Abstract
CRYPTOCHOME proteins are necessary for mammalian circadian rhythms and have many well-established biochemical roles within the molecular clock. While studies examining the effect of null Cry alleles have been informative, they have failed to dissect out the relative importance of, and the molecular mechanisms behind, the many roles of the CRY1 and CRY2 proteins. To address this, we created an allelic series of Cry mutants through random mutagenesis, followed by a cell-based screen to isolate mutants with aberrant repression of CLOCK-BMAL1. We identified 22 mutants with mutations resulting in single amino acid substitutions which cause a variety of deficiencies in different CRY functions. To illustrate the breadth and value of these new tools, we present an in-depth analysis of two of these mutants, CRY2G354D and CRY2G351D; the former shows deficiency in clock protein binding and is required for repression by both CRYs, while in contrast, the latter displays normal binding function but exhibits a CRY2-specific repression phenotype. Further, while overexpression of CRY2 in NIH 3T3 cells caused a dose-dependent decrease in rhythm amplitude, overexpression of CRY2G351D abolished rhythmicity. In summary, characterization of these unique alleles provides new opportunities for more-sophisticated insight into the multifaceted functions of the CRY proteins in circadian rhythms.
Organisms from cyanobacteria to humans have evolved circadian rhythms to anticipate the dramatic changes in environmental conditions that occur on a daily basis. The driving force behind the 24-h periodicity of these endogenous rhythms is a cell-autonomous molecular oscillator, composed of transcriptional/translational feedback loops (reviewed in reference 1).
In mammals, the Cryptochrome (CRY) proteins CRY1 and CRY2 play an integral role in the circadian clock by closing the core negative-feedback loop (9, 12, 28, 29). In 1999, two groups showed that mice lacking both Crys demonstrate a complete loss of rhythm in circadian wheel-running behavior when put in constant darkness (26, 28), suggesting an essential role for murine CRY proteins (mCRYs) in the core molecular mechanism. This year, it was shown that both CRY1 and CRY2 repressed CLOCK-BMAL1-mediated transcription much more potently than the mammalian PERIOD (PER) proteins in transiently transfected cells (9, 12). While it is known that CRYs are essential for circadian rhythms at both the behavioral (28, 29) and molecular (21) levels, many questions concerning how the various known actions of the CRY proteins contribute to their essential roles in the molecular mechanism of the circadian clock remain unanswered.
Mammalian CRY1 and CRY2 have several well-established biochemical roles in the molecular oscillator in addition to their role in repressing the CLOCK-BMAL1 heterodimer. Both CRY1 and CRY2 bind the PER proteins (9, 12), and this interaction affects PER's stability (30) and localization (12). While the CRY interaction domain on PER has been described, the regions important for PER binding on CRY have only begun to be elucidated (4). CRY protein stability in turn is regulated by FBXL3, an E3 ligase (2, 8, 23). A loss-of-function mutation in Fbxl3 results in reduced affinity for CRYs and aberrant period lengths in locomotor activity rhythms (8, 23). The relative importance of these various roles of CRYs in circadian rhythms and their impact on the repressive functions of CRYs have been difficult to dissect.
The potent repression of CLOCK-BMAL1 by CRYs has been partly characterized at a molecular level. The main body of the protein, called the photolyase homology region (PHR), has been shown to be sufficient to repress CLOCK-BMAL1 in Xenopus laevis. (31) and is well conserved between CRY1 and CRY2. While it has been shown that CRY binding with CLOCK-BMAL1 is required for repression of their transcriptional activity (21), the mechanism by which the repression occurs is not known. One hypothesis suggests that CRYs repress through facilitating chromatin remodeling by inhibiting histone acetyltransferases (7) or by recruiting the histone deacetylase mSin3b (17). In contrast, Ripperger et al. (19) proposed that repression by CRYs may occur through changing the affinity of the CLOCK-BMAL1 complex for its target DNA-binding sequence, the E-box. The relative importance of these or other mechanisms in the repressive function is still not clear.
Null mouse models for the CRYs have different and opposing impacts on period lengths of locomotor activity rhythms. Even though both CRY1 and CRY2 can strongly repress CLOCK-BMAL1 and are highly conserved (77% identical/86% similar) in their PHRs, Cry1−/− mice exhibit a short-period locomotor phenotype, while Cry2−/− mice exhibit a long period (15, 28, 29). Further, Liu et al. reported that the absence of Cry1 causes complete loss of rhythms in dissociated pacemaker neurons and in fibroblasts, while a long-period rhythmicity persists in the absence of Cry2 (13). These data, along with data from Oster et al. (18) showing that Per2Brdm1 Cry2−/− mice display robust rhythms while Per2Brdm1 Cry1−/− mice do not, suggest that CRY1 plays a different and, perhaps, more prominent role in the molecular oscillator. Collectively, these data indicate roles for CRY1 and CRY2 that are not redundant. The question remains of how two proteins, which are highly similar in domain structure and in biochemical function, have opposing effects on period length. Cry null mutants have demonstrated that these genes are necessary for rhythmicity; however, the molecular mechanisms behind the multifaceted functions of these proteins are unclear.
In order to elucidate the molecular determinants underlying the common and differential roles of the CRY proteins in the circadian clock, we performed random mutagenesis, followed by an unbiased cell-based screen to identify residues responsible for repression of CLOCK-BMAL1-mediated transcription. This screen, followed by biochemical secondary screens, led to the identification of numerous mutants for both Cry1 and Cry2 which were deficient in different aspects of CRY function. Since the biochemical mechanism by which Cry1 and Cry2 have opposing effects on clock function when they are individually knocked out in mice (26, 28) is not known, we focused our in-depth analysis on two mutants that were found in the CRY2 screen but not in the CRY1 screen. Collectively, characterization of these mutants has identified novel roles for specific residues in both the CRY1 and CRY2 functions and generated a valuable allelic series of Cry mutants which will facilitate future studies of the specific role of CRYs in the cellular circadian clock.
MATERIALS AND METHODS
Plasmids. (i) Wild-type constructs.
The coding sequence of each gene was amplified by PCR and then subcloned into various expression vectors. Bmal1 was subcloned into the p3XFLAG-CMV-14 vector (Sigma). Cry1 was subcloned into the pRluc-N vector (PerkinElmer). Cry2 was subcloned into the peGFP-C2 vector (Clontech) by fusing it to a C-terminal green fluorescent protein (GFP) tag. Both Cry1 and Cry2 were subcloned into the pCMV-tag3c and pCMV-tag2b vectors (Stratagene), with addition of an N-terminal Myc and Flag tag, respectively. All constructs were verified by diagnostic restriction digestion and sequencing. All point mutations were introduced by QuikChange site-directed mutagenesis (Stratagene) and verified by sequencing.
(ii) NLS-mCry PHR constructs.
The PHRs of CRY1 and CRY2 were amplified and subcloned into pCMV-tag2b, which had a heterologous nuclear localization signal (NLS) (PPKKKRKVEGEF) cloned in frame with the FLAG tag. A premature stop codon was introduced after the PHR (Cry1Y494* and Cry2Y512*).
(iii) Cry tail chimeras.
The Cry1 PHR-Cry2 tail and the Cry2 PHR-Cry1 tail chimeras were cloned into the pCMV-tag2b vector (Stratagene). To generate the Cry1 PHR-Cry2 tail chimera, the PHR of the Cry1 gene (encoding amino acids 1 to 494) and the C-terminal tail of Cry2 (encoding amino acids 513 to the stop codon) were amplified and were ligated using a KpnI site, added to the 3′ end of the Cry1 piece and to the 5′ end of the Cry2 piece. To generate the Cry2 PHR-Cry1 tail chimera, the PHR of the Cry2 gene (encoding amino acids 1 to 512) and the tail of Cry1 (encoding amino acids 495 to the stop codon) were amplified and then ligated via an EcoRV site at the junction.
Construction and screening of the CRY1 and CRY2 mutant libraries.
pCMV-Cry1-RenillaLuciferase and pCMV-Cry2-GFP were mutagenized with hydroxylamine, as previously described in the work of Sato et al. (21). The mutagenized plasmids were then purified with a QIAquick PCR purification kit, transformed into DH5α competent cells (Invitrogen), and plated on LB agar trays. Single colonies were picked by hand, transferred to 96-well sterile blocks (Macherey-Nagel) containing tryptone broth medium with 80 μg/ml kanamycin, and grown overnight at 37°C with shaking. Mutant clone DNA was isolated using the NucleoSpin Robot-96 plasmid miniprep kit (Macherey-Nagel). Once purified, mutant plasmids were quantitated in 96-well microtiter plates with a PowerWave XS spectrometer (Bio-Tek) and normalized to a final concentration of 20 ng/μl with a Hamilton Star liquid-handling robot. Ninety-six-well plates containing normalized mutant DNA were compressed into 384-well bioassay plates (Greiner Bio-One) by transferring 20 ng of mutant plasmid carrying Cry1 or Cry2 with a MiniTrak robot (PerkinElmer) into assigned 384-well plate coordinates. Reverse transfection of Cry1 and Cry2 mutant clones into HEK293 cells and screening for decreased transcriptional activation were performed, as previously described (21). Hits were defined as mutants exhibiting decreased repression, compared to that of wild-type Cry.
Identification of mutations in the Cry1 and Cry2 genes by sequencing.
The coding sequence of each hit was sequenced on both the plus and minus strands, using overlapping sequencing primers to generate a consensus sequence. The consensus sequences were then aligned with the wild-type coding sequence to identify point mutations.
Sequencing primers. (i) Cry1 primers.
The primers used to sequence the plus strand of Cry1 were as follows: pRluc-N F 518-535 (5′-ACGCAAATGGGCGGTAGG-3′), mCry1661F (5′-TTGGAAAGGCATTTGGAAAG-3′), and mCry11229F (5′-TTCACTGCTACTGCCCTGTG-3′). The primers used to sequence the minus strand of Cry1 were as follows: mCry1731R (5′-AGGGAGTTTGCATTCATTCG-3′) and pRluc-N R 680-663 (5′-CATGGTGGCTCTAGCCGG-3′).
(ii) Cry2 primers.
The primers used to sequence the plus strand of Cry2 were as follows: mCry2vectorF (5′-AGGGGCAATGACGCAAATG-3′), mCry2-484F (5′-CAGAAACCCCCCTTACCTAC-3′), and mCry2-913F (5′-TTTGGACAACTCCTGTGGCGAG-3′). The primers used to sequence the minus strand of Cry2 were as follows: mCry2-348R (5′-AAAGGTCAAACGGGTCAC-3′), mCry2-895R (5′-TGCTGTTCCTCTTCACCTTC-3′), mCry2-1425R (5′-AGCCTTCTGAACTGACTCGG-3′), and eGFP-N (5′-CGTCGCCGTCCAGCTCGACCAG-3′).
Cell culture, transient transfection, and Western blotting analysis.
HEK293, COS7, and NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and were transfected with expression plasmids, using Fugene (Roche), according to the manufacturer's instructions.
For Western blotting analysis, lysates were harvested using TGED buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 5% glycerol, 0.5 mM dithiothreitol) with protease inhibitors (Sigma) and 0.5% Triton X-100. Equal amounts of total protein were run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, followed by blotting. The proteins of interest were identified using the following primary antibodies, rabbit anti-GFP (Abcam), mouse anti-V5 (Invitrogen), mouse anti-FLAG (Sigma), or mouse antitubulin (Sigma), followed by the appropriate secondary antibody, either goat anti-rabbit antibody-peroxidase (KPL) or goat anti-mouse antibody-horseradish peroxidase (Chemicon). The bound antibody was visualized using a chemiluminescence Western blotting kit (Roche). In order to do densitometry, bands from each sample were detected and quantified using the Storm phosphorimager (GE Biosciences) and then ImageQuantTL software (GE Healthcare) and normalized to the tubulin signal.
Luciferase repression assay.
The luciferase reporter assay was carried out as previously described (27). Transcriptional activity was assessed with the dual-luciferase reporter assay system (Promega) by measuring the ratio of firefly luciferase activity to Renilla luciferase activity in each lysate.
Immunocytochemistry.
COS7 cells were transfected with 1 μg wild-type or mutant Cry2 and then allowed to incubate for 24 h before fixation. The cells were fixed with 4% paraformaldehyde for 15 min, stained with Hoechst's stain (Sigma), and then mounted, using Fluoromount G (Electron Microscopy Sciences). The cells were viewed using an Olympus inverted epifluorescence microscope (model IX-70). A blind count of 200 cells for each sample was performed, and the percentage of cells in each cell compartment—nuclear, cytoplasmic, or both—was calculated.
IP.
HEK293 cells were transfected with 150 ng null-Renilla Luciferase, 2 μg of Per-luciferase reporter gene, and either 1.5 μg of wild-type Cry2-Gfp or 3.0 μg of a mutant. After 24 h, the cells were harvested, lysed in TGED buffer (described above) with protease inhibitors (Sigma) and 0.5% Triton X-100, and then precleared for 20 min at 13,000 rpm. Eight hundred micrograms of total protein was mixed with 2 μl anti-GFP (Abcam) for 2 h at 4°C. Prepared beads from a protein G immunoprecipitation (IP) kit (Sigma) were added, and the column was incubated overnight at 4°C. The column was washed with TGED plus 0.5% Triton X-100. The bound proteins were eluted in 1× Laemmli's buffer and then analyzed by blotting, as described above.
CRY2 PHR structure model.
A homology model of the CRY2 PHR was generated by the SWISS-MODEL protein homology modeling server (10) by using the sequence of the CRY2 PHR and the coordinates of an Aspergillus nidulans cyclobutane pyrimidine dimer photolyase structure (11) (Protein Data Bank code 1OWL). This model was then visualized using Protein Explorer (14; www.proteinexplorer.org).
Real-time whole-well cycling cell assays with NIH 3T3 cells.
NIH 3T3 cells were seeded into 35-mm dishes and simultaneously transfected with Per2-luciferase, Clock, Bmal1, and various doses of either pCMV-Cry1-RenillaLuciferase, pCMV-Cry2-GFP, or mutant pCMV-Cry2-Gfp. After 48 h, the cells were synchronized with lumicycle medium (Dulbecco's modified Eagle's medium, 10% fetal bovine serum, 10 mM HEPES, 2 mM l-glutamine, 1× antimycotic antibiotic [Invitrogen], 0.1 mM luciferin, 100 nM dexamethasone). Their real-time bioluminescence was measured using a Lumicycle system (ActiMetrics). Traces were detrended by adjacent averaging (Origin 7.5), and then the data from 3.0 days were analyzed using a chi-square periodogram (P < 0.001) and Lumicycle Analysis software (ActiMetrics).
RESULTS
Random mutagenesis and cell-based functional screen of Crys.
To generate mutant Cry alleles, wild-type Cry1 and Cry2 clones were chemically mutagenized and mutant libraries of each were generated. These mutant libraries were then screened in a high-throughput, cell-based reporter assay in which each mutant's ability to repress CLOCK-BMAL1-mediated transcription was assessed via a Per1-Luc reporter construct (Fig. 1A and B). Wells containing the reporter, Clock, and Bmal1 all showed high luciferase activation, which was completely suppressed upon the addition of wild-type Cry, as previously reported (9, 12). While most of the ∼1,100 Cry mutants tested displayed wild-type-like repression, a small population of mutants exhibited decreased repression, and these clones were designated “hits.” Thirty-seven Cry1 and 39 Cry2 hits were selected and rescreened, and 30 of each were confirmed to have a deficient repressive ability (Table 1).
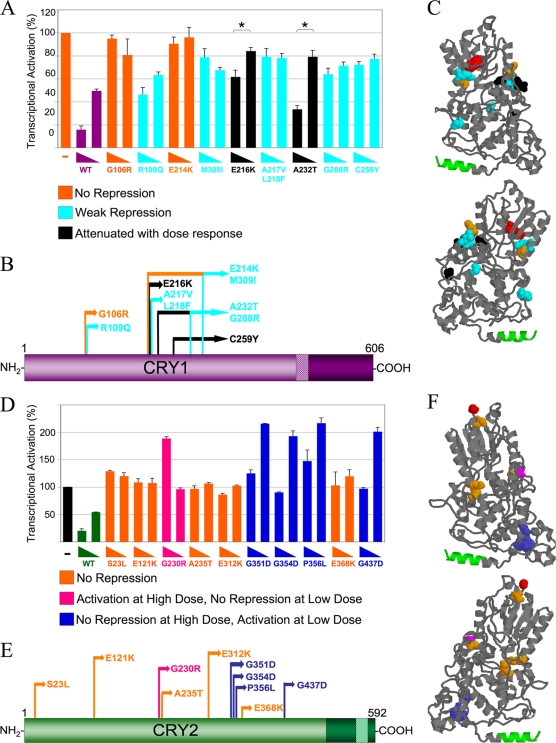
FIG. 1.
Functional screening of Cry mutant libraries reveals novel domains essential for repression of CLOCK-BMAL1-mediated transcription. Mutagenized Cry1 (yellow) (A) or Cry2 clones (red) (B) were transfected into HEK293 cells along with Clock, Bmal1, and an E-box-driven luciferase reporter plasmid in a 384-well format. Approximately 1,100 mutants were tested in duplicate. Controls, which consisted of Clock and Bmal1 plasmids only (red in panel A, yellow in panel B), reporter construct only (black), or Clock, Bmal1, and wild-type Cry (blue) were included. Luciferase assays were performed to measure transcriptional activation and then normalized to the mean of results for the wild-type Cry controls. All data shown are averages from two replicates. While most of the mutant proteins showed normal repressive activity, many showed decreased repression (brackets). In the schematics, CRY1 (C) and CRY2 (D) are drawn to scale. The PHRs of CRY1 and CRY2 (light purple and light green, respectively) and the C-terminal tails (dark purple and dark green, respectively) are represented. In CRY1, the coiled-coil domain is shown (closed arrow). The canonical NLS in CRY2 is shown (open arrow). Of the Cry mutants sequenced, seven had no changes in the coding sequence, 32 had mutations that resulted in premature stop codons (*), and 23 contained point mutations that resulted in amino acid substitutions (black arrows). Some of the mutants contained more than one mutation (double arrows).
TABLE 1.
Summary of random mutagenesis screen hit analysis
| Clone | No. of mutants rescreened | No. of mutants sequenced | No. of confirmed mutants witha:
|
|||
|---|---|---|---|---|---|---|
| No base pair change in CDS | Silent mutation | Truncation | Substitution | |||
| Cry1 mutant | 37 | 30 | 3 | 1 | 16 | 10 (7) |
| Cry2 mutant | 39 | 30 | 0 | 0 | 15 | 15 |
Of the approximately 1,100 mutant constructs in the Cry1 and Cry2 screens, 37 Cry1 and 39 Cry2 hits were selected for their defective repression and rescreened. Of those, 30 mutants of each were confirmed to exhibit decreased repressive ability. Each was sequenced to determine if there was a mutation in the coding sequence (CDS) that could account for their repression deficiency. The coding regions of some clones did not contain mutations, which implies that perhaps the vector or the promoter was mutated. In the Cry1 screen, some residues were mutated more than once, so we isolated 10 substitution mutants but only 7 different mutated residues, denoted by the parentheses.
Sequence analysis of the hits revealed that approximately half had premature stop codons, while 7 of the Cry1 mutants and 15 of the Cry2 mutants had one or more point mutations in the coding sequence (Fig. 1C and D; Table 1). All of the mutations that we identified fell within the sequence encoding the PHR of the CRY proteins. Further, the mutations clustered in regions of the PHR whose CRY functions have not been previously described, and in some cases, the same codons occurred in more than one clone. Although our screen was not saturating, detection of several mutations that cluster in the PHR of CRY2 that were not identified in the CRY1 screen suggest that CRY1 and CRY2 may have different molecular determinants of repression of the CLOCK-BMAL1 heterodimer.
The repressive activities of the mutants with amino acid substitutions were further tested in a dose-response reporter assay, using two subsaturating doses of Cry1 or Cry2 (Fig. 2). Point mutations identified in the mutagenesis screen were first reintroduced into wild-type Cry constructs to investigate the contribution of each amino acid change in repression, particularly in clones that had more than one change. The CRY1 mutants fell into groups that exhibited three different general profiles (Fig. 2A): no repression, weak repression, or attenuated repression that showed a significant dose response (P < 0.01).
FIG. 2.
Some CRY mutants display loss of repression dose-response profiles, while some CRY2 mutants cause increases in CLOCK-BMAL1-mediated transcription. (A) CRY1 dose-response assay. HEK293 cells were transfected with Per2-luciferase reporter, Clock, Bmal1, and 25 or 5 ng of either mutant or wild-type (WT) Cry1-Renilla Luciferase. Each data point is averaged from results of three to six replicates, with the error bars representing the standard errors of the means (SEM). The mutants exhibited three general dose-response profiles: no repression, weak repression, and attenuated repression with a significant dose response (P < 0.01). (B) Schematic of CRY1 mutants' primary structure. The domains are indicated as described in the legend of Fig. 1. Each mutant's profile is color coded (see the key for panel A). (C) Homology model of the CRY1 PHR. This model was generated using the SWISS-MODEL protein homology-modeling server (10; http://swissmodel.expasy.org/workspace/) based on the structure of A. nidulans photolyase (Protein Data Bank code 1OWL) (11) and visualized using Protein Explorer (14). The N-terminal-most residue (red) and the coiled-coil domain (green) are shown. The mutations are color coded by repression profile (see the key in panel A). (D) CRY2 dose-response assay. This experiment was performed as described for panel A, except that 150 or 20 ng of either mutant or wild-type Cry2-GFP was used. The mutants exhibited three different dose-response profiles: no repression, no repression at the low dose with activation at a high dose, and no repression at the high dose with activation at a low dose. (E) Schematic of CRY2 protein with mutant profiles indicated. (F) CRY2 3D homology model with mutant repression profiles. This model was generated as described for panel C but using the CRY2 sequence. The mutant residues are color coded as described in panel D.
In silico modeling was performed to determine the locations of these mutations in the predicted tertiary structure of CRY1. This analysis revealed that the mutations clustered around two physically associated areas of the PHR. The location of each mutation within the protein did not correlate with its repression profiles. In fact, closely clustered residues with identical substitutions showed different profiles (Fig. 2B and C). For example, CRY1E214K showed no repression, while CRY1E216K exhibited attenuated but significant repression that was dose dependent.
When the CRY2 mutants were then assayed in a similar dose-response experiment, several mutants exhibited a complete loss of repression, like those from the Cry1 screen. A subset of CRY2 mutants exhibited activation above levels seen in the CLOCK- and BMAL1-only control, acting more like transcriptional activators than repressors. Within this group, some mutants showed activation at the high dose, with no repression at the low dose, while others showed no repression at the high dose, with activation at the low dose. Four amino acid substitutions that caused no repression at the high dose but activation at the low dose clustered together on the three-dimensional (3D) homology model of CRY2 (Fig. 2F). This region was a hit in the Cry2 screen only.
Identification of a residue important for CRY2-specfic repression.
Since the biochemical underpinnings for the difference between the behavioral phenotypes of Cry1−/− and Cry2−/− mice are not known, we were particularly interested in two mutations that were found only in the CRY2 screen (G351D and G354D). The amino acids at both positions are conserved in CRY1 and CRY2 (Fig. 3A), and the mutations were expressed and properly localized to the nucleus (Fig. 3B). These mutants shared the same repression profile in the dose-response luciferase assay (no repression at the high dose, with activation at the low dose) and were predicted to be in a tight cluster in the protein's 3D structure (Fig. 3A), suggesting that they may be part of a domain required for repression only in CRY2.
FIG. 3.
CRY2G351D and CRY2G354D are expressed at wild-type levels and are localized to the nucleus. (A) CRY2 3D homology model highlighting two CRY2 residues mutated only in the CRY2 screen. This model was generated as described for Fig. 2. The N-terminal residue is shown in black, while the coiled-coil domain is shown in dark blue. The CRY2 mutants identified in the screen are shown in turquoise, except for the G351D and G354D mutants, drawn in magenta and light pink, respectively. Under the model is a protein alignment of residues G333 to W339 in CRY1 and G351 to W357 in CRY2. Conserved residues are represented in gray, with G351D and G354 highlighted. (B) Immunocytochemistry of CRY2 mutants. Either wild-type Cry2-GFP or mutant Cry2-GFP was transfected into COS7 cells. CRY2-GFP (wild type or mutant) is green, the nuclei are blue, and colocalization is light blue. A blind count of 200 cells for each sample was performed, and the percentage of cells in each cell compartment, nuclear (N), cytoplasmic (C), or both (N/C), was calculated. The graphs below each picture indicate the percentages of cells showing N, N/C, or C localization.
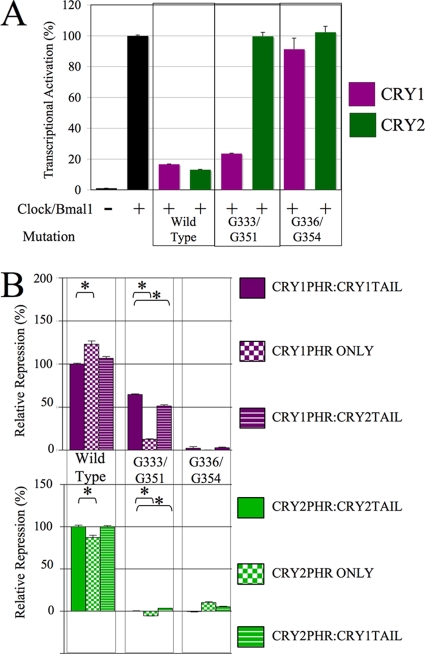
In order to test whether CRY2G351 and CRY2G354 were essential for repression by CRY2 only, the analogous mutations were made in CRY1 (G333D and G336D, respectively), and then the mutants were tested for repression in a luciferase reporter assay (Fig. 4A). While CRY1G336D exhibited a complete loss of repression identical to that of its CRY2 counterpart (CRY2G354D), CRY1G333D, whose mutant residue is in close proximity to the residue exhibiting the same change of glycine to aspartic acid, had little effect.
FIG. 4.
Identification of a repression domain unique to CRY2. (A) Mutations in CRY1 (G333D and G336D) and CRY2 (G51D and G354D) were each introduced and tested for their repression function in a luciferase assay. HEK293 cells were transfected with the Per-luciferase reporter, Clock, Bmal1, and 25 ng of either the wild-type or mutant Cry1 or Cry2, as indicated. The raw data were normalized such that the average level of activation of Clock, Bmal1, and the reporter control was equal to 100% activation. Each data point is averaged from the results of three replicates, with the error bars representing the SEM. (B) In the experiment whose results are shown, the G333D/G351D and G336D/G354D mutations were each introduced into six different constructs: wild-type CRY1 (CRY1PHR:CRY1TAIL), CRY1PHR with no tail (CRY1PHR ONLY), CRY1PHR:CRY2TAIL, wild-type CRY2 (CRY2PHR:CRY2TAIL), CRY2PHR with no tail (CRY2PHR ONLY), and CRY2PHR:CRY1TAIL. These constructs were then tested in a luciferase assay, as described for panel A, and the data were normalized such that the average repression of the wild-type CRYs was set equal to 100%. Each data point was averaged from three to nine replicates, with the error bars representing the SEM. The asterisks indicate groups that were significantly different from each other (P < 0.01).
To investigate the possibility that the C-terminal tail may affect the repression deficit exhibited by CRY2G351D, CRY1 and CRY2 mutants (G333 or G354 and G336 or G354, respectively) were tested in six different constructs: full-length wild-type CRY1 and CRY2, CRY1PHR and CRY2PHR (missing their C-terminal tails and fused to a heterologous NLS), and CRY1/CRY2 PHR/tail chimeras (CRY1PHR with the CRY2 tail [CRY1PHR:CRY2TAIL] and CRY2PHR with the CRY1 tail [CRY1PHR:CRY1TAIL]). Consistent expression levels of the resulting proteins were verified (data not shown). These constructs were then tested in a luciferase reporter assay (Fig. 4B). Wild-type CRY1 and CRY2, regardless of the presence or identity of the C-terminal tail, could repress normally. Interestingly, when the mutation G333D was made in CRY1PHR only, its ability to repress was greatly reduced. In contrast, removing or altering the C-terminal tail of CRY1G336D, CRY2G351D, or CRY2G354D had little to no effect on their repressive ability. These data show that the ability of CRY1 protein containing G333D to strongly repress is dependent on the presence of the C-terminal tail.
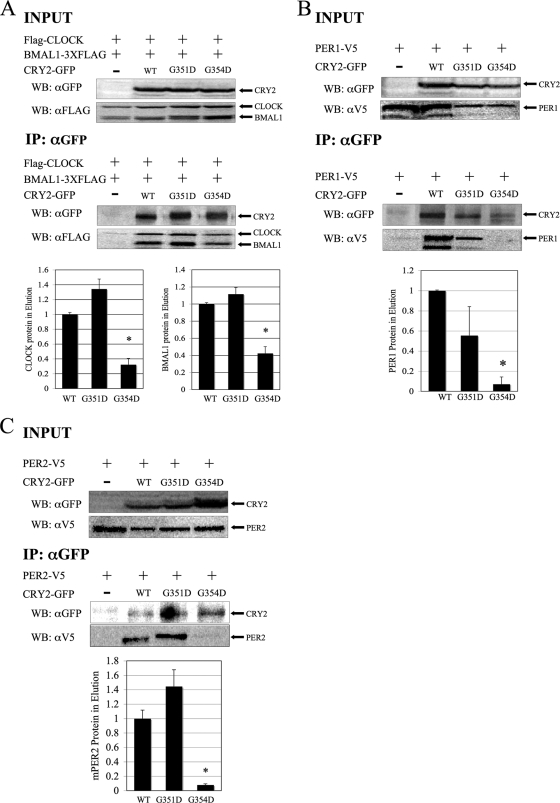
Binding to core circadian clock components is differentially disrupted in CRY2G354D and CRY2G351D.
To characterize the ability of the CRY2 mutants (G351D and G354D) to bind CLOCK and BMAL1, HEK293 cells were transfected with Flag-Clock, Bmal1-Flag3 (with a three-Flag tag), and either wild-type or mutant Cry2-GFP. The CRY2-GFP (mutant or wild type)-containing protein complexes were then immunoprecipitated, and the eluants were analyzed for the presence of CLOCK and BMAL1 by Western blot analyses. CRY2G354D, a residue important for repression by both CRY1 and CRY2, showed significantly decreased binding to CLOCK-BMAL1, PER1, and PER2 (Fig. 5). The region of CRY2 where this residue is located has not been previously implicated in protein-protein interactions with other clock proteins. On the other hand, CRY2G351D showed intact binding to both CLOCK-BMAL1 and both PERs. The data show that these two mutants are disrupted in two different actions of CRY proteins, protein-protein interaction and repression of CLOCK-BMAL1-mediated transcription, and that one residue is vital for the repressive function in both CRYs, while the other one is essential only for repression in CRY2.
FIG. 5.
CRY2G354D exhibits decreased binding to CLOCK-BMAL1, PER1, and PER2, while CRY2G354D retains strong binding. (A) Coimmunoprecipitation. HEK293 cells were transfected with Flag-Clock, Bmal1-Flag3 (3XFLAG), and Cry2-GFP constructs, as indicated. CRY2 proteins were immunoprecipitated, using anti-GFP in conjunction with a protein G IP kit. Copurified proteins were detected by Western blotting (WB) using rabbit anti-GFP (αGFP) or monoclonal mouse anti-FLAG M2 (αFLAG) antibodies. The images shown are representative of three independent experiments. Underneath, the quantitation of the eluted samples from coimmunoprecipitation is shown. The ratio of CRY2 to CLOCK or CRY2 to BMAL1 in the elution was calculated. The data were normalized such that the average amount of CLOCK and BMAL1 that wild-type CRY2 pulled down was set to 1. The error bars represent the SEM. The asterisks indicate groups that were significantly different from each other (P < 0.05). (B and C) Coimmunoprecipitation of CRY2 mutants with PER proteins. In this experiment, HEK293 cells were transfected with PER1-V5 (B) or PER2-V5 (C) and CRY2-GFP constructs, as indicated. Wild-type and mutant CRY2 proteins were pulled down, as described above. Copurified proteins were detected by Western blotting, using either rabbit anti-GFP or monoclonal mouse anti-V5 antibodies. The images shown are representative of three independent experiments.
The effect of wild-type CRY on the molecular clock.
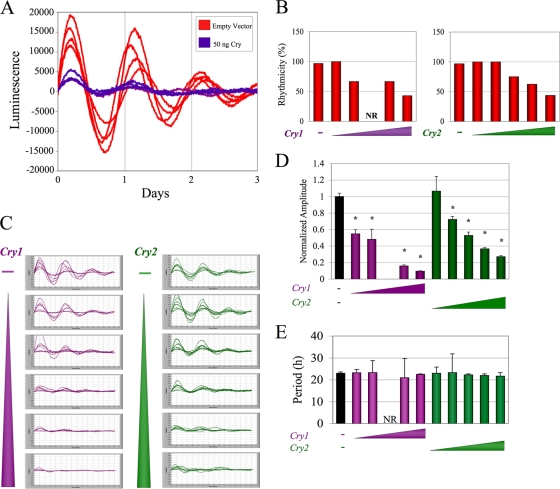
Because biochemical analysis of CRY2G351D and CRY2G354D revealed amino acids important for CRY2-specific repression and for clock protein binding, we next determined the effect of these mutants on circadian clock function. To accomplish this, NIH 3T3 cells were transfected with a Per2-Luciferase reporter, Clock, Bmal1, and various doses of wild-type or mutant Cry1 or Cry2 under a constitutive promoter. Following synchronization, oscillations in real-time bioluminescence were recorded. Before testing our Cry mutants in the system, it was first important to characterize the effect of constitutive CRY levels on rhythms. When cultures were transfected with constitutively expressed wild-type Cry, most of the cultures exhibited significant rhythmicity, with a dose-dependent effect of Cry expression on a functioning oscillator (Fig. 6A and B), consistent with a previous publication (24). At one intermediate dose, none of the cultures transfected exhibited significant rhythms. Even at the highest dose of Cry, approximately 40% of wells showed robust rhythms. In addition, the NIH 3T3 cells expressing increased CRY levels exhibited significantly decreased amplitude in a dose-dependent manner (P < 0.001) (Fig. 6C). Although the amplitude was very sensitive to the level of constitutive CRY, there was a clear persistence of low-amplitude rhythmicity (Fig. 6D). We then measured the period lengths of these cultures and found no significant change in average period at any dose (analysis of variance [ANOVA], P < 0.26) (Fig. 6E). These results indicate that although rhythmic CRY levels are not required for rhythm propagation or normal period length, they do contribute to rhythm amplitude in a dose-dependent way, as previously noted (24, 25). Thus, this overexpression system is useful to assess the effect of our CRY2 mutants on the molecular clock.
FIG. 6.
Constitutive Cry expression does not abolish rhythmicity or alter period length but decreases rhythm amplitude. (A) Bioluminescence traces of cycling cells. NIH 3T3 cells were transfected with Per2-luciferase, Clock, and Bmal1 and then either 50 ng Cry1 or an empty vector. (B) Quantitation of culture rhythmicity. Cultures were transfected with constructs as indicated for panel A and also with various doses (1 to 100 ng) of Cry1 or Cry2. After synchronization, bioluminescence was recorded. Traces were detrended and analyzed using a chi-square periodogram (P < 0.001). Cultures were considered rhythmic if the periodogram analysis yielded one significant peak in the range of 15 to 35 h. The bars represent the averages of results from 4 to 12 culture dishes. (C) Records of rhythmic cultures from the experiment whose results are shown in panel B. The triangles represent increasing doses of Cry, with the highest dose at the bottom. (D) Quantification of rhythm amplitude. The amplitudes of the rhythms of the data in panel C were determined. While the cultures transfected with 10 ng of Cry1 exhibited some fluctuations of luciferase activity, no significant period length was detected. Amplitudes of all cultures were normalized to the average amplitude of cultures without Cry within the same experiment. Cultures transfected with all doses of Cry1 and all doses of Cry2, except 1 ng, exhibited rhythms with significantly decreased amplitude (ANOVA, P = 1.34 × 10−19; *, P < 0.0001). (E) The period of each rhythmic culture shown in panel C was determined by chi-square analysis. No significant difference in period was detected (ANOVA; P < 0.26). NR, not rhythmic.
We next sought to test the effects of overexpression of mutant Cry alleles on the molecular clock. When either Cry2G351D or Cry2G354D was expressed at a low dose in synchronized NIH 3T3 cells as described above, all cultures exhibited strong rhythms, with a period and amplitude that were not statistically different from those of cells expressing a comparable dose of wild-type Cry2 (Fig. 7). In addition, expression of Cry2G354D at a high dose had the same effect on rhythmicity as overexpression of wild-type Cry2 but had an effect on amplitude less severe than the effect of the wild type, consistent with its decreased repressor capacity (Fig. 7A to C). However, when Cry2G351D was tested in the cycling cell paradigm, none of the cultures exhibited significant rhythmicity (Fig. 7).
FIG. 7.
Expression of CRY2G351D causes complete loss of rhythmicity of a functional molecular clock. NIH 3T3 cultures were transfected and synchronized as described for Fig. 6, and real-time bioluminescence was recorded. (A) Detrended traces of cultures expressing 100 ng of wild-type (WT) Cry2, Cry2G351D, or Cry2G354D. (B) The rhythmicity of each culture was determined, as described for Fig. 6 and then classified as rhythmic, not rhythmic, or irregular, meaning that it was assigned a period that did not appropriately fit the trace. (C) Amplitude analysis of rhythmic cultures expressing mutant CRY2. Cultures transfected with reporter, Clock, Bmal1, and either filler DNA (black) or a low or high dose of Cry2 (wild type or mutant) were synchronized, and their light output was measured. The traces were detrended and the amplitude was determined, as described for Fig. 6. The average amplitude of cultures expressing Cry2 with different mutations is illustrated. The error bars represent the SEM. None of the cultures expressing a high dose of Cry2G351D were rhythmic, so the amplitude could not be determined. At the high dose, cultures expressing Cry2G354D showed rhythms with significantly higher amplitude than that of wild-type Cry2 (P < 0.006). (D) Period analysis of rhythmic cultures expressing high doses of CRY2. The period of the rhythms from the culture expressing a high dose of Cry2 or Cry2G354D were determined and plotted, as described for Fig. 6. NR, not rhythmic.
DISCUSSION
CRY1 and CRY2 are integral components of the circadian clock and strong transcriptional repressors of the CLOCK-BMAL1 complex. To define the molecular mechanisms of repression by CRY1 and CRY2, we generated and screened libraries of randomly mutagenized CRY1 and CRY2 clones and identified functionally important residues within each protein crucial for their repression of CLOCK-BMAL1 that have different roles in specific actions of CRY1 and/or CRY2. Utilizing this unbiased approach to identify single point mutations that affected the functionality of both CRYs has led to the identification of novel roles for specific residues. Furthermore, random mutagenesis as opposed to deletion of large portions of the protein has allowed us to explore the structure/function of CRYs without causing gross changes in protein structure and thus affecting many aspects of CRY function.
As a result of our random mutagenesis screen, we generated many novel mutant Cry alleles. The residue changes corresponding with defects in transcriptional repression were confined to the PHR, and no changes were observed in the C-terminal tail, which has been shown to be important for nuclear localization in mammals (4, 20) and Xenopus (31). This is in line with data from Chaves et al. (4), who reported that interaction of mCRY1 with BMAL1 via the coiled-coil domain in its PHR could facilitate CRY's nuclear localization, even in the event that its NLSs are mutated. Therefore, since both mCry and Bmal1 are endogenously expressed in the cells used for the screen, it is likely that any mutants with disruptions in the NLS maintained wild-type-like repression in the screen.
When the repressive ability of CRY1 mutants were tested in a dose-response assay, we observed deficiencies in repression that fell into three general categories: no repression, weak repression, and attenuated repression that showed a significant dose response. It is interesting that some of the mutants show partial repression that is independent of dose but that others show partial repression that changes with dose. We speculate that the dose-independent mutants may function in a dominant fashion. While all of the CRY1 mutants showed impaired repression compared to that of wild-type CRY1, many showed partial loss of function. In contrast, we did not observe partial-repression phenotypes in any of our CRY2 mutants. In fact, some of them actually caused transcriptional activation over and above the activation seen in the lysates transfected with Clock, Bmal1, and reporter only. We favor the interpretation that these mutant proteins are gain-of-function mutants with transcriptional-activation activity. The fact that the mutants from the CRY1 and CRY2 screens demonstrated repression profiles distinct from each other was unexpected, considering the high sequence similarly between the two proteins, even in the context of a nonsaturating screen.
In silico modeling of the 3D location of the mutants on the predicted tertiary structure of the CRY2 protein drew our attention to two residues (G351 and G354) that were hits only in the CRY2 screen, shared the same repression profile, and were predicted to be in close proximity on the protein's predicted tertiary structure. Despite the nonsaturating nature of the screen, the fact that these residues were conserved between CRY1 and CRY2 but were identified only in the CRY2 screen suggested to us that perhaps these residues were differentially utilized in the mechanism of repression by CRY2 but not CRY1. In fact, when the analogous mutations were made in the CRY1 protein, one of them (CRY1G336D/CRY2G354D) resulted in complete loss of repression in both CRY1 and CRY2 and is therefore not specific to CRY2 repression, while the other (CRY1G333D/CRY2G351D) rendered CRY2, but not CRY1, unable to repress. This differential effect may represent a biochemical mechanism by which CRY2, but not CRY1, represses the CLOCK-BMAL1 complex. Until now, the only biochemical differences seen between CRY1 and CRY2 were subtle differences in the levels of potency of their repression and binding to some core clock components (9, 22) and differential nuclear localization mechanisms in Xenopus and mammals (4, 20, 31). Our data support the notion that the opposing period length phenotypes in Cry1−/− and Cry2−/− mice may be due to innate biochemical differences between the two proteins, as opposed to the idea that CRY proteins are biochemically redundant and the differential behavioral phenotypes can be attributed to differences in timing of their expression.
Between CRY1 and CRY2, the N terminus is highly conserved and only the C-terminal tails differ in size and amino acid composition. Since the C-terminal tails of CRY1 and CRY2 are the most obvious structural difference between the two proteins, we hypothesized the differential effects of the same mutation in each protein might be due to functional interplay with the C-terminal tail. Interestingly, when mutation G333D/G351D was introduced into Cry1 constructs consisting of the Cry1 PHR only (without a C-terminal tail) or the Cry1 PHR fused to the Cry2 C-terminal tail, their ability to repress was significantly decreased. The effect was most severe when there was no C-terminal tail. These data strongly suggest that the ability of CRY1 to retain its repressive ability when mutated is dependent on the presence of a C-terminal tail. The ability of the C-terminal tail to affect CRY function has been previously reported for Drosophila melanogaster (3, 6), where the C-terminal tail of the CRY protein functionally interacts with the N-terminal portion to regulate its activity (6). When Drosophila CRY lacks its C-terminal tail, it is constitutively active (6). Perhaps CRY1G333D loses its ability to repress normally when the C-terminal tail is altered or removed because the tail interacts with the PHR to facilitate its mechanism of repression. While our studies clearly demonstrate a functional interaction between the N- and C-terminal portions of the mammalian CRY proteins, the presence of a physical interaction has yet to be shown.
In order to explain the altered repression seen when G351D or G354D were introduced into CRY2, both mutants were tested for their ability to bind CLOCK-BMAL1 and both PER proteins. Previous studies have shown that the ability of CRY to bind CLOCK-BMAL1 is required for its repression of CLOCK-BMAL1-mediated transcription (4, 21). CRY2G354D showed decreased binding to CLOCK-BMAL1, which may partially account for its decreased repression. In addition, this mutant was also highly deficient in binding of PER1 and PER2. Although the CRY-binding domain of PER localizes to its C terminus (16), the PER-binding domain on CRY has not been well defined. These data identify a new residue in CRY2 important for protein-protein interaction. Whether this residue influences the function of the coiled-coil binding domain in a manner analogous to the one identified in CRY1 (4) or whether it is an essential residue in a novel binding domain remains to be determined. In addition, it is not clear from our data whether the mutants' decreased PER binding causes their defective repression. Data showing that CRY can repress CLOCK-BMAL1 in transient-transfection luciferase assays without coexpression of PER suggest that PER may not be required for CRY's repression (9, 12). In contrast to CRY2G354D, CRY2G351D retained its ability to bind CLOCK-BMAL1 and both PERs, which is surprising in light of its complete inability to repress. Collectively, these data show that PER binding by CRY proteins is not necessarily predictive of repression. The fact that CRY2G351D can bind the major clock components but still cannot repress CLOCK-BMAL1 suggests a repression-specific role for this residue in regulating the CLOCK/BMAL1 heterodimer. Therefore, we speculate that this CRY2 loss-of-function mutant functions downstream of its interaction with CLOCK, BMAL1, or the PERs.
We also examined the effect of overexpression of these mutants on circadian clock function in an autonomous cellular model, NIH 3T3 cells. While expression of CRY2 protein (wild type or mutants) did not cause any detectable change in period, the amplitude of the rhythms was sensitive to overexpression of all Cry2 constructs tested, but to various degrees. Expression of CRY2G354D, deficient in its ability to bind CLOCK, BMAL1, and PER, caused a significant increase in rhythm amplitude over that of wild-type CRY2, consistent with its partial loss-of-function phenotype. Interestingly, expression of CRY2G351D, which demonstrates intact interaction with CLOCK, BMAL1, and PERs, had the most severe effect on rhythms, causing complete loss of circadian rhythmicity in kinetic imaging assays. CRY2G351D, which cannot repress but can still bind PER, has a more severe effect on amplitude than wild-type CRY2. In total, our data suggest that CRY2G351D acts as a strong dominant negative in the context of a functional circadian clock. The fact that overexpression of constitutive CRY2G351D leads to the most severe effect on molecular rhythms may be due in part to its ability to bind CLOCK-BMAL1 and the PER proteins and, perhaps in doing so, alter the robustness of the molecular clock in a dominant fashion. Therefore, future work comparing the circadian behavior of a CRY2G351D knock-in mouse to that of the Cry2 null mouse will likely lend valuable insight into the role of CRY2, as opposed to CRY1, in circadian rhythms.
The question remains of how identical amino acid substitutions in two clustered residues in CRY2 can lead to such distinct effects on CRY function. Both residues in CRY2 are mutated from glycine, an amino acid with a noncharged side group, to aspartic acid, which has an acidic side chain. Our in silico modeling predicts that this difference may be due to the locations of these residues. G351 is closer to the surface of the protein, while G354 is less superficial and localized closer to the coiled-coil domain, which has previously been shown to be involved in binding to CLOCK-BMAL1 and the PER proteins (4). Our immunoprecipitation data demonstrated that the G354D mutation disrupts CRY2's binding to CLOCK-BMAL1, PER1, and PER2 (Fig. 5), which is similar in phenotype to when the coiled-coil domain is deleted from CRY1 (4). On the other hand, G351D, which is located on a face of the protein different from this previously described interaction domain, does not show disrupted binding to these core clock components.
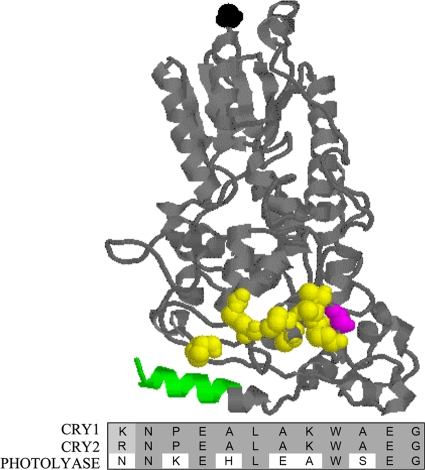
Another interesting issue that our data raise is how mutation of this conserved residue, G333/G351 in the CRY proteins, results in much stronger loss of repression in CRY2 than in CRY1. G333/G351 is immediately upstream of a small patch of 13 residues that are highly conserved between repressive CRYs, but not in Photolyase, a close family member that cannot repress CLOCK-BMAL1 (Fig. 8). Many of the residues that are neutral or basic in repressive CRYs are acidic or neutral in Photolyase. G351D introduces an acidic residue that confers a negative charge and may change CRY1 and CRY2 to be more like Photolyase. These data suggest that this small region may be vital for the ability of CRYs to repress through interactions of polar amino acids.
FIG. 8.
CRY2G354 is immediately upstream of a region of amino acids conserved in repressive CRYs but not Photolyase. CRY2 3D homology model. This model was generated as described for Fig. 2. The N-terminal residue is shown in black, while the coiled-coil domain is shown in green. CRY2G351 is drawn in magenta, while the residues in the region adjacent to G351 that are conserved in all repressive CRYs, but not Photolyase, are shown in yellow. Under the model is a protein alignment of these residues in CRY1, CRY2, and Photolyase. Conserved residues are shaded in gray.
One hypothesis to explain the differential impact of this region on CRY2, but not CRY1, function is that this residue is involved in recruitment of a phosphatase to the promoter by CRY2. Dardente et al. (5) reported that only CRY2 causes stabilization of the unphosphorylated, transcriptionally inactive form of BMAL1. One possibility is that CRY2 may recruit to the promoter a phosphatase that is able to dephosphorylate BMAL1, leading to its repression. The mutation G351D may change CRY2's ability to bind the phosphatase. Another possible explanation is that this amino acid impacts chromatin remodeling. Several groups have shown that chromatin remodeling is integral to molecular clock function. Naruse et al. (17) showed that CRY1 binds to mSin3B, a histone deacetylase, and that this leads to downregulation of the Per1 gene. Perhaps the residue we have isolated is involved in recruitment of histone deacetylases to the promoter. It is possible that CRY2, compared to CRY1, may bind different corepressors via different domains, explaining why mutation G351 has different effects on CRY1 and CRY2.
In conclusion, our unbiased mutagenesis screen of the CRY proteins has generated an invaluable allelic series of mutant Crys that can be used to elucidate the multifaceted roles of CRY1 and CRY2 in the molecular clock. In addition, through careful analysis of two particular mutants, we identified a novel residue that is important for CRY2's protein-protein interaction with other clock proteins and another residue that has functionally distinct roles in CRY1 and CRY2. This work has made progress in elucidating the molecular mechanisms underlying the differential roles of the CRY proteins in the molecular circadian clock and will form the basis for future studies that will integrate our new biochemical knowledge about the role of the CRY proteins with other clock components to allow a clearer picture of the molecular timekeeping mechanism to emerge.
Acknowledgments
This work was supported by grants from the National Institute of Mental Health (R01 MH-074924), the Silvio O. Conte Center for Neuroscience (P50 MH074924), and the Jack Kent Cooke Foundation.
We also thank Karla Marz for assistance in generating and visualizing the CRY1 and CRY2 3D homology models.
Footnotes
Published ahead of print on 17 August 2009.
REFERENCES
- 1.Bell-Pedersen, D., V. M. Cassone, D. J. Earnest, S. S. Golden, P. E. Hardin, T. L. Thomas, and M. J. Zoran. 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6:544-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busino, L., F. Bassermann, A. Maiolica, C. Lee, P. M. Nolan, S. I. Godinho, G. F. Draetta, and M. Pagano. 2007. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316:900-904. [DOI] [PubMed] [Google Scholar]
- 3.Busza, A., M. Emery-Le, M. Rosbash, and P. Emery. 2004. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science 304:1503-1506. [DOI] [PubMed] [Google Scholar]
- 4.Chaves, I., K. Yagita, S. Barnhoorn, H. Okamura, G. T. van der Horst, and F. Tamanini. 2006. Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol. Cell. Biol. 26:1743-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dardente, H., E. E. Fortier, V. Martineau, and N. Cermakian. 2007. Cryptochromes impair phosphorylation of transcriptional activators in the clock: a general mechanism for circadian repression. Biochem. J. 402:525-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dissel, S., V. Codd, R. Fedic, K. J. Garner, R. Costa, C. P. Kyriacou, and E. Rosato. 2004. A constitutively active cryptochrome in Drosophila melanogaster. Nat. Neurosci. 7:834-840. [DOI] [PubMed] [Google Scholar]
- 7.Etchegaray, J. P., C. Lee, P. A. Wade, and S. M. Reppert. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421:177-182. [DOI] [PubMed] [Google Scholar]
- 8.Godinho, S. I., E. S. Maywood, L. Shaw, V. Tucci, A. R. Barnard, L. Busino, M. Pagano, R. Kendall, M. M. Quwailid, M. R. Romero, J. O'Neill, J. E. Chesham, D. Brooker, Z. Lalanne, M. H. Hastings, and P. M. Nolan. 2007. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316:897-900. [DOI] [PubMed] [Google Scholar]
- 9.Griffin, E. A., Jr., D. Staknis, and C. J. Weitz. 1999. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286:768-771. [DOI] [PubMed] [Google Scholar]
- 10.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss- PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 11.Kort, R., H. Komori, S. Adachi, K. Miki, and A. Eker. 2004. DNA apophotolyase from Anacystis nidulans: 1.8 Å structure, 8-HDF reconstitution and X-ray-induced FAD reduction. Acta Crystallogr. D 60:1205-1213. [DOI] [PubMed] [Google Scholar]
- 12.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193-205. [DOI] [PubMed] [Google Scholar]
- 13.Liu, A. C., D. K. Welsh, C. H. Ko, H. G. Tran, E. E. Zhang, A. A. Priest, E. D. Buhr, O. Singer, K. Meeker, I. M. Verma, F. J. Doyle III, J. S. Takahashi, and S. A. Kay. 2007. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129:605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martz, E. 2002. Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem. Sci. 27:107-109. [DOI] [PubMed] [Google Scholar]
- 15.Miyamoto, Y., and A. Sancar. 1999. Circadian regulation of cryptochrome genes in the mouse. Brain Res. Mol. Brain Res. 71:238-243. [DOI] [PubMed] [Google Scholar]
- 16.Miyazaki, K., M. Mesaki, and N. Ishida. 2001. Nuclear entry mechanism of rat PER2 (rPER2): role of rPER2 in nuclear localization of CRY protein. Mol. Cell. Biol. 21:6651-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naruse, Y., K. Oh-hashi, N. Iijima, M. Naruse, H. Yoshioka, and M. Tanaka. 2004. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 24:6278-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oster, H., A. Yasui, G. T. J. van der Horst, and U. Albrecht. 2002. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 16:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ripperger, J. A., and U. Schibler. 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38:369-374. [DOI] [PubMed] [Google Scholar]
- 20.Sakakida, Y., Y. Miyamoto, E. Nagoshi, M. Akashi, T. J. Nakamura, T. Mamine, M. Kasahara, Y. Minami, Y. Yoneda, and T. Takumi. 2005. Importin alpha/beta mediates nuclear transport of a mammalian circadian clock component, mCRY2, together with mPER2, through a bipartite nuclear localization signal. J. Biol. Chem. 280:13272-13278. [DOI] [PubMed] [Google Scholar]
- 21.Sato, T. K., R. G. Yamada, H. Ukai, J. E. Baggs, L. J. Miraglia, T. J. Kobayashi, D. K. Welsh, S. A. Kay, H. R. Ueda, and J. B. Hogenesch. 2006. Feedback repression is required for mammalian circadian clock function. Nat. Genet. 38:312-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shearman, L. P., S. Sriram, D. R. Weaver, E. S. Maywood, I. Chaves, B. Zheng, K. Kume, C. C. Lee, G. T. van der Horst, M. H. Hastings, and S. M. Reppert. 2000. Interacting molecular loops in the mammalian circadian clock. Science 288:1013-1019. [DOI] [PubMed] [Google Scholar]
- 23.Siepka, S. M., S. H. Yoo, J. Park, W. Song, V. Kumar, Y. Hu, C. Lee, and J. S. Takahashi. 2007. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129:1011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda, H. R., W. Chen, A. Adachi, H. Wakamatsu, S. Hayashi, T. Takasugi, M. Nagano, K. Nakahama, Y. Suzuki, S. Sugano, M. Iino, Y. Shigeyoshi, and S. Hashimoto. 2002. A transcription factor response element for gene expression during circadian night. Nature 418:534-539. [DOI] [PubMed] [Google Scholar]
- 25.Ueda, H. R., S. Hayashi, W. Chen, M. Sano, M. Machida, Y. Shigeyoshi, M. Iino, and S. Hashimoto. 2005. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37:187-192. [DOI] [PubMed] [Google Scholar]
- 26.van der Horst, G. T. J., M. Muijtijens, K. Kobayashi, R. Takano, S.-I. Kanno, M. Takao, J. de Wit, A. Verkerk, A. P. M. Eker, D. van Leenen, R. Buijs, D. Bootsma, J. H. J. Hoeijmakers, and A. Yasui. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627-630. [DOI] [PubMed] [Google Scholar]
- 27.van der Schalie, E. A., F. E. Conte, K. E. Marz, and C. B. Green. 2007. Structure/function analysis of Xenopus cryptochromes 1 and 2 reveals differential nuclear localization mechanisms and functional domains important for interaction with and repression of CLOCK-BMAL1. Mol. Cell. Biol. 27:2120-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vitaterna, M. H., C. P. Selby, T. Todo, H. Niwa, C. Thompson, E. M. Fruechte, K. Hitomi, R. J. Thresher, T. Ishikawa, J. Miyazaki, J. S. Takahashi, and A. Sancar. 1999. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. USA 96:12114-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitmore, D., and P. Sassone-Corsi. 1999. Cryptic clues to clock function. Nature 398:557-558. [DOI] [PubMed] [Google Scholar]
- 30.Yagita, K., F. Tamanini, M. Yasuda, J. H. Hoeijmakers, G. T. van der Horst, and H. Okamura. 2002. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 21:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, H., F. Conte, and C. B. Green. 2003. Nuclear localization and transcriptional repression are confined to separable domains in the circadian protein CRYPTOCHROME. Curr. Biol. 13:1653-1658. [DOI] [PubMed] [Google Scholar]