B. S. Tseng, H. L. Dormann, B. M. Ueberheide, B. A. Garcia, J. Shabanowitz, D. F. Hunt, H. Funabiki, and C. D. Allis. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438:1116-1122. [DOI] [PubMed] [Google Scholar]
- 28.Freund, A., V. Jolivel, S. Durand, N. Kersual, D. Chalbos, C. Chavey, F. Vignon, and G. Lazennec. 2004. Mechanisms underlying differential expression of interleukin-8 in breast cancer cells. Oncogene 23:6105-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujioka, S., J. Niu, C. Schmidt, G. M. Sclabas, B. Peng, T. Uwagawa, Z. Li, D. B. Evans, J. L. Abbruzzese, and P. J. Chiao. 2004. NF-κB and AP-1 connection: mechanism of NF-κB-dependent regulation of AP-1 activity. Mol. Cell. Biol. 24:7806-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerlo, S., G. Haegeman, and W. Vanden Berghe. 2008. Transcriptional regulation of autocrine IL-6 expression in multiple myeloma cells. Cell Signal. 20:1489-1496. [DOI] [PubMed] [Google Scholar]
- 31.Guo, L., J. Hu-Li, and W. E. Paul. 2005. Probabilistic regulation in TH2 cells accounts for monoallelic expression of IL-4 and IL-13. Immunity 23:89-99. [DOI] [PubMed] [Google Scholar]
- 32.Hauser, A. T., and M. Jung. 2008. Targeting epigenetic mechanisms: potential of natural products in cancer chemoprevention. Planta Med. 74:1593-1601. [DOI] [PubMed] [Google Scholar]
- 33.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39:311-318. [DOI] [PubMed] [Google Scholar]
- 34.Hollander, G. A., S. Zuklys, C. Morel, E. Mizoguchi, K. Mobisson, S. Simpson, C. Terhorst, W. Wishart, D. E. Golan, A. K. Bhan, and S. J. Burakoff. 1998. Monoallelic expression of the interleukin-2 locus. Science 279:2118-2121. [DOI] [PubMed] [Google Scholar]
- 35.Holloway, A. F., S. Rao, X. Chen, and M. F. Shannon. 2003. Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor kappaB proteins. J. Exp. Med. 197:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichikawa, H., Y. Takada, S. Shishodia, B. Jayaprakasam, M. G. Nair, and B. B. Aggarwal. 2006. Withanolides potentiate apoptosis, inhibit invasion, and abolish osteoclastogenesis through suppression of nuclear factor-kappaB (NF-kappaB) activation and NF-kappaB-regulated gene expression. Mol. Cancer Ther. 5:1434-1445. [DOI] [PubMed] [Google Scholar]
- 37.Inouye, S., M. Fujimoto, T. Nakamura, E. Takaki, N. Hayashida, T. Hai, and A. Nakai. 2007. Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J. Biol. Chem. 282:33210-33217. [DOI] [PubMed] [Google Scholar]
- 38.Ito, T., M. Yamauchi, M. Nishina, N. Yamamichi, T. Mizutani, M. Ui, M. Murakami, and H. Iba. 2001. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J. Biol. Chem. 276:2852-2857. [DOI] [PubMed] [Google Scholar]
- 39.Jiang, C., and B. F. Pugh. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10:161-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson, B. V., A. G. Bert, G. R. Ryan, A. Condina, and P. N. Cockerill. 2004. Granulocyte-macrophage colony-stimulating factor enhancer activation requires cooperation between NFAT and AP-1 elements and is associated with extensive nucleosome reorganization. Mol. Cell. Biol. 24:7914-7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaileh, M., W. Vanden Berghe, A. Heyerick, J. Horion, J. Piette, C. Libert, D. De Keukeleire, T. Essawi, and G. Haegeman. 2007. Withaferin a strongly elicits IkappaB kinase beta hyperphosphorylation concomitant with potent inhibition of its kinase activity. J. Biol. Chem. 282:4253-4264. [DOI] [PubMed] [Google Scholar]
- 42.Kirschmann, D. A., R. A. Lininger, L. M. Gardner, E. A. Seftor, V. A. Odero, A. M. Ainsztein, W. C. Earnshaw, L. L. Wallrath, and M. J. Hendrix. 2000. Down-regulation of HP1Hsalpha expression is associated with the metastatic phenotype in breast cancer. Cancer Res. 60:3359-3363. [PubMed] [Google Scholar]
- 43.Koga, K., G. Takaesu, R. Yoshida, M. Nakaya, T. Kobayashi, I. Kinjyo, and A. Yoshimura. 2009. Cyclic adenosine monophosphate suppresses the transcription of proinflammatory cytokines via the phosphorylated c-Fos protein. Immunity 30:372-383. [DOI] [PubMed] [Google Scholar]
- 44.Koutroubas, G., M. Merika, and D. Thanos. 2008. Bypassing the requirements for epigenetic modifications in gene transcription by increasing enhancer strength. Mol. Cell. Biol. 28:926-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 46.Krappmann, D., E. Wegener, Y. Sunami, M. Esen, A. Thiel, B. Mordmuller, and C. Scheidereit. 2004. The IκB kinase complex and NF-κB act as master regulators of lipopolysaccharide-induced gene expression and control subordinate activation of AP-1. Mol. Cell. Biol. 24:6488-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 48.Lin, W. W., and M. Karin. 2007. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Investig. 117:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lomberk, G., D. Bensi, M. E. Fernandez-Zapico, and R. Urrutia. 2006. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8:407-415. [DOI] [PubMed] [Google Scholar]
- 50.Lu, T., S. S. Sathe, S. M. Swiatkowski, C. V. Hampole, and G. R. Stark. 2004. Secretion of cytokines and growth factors as a general cause of constitutive NFkappaB activation in cancer. Oncogene 23:2138-2145. [DOI] [PubMed] [Google Scholar]
- 51.Luo, J. L., W. Tan, J. M. Ricono, O. Korchynskyi, M. Zhang, S. L. Gonias, D. A. Cheresh, and M. Karin. 2007. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature 446:690-694. [DOI] [PubMed] [Google Scholar]
- 52.Maniotis, A. J., K. Valyi-Nagy, J. Karavitis, J. Moses, V. Boddipali, Y. Wang, R. Nunez, S. Setty, Z. Arbieva, M. J. Bissell, and R. Folberg. 2005. Chromatin organization measured by AluI restriction enzyme changes with malignancy and is regulated by the extracellular matrix and the cytoskeleton. Am. J. Pathol. 166:1187-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mantovani, A., P. Allavena, A. Sica, and F. Balkwill. 2008. Cancer-related inflammation. Nature 454:436-444. [DOI] [PubMed] [Google Scholar]
- 54.Marban, C., S. Suzanne, F. Dequiedt, S. de Walque, L. Redel, C. Van Lint, D. Aunis, and O. Rohr. 2007. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 26:412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mateescu, B., P. England, F. Halgand, M. Yaniv, and C. Muchardt. 2004. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 5:490-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natoli, G., S. Saccani, D. Bosisio, and I. Marazzi. 2005. Interactions of NF-kappaB with chromatin: the art of being at the right place at the right time. Nat. Immunol. 6:439-445. [DOI] [PubMed] [Google Scholar]
- 57.Neve, R. M., K. Chin, J. Fridlyand, J. Yeh, F. L. Baehner, T. Fevr, L. Clark, N. Bayani, J. P. Coppe, F. Tong, T. Speed, P. T. Spellman, S. DeVries, A. Lapuk, N. J. Wang, W. L. Kuo, J. L. Stilwell, D. Pinkel, D. G. Albertson, F. M. Waldman, F. McCormick, R. B. Dickson, M. D. Johnson, M. Lippman, S. Ethier, A. Gazdar, and J. W. Gray. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10:515-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ng, K. W., P. Ridgway, D. R. Cohen, and D. J. Tremethick. 1997. The binding of a Fos/Jun heterodimer can completely disrupt the structure of a nucleosome. EMBO J. 16:2072-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozanne, B. W., H. J. Spence, L. C. McGarry, and R. F. Hennigan. 2006. Invasion is a genetic program regulated by transcription factors. Curr. Opin. Genet. Dev. 16:65-70. [DOI] [PubMed] [Google Scholar]
- 60.Ozsolak, F., J. S. Song, X. S. Liu, and D. E. Fisher. 2007. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 25:244-248. [DOI] [PubMed] [Google Scholar]
- 61.Pietersen, A. M., and M. van Lohuizen. 2008. Stem cell regulation by polycomb repressors: postponing commitment. Curr. Opin. Cell Biol. 20:201-207. [DOI] [PubMed] [Google Scholar]
- 62.Ramirez-Carrozzi, V., A. Nazarian, C. Sarah, L. Gore, R. Sridharan, A. Imbalzano, and S. Smale. 2006. Selective and antagonistic functions of SWI/SNF and Mi-2[beta] nucleosome remodeling complexes during an inflammatory response. Genes Dev. 20:282-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rieber, M., and M. Strasberg-Rieber. 2007. Different chromatin organization in benign and malignant cells revealed by unequal nuclease sensitivity between tumor and normal cell genomes. Am. J. Pathol. 170:787-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20:1405-1428. [DOI] [PubMed] [Google Scholar]
- 65.Schones, D. E., K. Cui, S. Cuddapah, T. Y. Roh, A. Barski, Z. Wang, G. Wei, and K. Zhao. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132:887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Serra, C., D. Palacios, C. Mozzetta, S. V. Forcales, I. Morantte, M. Ripani, D. R. Jones, K. Du, U. S. Jhala, C. Simone, and P. L. Puri. 2007. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell 28:200-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi, B., J. Liang, X. Yang, Y. Wang, Y. Zhao, H. Wu, L. Sun, Y. Zhang, Y. Chen, R. Li, Z. Zhang, M. Hong, and Y. Shang. 2007. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell. Biol. 27:5105-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sliva, D. 2008. Suppression of cancer invasiveness by dietary compounds. Mini Rev. Med. Chem. 8:677-688. [DOI] [PubMed] [Google Scholar]
- 69.Song, L., J. Li, M. Hu, and C. Huang. 2008. Both IKKalpha and IKKbeta are implicated in the arsenite-induced AP-1 transactivation correlating with cell apoptosis through NF-kappaB activity-independent manner. Exp. Cell Res. 314:2187-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Srinivasan, S., R. S. Ranga, R. Burikhanov, S. S. Han, and D. Chendil. 2007. Par-4-dependent apoptosis by the dietary compound withaferin A in prostate cancer cells. Cancer Res. 67:246-253. [DOI] [PubMed] [Google Scholar]
- 71.Stan, S. D., E. R. Hahm, R. Warin, and S. V. Singh. 2008. Withaferin A causes FOXO3a- and Bim-dependent apoptosis and inhibits growth of human breast cancer cells in vivo. Cancer Res. 68:7661-7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stein, B., A. S. Baldwin, Jr., D. W. Ballard, W. C. Greene, P. Angel, and P. Herrlich. 1993. Cross-coupling of the NF-kappa B p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 12:3879-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Surh, Y. J. 2003. Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer. 3:768-780. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki, T., and N. Miyata. 2006. Epigenetic control using natural products and synthetic molecules. Curr. Med. Chem. 13:935-958. [DOI] [PubMed] [Google Scholar]
- 75.Tirosh, I., and N. Barkai. 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 18:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vanden Berghe, W., K. De Bosscher, E. Boone, S. Plaisance, and G. Haegeman. 1999. The nuclear factor-kappaB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J. Biol. Chem. 274:32091-32098. [DOI] [PubMed] [Google Scholar]
- 77.Vanden Berghe, W., N. Dijsselbloem, L. Vermeulen, M. Ndlovu, E. Boone, and G. Haegeman. 2006. Attenuation of mitogen- and stress-activated protein kinase-1-driven nuclear factor-κB gene expression by soy isoflavones does not require estrogenic activity. Cancer Res. 66:4852-4862. [DOI] [PubMed] [Google Scholar]
- 78.Vanden Berghe, W., M. Ndlovu, R. Hoya-Arias, N. Dijsselbloem, S. Gerlo, and G. Haegeman. 2006. Keeping up NF-kB appearances: epigenetic control of immunity or inflammation triggered epigenetics. Biochem. Pharmacol. doi: 10.1016/j.bcp.2006.07.012. [DOI] [PubMed]
- 79.Vanden Berghe, W., S. Plaisance, E. Boone, K. De Bosscher, M. L. Schmitz, W. Fiers, and G. Haegeman. 1998. p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor-kappaB p65 transactivation mediated by tumor necrosis factor. J. Biol. Chem. 273:3285-3290. [DOI] [PubMed] [Google Scholar]
- 80.Van Laere, S., I. Van der Auwera, G. G. Van den Eynden, S. B. Fox, F. Bianchi, A. L. Harris, P. van Dam, E. A. Van Marck, P. B. Vermeulen, and L. Y. Dirix. 2005. Distinct molecular signature of inflammatory breast cancer by cDNA microarray analysis. Breast Cancer Res. Treat 93:237-246. [DOI] [PubMed] [Google Scholar]
- 81.Van Lint, C., J. Ghysdael, P. Paras, Jr., A. Burny, and E. Verdin. 1994. A transcriptional regulatory element is associated with a nuclease-hypersensitive site in the pol gene of human immunodeficiency virus type 1. J. Virol. 68:2632-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van't Veer, L. J., H. Dai, M. J. van de Vijver, Y. D. He, A. A. Hart, M. Mao, H. L. Peterse, K. van der Kooy, M. J. Marton, A. T. Witteveen, G. J. Schreiber, R. M. Kerkhoven, C. Roberts, P. S. Linsley, R. Bernards, and S. H. Friend. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530-536. [DOI] [PubMed] [Google Scholar]
- 83.Verdin, E. 1991. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J. Virol. 65:6790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verdin, E., P. Paras, Jr., and C. Van Lint. 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 12:3249-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vermeulen, L., G. De Wilde, P. V. Damme, W. Vanden Berghe, and G. Haegeman. 2003. Transcriptional activation of the NF-kappaB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J. 22:1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vicent, G. P., C. Ballare, A. S. Nacht, J. Clausell, A. Subtil-Rodriguez, I. Quiles, A. Jordan, and M. Beato. 2006. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell 24:367-381. [DOI] [PubMed] [Google Scholar]
- 87.Voncken, J. W., H. Niessen, B. Neufeld, U. Rennefahrt, V. Dahlmans, N. Kubben, B. Holzer, S. Ludwig, and U. R. Rapp. 2005. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J. Biol. Chem. 280:5178-5187. [DOI] [PubMed] [Google Scholar]
- 88.Wang, X., K. Belguise, N. Kersual, K. H. Kirsch, N. D. Mineva, F. Galtier, D. Chalbos, and G. E. Sonenshein. 2007. Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat. Cell Biol. 9:470-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang, Y., J. G. Klijn, Y. Zhang, A. M. Sieuwerts, M. P. Look, F. Yang, D. Talantov, M. Timmermans, M. E. Meijer-van Gelder, J. Yu, T. Jatkoe, E. M. Berns, D. Atkins, and J. A. Foekens. 2005. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365:671-679. [DOI] [PubMed] [Google Scholar]
- 90.Winters, M. 2006. Ancient medicine, modern use: Withania somnifera and its potential role in integrative oncology. Altern. Med. Rev. 11:269-277. [PubMed] [Google Scholar]
- 91.Wolter, S., A. Doerrie, A. Weber, H. Schneider, E. Hoffmann, J. von der Ohe, L. Bakiri, E. F. Wagner, K. Resch, and M. Kracht. 2008. c-Jun controls histone modifications, NF-κB recruitment, and RNA polymerase II function to activate the ccl2 gene. Mol. Cell. Biol. 28:4407-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zerbini, L. F., Y. Wang, J. Y. Cho, and T. A. Libermann. 2003. Constitutive activation of nuclear factor kappaB p50/p65 and Fra-1 and JunD is essential for deregulated interleukin 6 expression in prostate cancer. Cancer Res. 63:2206-2215. [PubMed] [Google Scholar]
- 93.Zhang, W., H. Deng, X. Bao, S. Lerach, J. Girton, J. Johansen, and K. M. Johansen. 2006. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development 133:229-235. [DOI] [PubMed] [Google Scholar]
- 94.Zhou, Y., C. Yau, J. W. Gray, K. Chew, S. H. Dairkee, D. H. Moore, U. Eppenberger, S. Eppenberger-Castori, and C. C. Benz. 2007. Enhanced NFkappaB and AP-1 transcriptional activity associated with antiestrogen resistant breast cancer. BMC Cancer 7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]