Abstract
hnRNP A1 binds to RNA in a cooperative manner. Initial hnRNP A1 binding to an exonic splicing silencer at the 3′ end of human immunodeficiency virus type 1 (HIV-1) tat exon 3, which is a high-affinity site, is followed by cooperative spreading in a 3′-to-5′ direction. As hnRNP A1 propagates toward the 5′ end of the exon, it antagonizes binding of a serine/arginine-rich (SR) protein to an exonic splicing enhancer, thereby inhibiting splicing at that exon's alternative 3′ splice site. tat exon 3 and the preceding intron of HIV-1 pre-mRNA can fold into an elaborate RNA secondary structure in solution, which could potentially influence hnRNP A1 binding. We report here that hnRNP A1 binding and splicing repression can occur on an unstructured RNA. Moreover, hnRNP A1 can effectively unwind an RNA hairpin upon binding, displacing a bound protein. We further show that hnRNP A1 can also spread in a 5′-to-3′ direction, although when initial binding takes place in the middle of an RNA, spreading preferentially proceeds in a 3′-to-5′ direction. Finally, when two distant high-affinity sites are present on the same RNA, they facilitate cooperative spreading of hnRNP A1 between the two sites.
The coding sequences of many eukaryotic genes are interrupted by noncoding introns, which are also present in the primary transcripts, or pre-mRNAs. The introns must be precisely removed, and the coding exons joined, to allow translation of functional proteins. Pre-mRNA splicing, a nuclear process, can be constitutive or alternative. Constitutive splicing is the removal of introns by joining together all the adjacent exons in the order of their arrangement. In constitutive splicing, a single protein is produced from a single pre-mRNA, regardless of where and when the gene is expressed. In alternative splicing, variable use of splice sites allows two or more mature mRNAs to be generated from the same pre-mRNA. For example, an entire exon or part of an exon can be included or skipped in different spliced mRNAs. Alternative splicing is a prevalent way by which many eukaryotes diversify the number of proteins produced from a single pre-mRNA transcript (57, 62).
Analysis of the human genome indicated that more than 74% of human genes encode at least two isoforms by alternative splicing (27, 28, 35, 48). An extreme example of alternative splicing is the Drosophila Dscam gene, in which a single pre-mRNA transcript apparently encodes 38,016 protein isoforms through combinatorial alternative splicing events (21, 54). Alternative splicing can in many cases be subject to regulation, for example, in a cell-type-specific manner, during embryonic development, or in response to signaling pathways.
Retroviruses such as human immunodeficiency virus type 1 (HIV-1) also depend on alternative splicing to produce all of the viral proteins from a single primary transcript (59). The unspliced transcript is necessary for viral replication, packaging into virions, and translation of several proteins, whereas other viral proteins are generated from partially spliced or fully spliced transcripts. Special mechanisms allow these incompletely spliced transcripts to be exported to the cytoplasm for translation (13).
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are trans-acting factors that bind to exonic splicing silencers (ESSs) or intronic splicing silencers, usually to inhibit the use of particular splice sites during regulated splicing events. There are some instances in which hnRNPs promote splicing instead of inhibiting it (8, 40, 49). The most common feature of hnRNPs is the presence of two or more RNA binding domains and an auxiliary domain believed to be responsible for protein-protein, RNA-protein, and single-stranded DNA-protein interactions. Most of these hnRNPs can also form homophilic interactions and heterophilic interactions with other hnRNPs (9, 12, 46). One of the most abundant hnRNPs is hnRNP A1 (17, 23). hnRNP A1 has been implicated in many alternative splicing events in human and several other eukaryotes (1, 2, 4, 5, 7, 25, 30, 43, 66). Human hnRNP A1 is a 320-amino-acid protein, of which the 196-amino-acid N-terminal domain comprises two RNA recognition motifs (RRMs) (39). The 124-amino-acid C-terminal domain is glycine rich and is believed to be responsible for cooperative binding, leading to repression of splicing (16, 55). At present, there are no available structures of intact hnRNP A1, but there are high-resolution crystal structures of its N-terminal domain spanning RRM1 and RRM2, which is known as unwinding protein 1 (UP1) (16, 55, 61, 63).
The manner in which hnRNP A1 controls alternative splicing is still not fully understood. A study from our lab focusing on splicing of exon 3 of the HIV-1 tat pre-mRNA showed an antagonistic effect of an ESS element, ESS3, mediated by hnRNP A1, vis-à-vis another cis-acting splicing regulatory element, known as an exonic splicing enhancer (ESE) (67). ESEs enhance splicing or promote inclusion of a particular exon through the binding of one or more activator proteins, such as members of the serine/arginine-rich (SR) family, which in turn recruit other components of the splicing machinery to the 5′ and 3′ splice sites (26, 37). SR proteins have one or two RRMs at their N terminus, which interact with the RNA (3, 6, 19, 33, 37, 68). The C-terminal domain of each SR protein comprises a highly conserved arginine/serine-rich (RS) domain; however, this domain is not always necessary for splicing (56, 66). SR proteins are important for the recognition of splice sites and act at the earliest stages of spliceosome assembly, as well as at later stages of splicing (10, 31, 58, 60). SR proteins have other functions in splicing and gene expression besides binding to ESEs, and they are essential for constitutive splicing (26). Even in the case of introns with strong splice sites, in which an ESE might not be required, SR proteins are essential for recognition of the splice sites and recruitment of the splicing machinery (18, 22, 26, 37, 43, 60, 66).
Initial high-affinity binding of hnRNP A1 to ESS3 is followed by its cooperative spreading along tat exon 3, which allows hnRNP A1 to displace the SR protein SC35 from its cognate ESE, thereby preventing splicing of tat exon 3 (67). That same study also showed that when another SR protein, SF2/ASF, binds to its cognate ESE, hnRNP A1 cannot effectively displace it, and therefore, there is inclusion of tat exon 3 (67). The net effect depends in part on the strength of the SR protein interaction with its cognate ESE and presumably on the nuclear abundance of particular SR proteins and hnRNP A1 in a given cell type.
There is increased expression of hnRNP A1 or SR proteins in some tumors and tumor cell lines compared to in normal cells and tissues (20, 29, 50). Putting all this information together presents a strong case for studying how cooperative binding of hnRNP A1 leads to alternative splicing of a specific exon. Understanding cooperative binding of hnRNP A1 in the context of HIV tat and other model substrates is expected to shed light on the mechanisms of alternative splicing in general.
The present study addresses the mechanism of hnRNP A1 cooperative binding to RNA. We show that hnRNP A1 cooperative binding results in unwinding of RNA secondary structure. After binding to a high-affinity site, hnRNP A1 spreads preferentially, though not exclusively, in a 3′-to-5′ direction and can displace other bound proteins from the RNA to repress splicing.
MATERIALS AND METHODS
Transcripts.
All pre-mRNA transcripts for in vitro splicing were 5′ capped and labeled by in vitro transcription in the presence of [α-32P]UTP from PCR templates with a T7 phage promoter (41). The PCR primers used to generate transcription templates for all constructs used for in vitro splicing are listed in Table S1 in the supplemental material. The template for PCR was the linearized plasmid pSP64-HβΔ6 (32) except for constructs NβMS2ESS, NβMS2ESSm, NβMS2*ESS, and NβMS2*ESSm, whose PCR templates were NβMS2ΔESS and NβMS2*ΔESS. All the model RNA transcripts were transcribed in the presence of [α-32P]CTP from the corresponding antisense oligonucleotides with a T7 phage promoter annealed to a T7 sense oligonucleotide as described previously (45). All unlabeled model RNA transcripts were similarly transcribed from synthetic oligonucleotide templates, using a T7-MEGAshortscript kit (Ambion catalog no. 1354), followed by 5′ end labeling with [γ-32P]ATP (53). For antisense oligonucleotides corresponding to Fig. 3C, see Table S2 in the supplemental material.
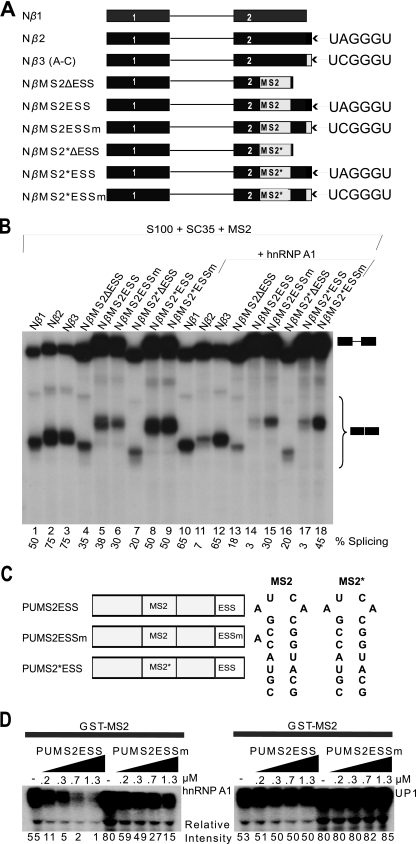
FIG. 3.
Unwinding of RNA secondary structure and splicing inhibition by hnRNP A1. (A) β-Globin minigenes with or without MS2 or MS2 mutant (MS2*) hairpins. (B) In vitro splicing of capped Nβ-globin minigene transcripts (1.15 nM) in S100 extract complemented with 0.4 μM SC35, in the presence of 1.73 μM GST-MS2 coat protein, and with or without 0.6 μM hnRNP A1. (C) Poly(U) MS2 constructs to test the unwinding activity of hnRNP A1 as it binds cooperatively. The sequences and secondary structures of MS2 and MS2* are shown next to the construct diagrams. (D) GST-MS2 pulldowns. Labeled RNA (15 nM) was first incubated with GST-MS2 protein (1.5 μM), followed by incubation with increasing concentrations of recombinant hnRNP A1 (left panel; 0 to 1.3 μM) or UP1 (right panel; 0 to 1.3 μM) and then incubation with GST-agarose beads. After washing, bound RNA was eluted, separated by denaturing PAGE, and detected by autoradiography. Band intensities were measured on a phosphorimager, and normalized values relative to the lowest band intensity are shown below the gels.
Recombinant proteins.
Untagged human hnRNP A1 was expressed in Escherichia coli and purified as described previously (44). Purified glutathione S-transferase (GST)-MS2 protein expressed in E. coli was a gift from Zuo Zhang. Purified human SC35 expressed in baculovirus was a gift from Michelle Hastings. Purified UP1 expressed in E. coli was a gift from Qingshuo Zhang.
In vitro splicing assays.
S100 extract from HeLa cells was prepared as described previously (42). In vitro splicing reactions were carried out in a final volume of 12.5 μl with 15 fmol (1.15 nM) of 32P-labeled, 7CH3-GpppG-capped T7 RNA transcripts and 35% (vol/vol) S100 extract, with a final concentration of 0.4 μM SC35, in the presence or absence of hnRNP A1 at a final concentration of 0.6 μM and in the presence or absence of GST-MS2. All the 32P-labeled RNAs in Fig. 2A were first incubated with GST-MS2 at a final concentration of 1.73 μM in standard splicing buffer (41) for 15 min at room temperature or at 30°C before the addition of extract mix with or without hnRNP A1 and further incubation at 30°C for 2 h as described previously (41).
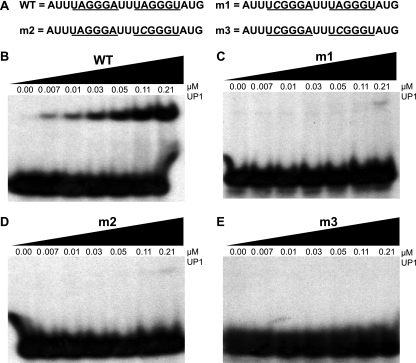
FIG. 2.
EMSA with UP1 and short RNAs with high-affinity sites. (A) Sequences of the 20-mer RNAs used for the binding assays. The synthetic RNAs were 5′ end labeled with 32P. The underlined sequences show the high-affinity hnRNP A1 hexamer motifs. The mutated nucleotide is shown in italic. The WT RNA has both motifs intact, and the m1, m2, and m3 mutants have a point mutation in the 5′ motif, the 3′ motif, or both motifs, respectively. (B) EMSA of WT RNA with the indicated final concentrations of purified recombinant UP1 (0 to 0.21 μM). (C) EMSA with m1 RNA. (D) EMSA with m2 RNA. (E) EMSA with m3 RNA. The final concentration of 32P-labeled RNA in these experiments was 16 nM.
UV cross-linking.
All UV cross-linking assays were performed in a Spectronics XL1000 instrument at 0.48 J/cm2 under splicing-reaction conditions as described previously (66, 67) except that the buffer was slightly modified by addition of final concentrations of 0.7 mg/ml heparin, 0.1 mg/ml yeast tRNA, and 0.07 mg/ml bovine serum albumin with 8 to 32 nM 32P-labeled RNA.
GST-MS2 pulldowns.
32P-labeled RNAs used in the GST-MS2 pulldowns were first incubated at 95°C for 3 to 5 min and then allowed to refold at room temperature for 1 to 2 min in cross-linking buffer as described above. Renatured RNAs were then allowed to form complexes with GST-MS2 at a 1.5 μM final concentration at 30°C for 20 min, after which increasing amounts (0.2, 0.3, 0.7, and 1.3 μM) of hnRNP A1 or UP1 were added, with further incubation for 20 min at 30°C. Glutathione-agarose beads were added and incubated at 4°C for 1 h, followed by washing the beads and elution and extraction of the RNA as described previously (65).
RNA footprinting.
Hydroxyl radical and RNase A footprinting experiments were done as described previously (11) with a 16 nM final concentration of 5′ 32P-labeled RNA and 1, 2, 4, and 8 pmol (0.05, 0.1, 0.2, and 0.4 μM) of recombinant hnRNP A1.
Gel shift assay.
The gel shift assay was performed as described previously (64), using 32P-5′-end-labeled 20-nucleotide (nt) RNAs. The labeled RNAs were incubated with increasing amounts of purified recombinant UP1 (64) on ice for 20 min in 18.8-μl reaction mixtures consisting of 16.2 mM MgCl2, 76.6 mM KCl, 1.1 mg/ml tRNA, and 0.1 mg/ml bovine serum albumin. The samples were loaded on a prechilled 5% (wt/vol) native polyacrylamide Tris-glycine gel (acrylamide/bisacrylamide ratio, 62:1) and run at 170 V for 45 min at room temperature. The fraction of bound RNA was calculated to plot binding curves using Prism software v.5 (GraphPad), and apparent equilibrium binding constant (Kd) values were calculated from the following equation: Y = Bmax × X/(Kd + X) + NS × X + background. Bmax is the maximum specific binding in the same units as Y (percent bound RNA). Kd is in the same units as X (μM). NS is the slope of nonspecific binding in Y units divided by X units. The background is the amount of nonspecific binding with no added radiolabeled RNA. This analysis assumes that only a small fraction of radiolabeled RNA binds, which means that the concentration that we added is virtually identical to the free concentration.
Phosphorimager analysis.
After electrophoretic separation, radioactive bands were detected and quantitated using a FLA5100 phosphorimager and Multigauge software v.2.3 (Fujifilm).
RESULTS
Cooperative binding of hnRNP A1 does not require RNA secondary structure.
Inhibition of splicing of exon 3 of an HIV-1 tat23 minigene occurs through cooperative binding of hnRNP A1, such that multiple molecules bind by spreading from a high-affinity binding site (ESS3) at the 3′ end of the RNA toward the 5′ end (67). The tat pre-mRNA can adopt an intricate secondary structure in solution, and it has been proposed that hnRNP A1 binding and silencing involve cooperative binding to these structured regions, rather than spreading along single-stranded RNA (14, 38). However, UP1, as its name indicates, can unwind RNA or DNA secondary and higher-order structures (24, 64). hnRNP A1 facilitates annealing of complementary nucleic acid strands below their melting temperature (Tm); on the other hand, when hnRNP A1 binds to duplex DNA, it lowers the Tm, thereby facilitating duplex unwinding; and at a temperature above the new Tm, hnRNP A1 can also maintain an equilibrium between single- and double-stranded DNA (51, 52). However, hnRNP A1 had not been shown to be capable of unwinding RNA secondary structure.
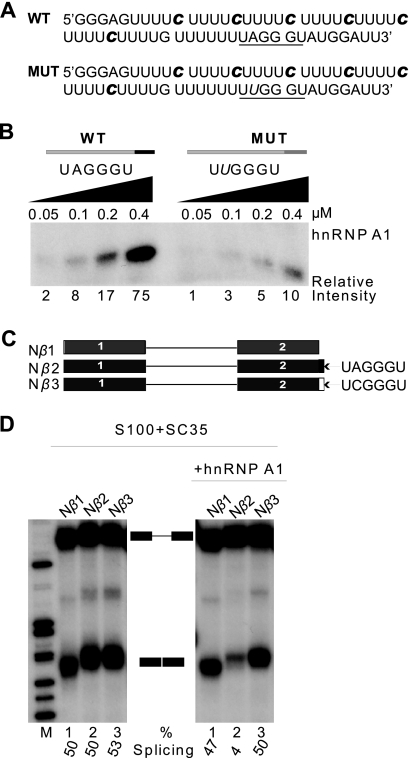
To address the potential involvement of RNA secondary structure in hnRNP A1 cooperative binding, we generated by in vitro transcription RNA comprised mainly of oligo(U) tracts, with 32P-labeled C at every fifth nucleotide position. We chose this nucleotide composition because hnRNP A1 has low affinity for poly(U) and poly(C) (1). Near the 3′ end of the RNA, we placed a high-affinity hnRNP A1 binding site, UAGGGU, as determined by systematic evolution of ligands by exponential enrichment (SELEX) (5) (Fig. 1A). Based on its composition and sequence, this RNA cannot form secondary structures, at least by conventional base pairing. To reduce other potential higher-order structures, the RNA was denatured at 95°C and rapidly cooled before incubation with recombinant hnRNP A1 at different concentrations. The complex formed between this RNA and hnRNP A1 was subjected to UV cross-linking, followed by digestion with RNases A and T1, separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and detection by autoradiography (Fig. 1B; see Fig. S1B in the supplemental material). The transfer of label to hnRNP A1 can be detected after nuclease digestion, because the protein spreads along the RNA from the high-affinity site (67). This was confirmed by the reduction in labeled hnRNP A1 when the high-affinity site was mutated at a single nucleotide, from UAGGGU (wild type [WT]) to UUGGGU (MUT). We conclude that unstructured RNA is compatible with cooperative binding of hnRNP A1.
FIG. 1.
hnRNP A1 cooperative binding does not require RNA secondary structure. (A) Sequences of WT and MUT RNAs for UV cross-linking experiments. The underlined hexanucleotide is a high-affinity hnRNP A1 binding site: UAGGGU is the WT version, and UUGGGU is the inactive, mutant version, with the mutated nucleotide shown in italic. The radiolabeled cytidines incorporated by in vitro transcription are indicated in bold italic. (B) UV cross-linking with 16 nM WT and MUT RNAs from panel A in the presence of increasing concentrations of recombinant hnRNP A1 (0.5 to 4 μM). The cross-linked products were digested with RNases A and T1, separated by SDS-PAGE, and detected by autoradiography. Band intensities were measured on a phosphorimager, and normalized values relative to the lowest band intensity are shown below the gel. (C) β-Globin minigene transcripts for in vitro splicing assays. The pre-mRNAs comprise 108 nt of exon 1, the 130-nt first intron, and 108 nt of exon 2. Nβ2 has an additional 6-nt ESS at the 3′ end; Nβ3 has a mutant version of the ESS (ESSm). (D) Splicing of 1.15 nM 7CH3-GpppG-capped pre-mRNAs from panel C in HeLa S100 extract complemented with 0.4 μM SC35, in the presence or absence of 0.6 μM hnRNP A1. The splicing efficiency [mRNA/(pre-mRNA + mRNA) × 100%] is shown below the autoradiogram.
To verify that the SELEX winner UAGGGU can act as an ESS, we constructed a β-globin minigene, Nβ2, comprising the last 101 nt of exon 1 and the first 101 nt of exon 2, and inserted UAGGGU at the 3′ end of exon 2, followed by a BamHI site (Fig. 1C). A similar control minigene, Nβ3, has a single point mutation changing UAGGGU to UCGGGU, which abrogates hnRNP A1 binding (as does UUGGGU; see below and Fig. 1B). Finally, Nβ1 is the parental minigene without an inserted hexamer. Labeled pre-mRNAs transcribed from these minigenes were spliced in HeLa cell cytosolic extract (S100) complemented with recombinant SC35, in the presence or absence of recombinant hnRNP A1 (which is limiting in S100 extract [42]) (Fig. 1D). The results show that splicing repression requires an intact ESS (cf. lanes 1 to 3 in right panel) and a sufficient amount of hnRNP A1 (cf. lanes 2 in right and left panels). Moreover, the A-to-C point mutation at position 2 of the ESS is sufficient to abolish splicing silencing (cf. lanes 2 and 3 in right panel), and likewise, another mutation in the ESS, A to U at position 2, also abolishes hnRNP A1 cooperative binding (Fig. 1B; see Fig. S1B in the supplemental material).
Note that we used two different hnRNP A1 binding site mutants, UUGGGU and UCGGGU; the latter disrupted hnRNP A1 binding to a greater extent than the former (data not shown). However, we used UUGGGU for cross-linking experiments to avoid introducing a labeled C nucleotide into the hnRNP A1 binding site. On the other hand, because this was not a consideration for the splicing experiments, we used the more disruptive UCGGGU mutation for the splicing assays. Additional mutations we tested that also effectively disrupted the hnRNP A1 binding site were UACGGU and UAUGGU (data not shown).
The effect of the A-to-C point mutation on binding is also shown in a gel shift assay (Fig. 2; see Fig. S2 in the supplemental material). In this case we used short RNAs and UP1, rather than hnRNP A1, so as to measure initial binding without further cooperative spreading. However, we found that in this type of experiment with short oligonucleotides, two high-affinity motifs are required for tight binding, which is consistent with previous hnRNP A1 SELEX data (5) and with structural and binding data for UP1 interaction with single-stranded telomeric DNA repeats (64). Our electrophoretic mobility shift assay (EMSA) data show that UP1 binds to 20-mer RNA containing UAGGGA and UAGGGU motifs separated by 2 nt, with an apparent Kd of ∼0.1 μM. Mutating the first motif to UCGGGA and/or the second motif to UCGGGU made UP1 binding essentially undetectable (Fig. 2; see Fig. S2 in the supplemental material).
hnRNP A1 binding results in unwinding of RNA secondary structure.
To further test whether or not cooperative binding of hnRNP A1 involves RNA secondary structure, we took advantage of an RNA with known secondary structure, namely, a natural hairpin that binds bacteriophage MS2 coat protein (22, 36). We inserted the MS2 hairpin in the middle of exon 2 of the β-globin minigene and included the ESS at the 3′ end. As a control, we inserted a hairpin with deletion of a single bulged nucleotide to abolish MS2 protein binding (22) (constructs MS2 and MS2* in Fig. 3A). We expected that tight binding by MS2 to the WT construct, but not to the mutant construct, would block hnRNP A1 propagation along the exon and therefore prevent splicing repression. In addition, omitting the MS2 coat protein should allow us to determine whether both hairpins would block the spreading of hnRNP A1.
The results we obtained were unexpected: we observed inhibition of splicing in the construct with the MS2 hairpin loop and the ESS, in the presence of MS2 coat protein (Fig. 3B, cf. lanes 14 and 15 with lanes 17 and 18; see Fig. S3A in the supplemental material). There are several possible explanations for this result: first, RNA secondary structure may actually facilitate cooperative binding of hnRNP A1 (14, 38) despite the presence of bound MS2 coat protein; second, hnRNP A1 may unwind the hairpin, displacing the tightly bound MS2 coat protein, and cooperatively spread along the exon to repress splicing; third, bound MS2 coat protein may permit or perhaps facilitate cooperative binding of hnRNP A1, although this seems improbable; and fourth, in spreading along the exon, hnRNP A1 may somehow bypass the hairpin with bound MS2 coat protein.
To distinguish among these possibilities, we used GST pulldowns to measure whether MS2 coat protein is displaced by hnRNP A1 cooperative binding. We made four artificial RNA transcripts composed mainly of oligo(U) with 32P-labeled C every fifth nucleotide, with an MS2 hairpin in the middle and a high-affinity hnRNP A1 binding site at the 3′ end (Fig. 3C). Each construct is either 90 or 91 nt long, depending on whether it has an MS2 or MS2* version of the hairpin. Each RNA construct was denatured at 95°C and allowed to refold at room temperature before addition of GST-MS2 protein. The results clearly demonstrate that hnRNP A1 displaces bound GST-MS2 protein, presumably by unwinding the stem-loop and/or by physical displacement (Fig. 3D, left panel; see Fig. S3B in the supplemental material). In a similar experiment, hnRNP A1 cooperative binding did not result in displacement of SF2/ASF from its ESE to inhibit splicing (data not shown); this result is consistent with the tight binding of SF2/ASF to its cognate ESE compared to SC35 (67).
As the amount of hnRNP A1 protein increases, the amount of GST-MS2 protein displaced increases, and this effect largely depends on the initial binding of hnRNP A1 to the high-affinity site (in Fig. 3D cf. the first five lanes [PUMS2ESS] [WT] with the last five lanes [PUMS2ESSm], in which a single point mutation abrogates the high-affinity hnRNP A1 binding site). The control RNA with the MS2* hairpin failed to bind MS2 coat protein, as expected (data not shown). A similar experiment was done with the same four RNA constructs and UP1 protein, which cannot undergo cooperative binding (67). As expected, UP1 was unable to displace GST-MS2 protein (Fig. 3D, right panel; see Fig. S3C in the supplemental material).
Cooperative binding of hnRNP A1 can also proceed from 5′ to 3′ to inhibit splicing.
Previous studies of hnRNP A1 cooperative binding focused on oligomerization in a 3′-to-5′ direction, after initial binding to an ESS at the 3′ end of a pre-mRNA (14, 15, 38, 67). To determine if hnRNP A1 can also spread in a 5′-to-3′ direction, we generated an artificial RNA comprising mainly oligo(U) tracts, with labeled 32P C at every fifth position, and a high-affinity hnRNP A1 SELEX winner sequence, UAGGGU (5), at the 5′ end (Fig. 4A). We also made a control RNA with a mutated hnRNP A1 binding sequence, UUGGGU. UV cross-linking of these RNAs after incubation with increasing concentrations of recombinant hnRNP A1 was followed by digestion with RNases A and T1, SDS-PAGE, and autoradiography (Fig. 4B; see Fig. S4A in the supplemental material). This experiment shows that cooperative binding of hnRNP A1 can proceed in a 5′-to-3′ direction.
FIG. 4.
hnRNP A1 cooperative binding spreading from the 5′ end to the 3′ end of exon 1 inhibits splicing. (A) Sequences of synthetic WT and mutant (MUT) RNAs for UV cross-linking experiments. The underlined hexanucleotide is a high-affinity hnRNP A1 binding site: UAGGGU is the WT version, and UUGGGU is the inactive, mutant version, with the mutated nucleotide shown in italic. The radiolabeled cytidines are indicated in bold italic. (B) UV cross-linking with WT and MUT RNAs (16 nM) from panel A in the presence of increasing concentrations of recombinant hnRNP A1 (0.05 to 0.4 μM). Detection and quantitation of the cross-linked products were as for Fig. 1B. (C) NSβ-globin minigene transcripts for in vitro splicing assays. The pre-mRNAs comprise 58 nt of exon 1, the 130-nt first intron, and 108 nt of exon 2. NSβ2, -3, -4, and -5 have in addition a 6-nt ESS or mutant ESSm at either the 5′ end of exon 1 or the 3′ end of exon 2; NSβ6 has the 6-nt ESS at both the 5′ end of exon 1 and the 3′ end of exon 2. (D) In vitro splicing of capped pre-mRNAs (1.15 nM) from panel C in S100 extract complemented with SC35 (0.4 μM), in the presence or absence of 0.6 μM hnRNP A1. The splicing efficiency (calculated as for Fig. 1D) is shown below the autoradiogram.
To test the effect of 5′-to-3′ hnRNP A1 cooperative binding on splicing, we designed short β-globin-derived minigene constructs with a 64-nt exon 1 and a 109-nt exon 2. We engineered a high-affinity hnRNP A1 binding sequence, UAGGGU (ESS), at the 5′ end of exon 1 by PCR; controls including a mutant hnRNP A1 motif, UCGGGU (ESSm), and a construct without the hnRNP A1 binding site were similarly made by PCR (Fig. 4C). In vitro splicing of pre-mRNA transcribed from these constructs in S100 extract complemented with SC35, with or without addition of hnRNP A1, is shown in Fig. 4D (see Fig. S4B in the supplemental material). Cooperative binding of hnRNP A1 propagating in a 5′-to-3′ direction in exon 1 inhibited splicing (cf. lanes 4 to 5 in the right panel and lanes 4 and 5 in the left panel).
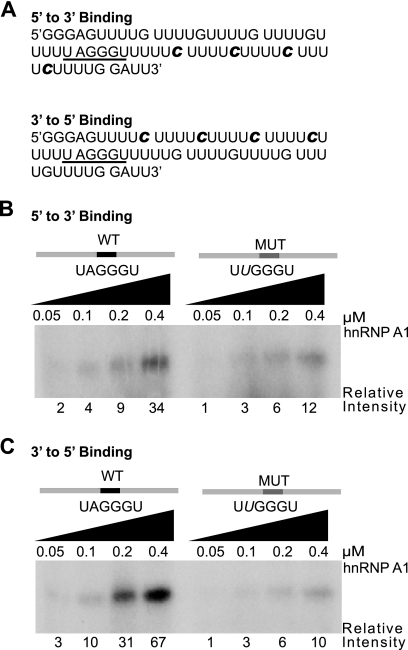
hnRNP A1 preferentially spreads in a 3′-to-5′ direction.
We next sought to determine whether hnRNP A1 can undergo cooperative binding with bidirectional spreading. To this end, we generated two RNAs with an hnRNP A1 high-affinity binding site in the middle (Fig. 5A). In the first construct, four nucleotides (cytosines) at every fifth position 3′ of the hnRNP A1 binding site were radiolabeled, whereas the sequences 5′ of the binding site were unlabeled. In the second construct, the labeled and unlabeled regions were reversed. Control substrates with a mutant hnRNP A1 binding site were also generated. UV cross-linking, digestion with RNases A and T1, and SDS-PAGE analysis were carried out as for Fig. 1. The ratio of WT to MUT intensities was greater for the 3′-to-5′ substrate (Fig. 5C; see Fig. S5B in the supplemental material) than for the 5′-to-3′ substrate (Fig. 5B; see Fig. S5A in the supplemental material), indicating preferential spreading of hnRNP A1 toward the 5′ end. A twofold reduction in the concentration of RNA and protein was enough to abrogate 5′-to-3′, but not 3′-to-5′, cooperative binding (data not shown). In light of this evidence, we conclude that hnRNP A1 5′-to-3′ cooperative binding is weaker than 3′-to-5′ binding.
FIG. 5.
Directionality of hnRNP A1 cooperative spreading. (A) Sequences of synthetic RNA transcripts for UV cross-linking experiments. The radiolabeled cytidines are indicated in bold italic. The underlined hexanucleotide is a high-affinity hnRNP A1 binding site (ESS). The top transcript (5′ to 3′ binding) has radiolabeled cytidines downstream of the ESS, whereas in the bottom transcript (3′ to 5′ binding), they are upstream of the ESS. (B) UV cross-linking with 16 nM WT transcript (5′ to 3′ binding) or its ESSm (MUT) version in the presence of increasing recombinant hnRNP A1 (0.05 to 0.4 μM). Detection and quantitation of the cross-linked products were as for Fig. 1B. (C) As for panel B but with 3′ to 5′ binding WT and MUT transcripts.
Determining the extent of spreading of hnRNP A1 along the RNA.
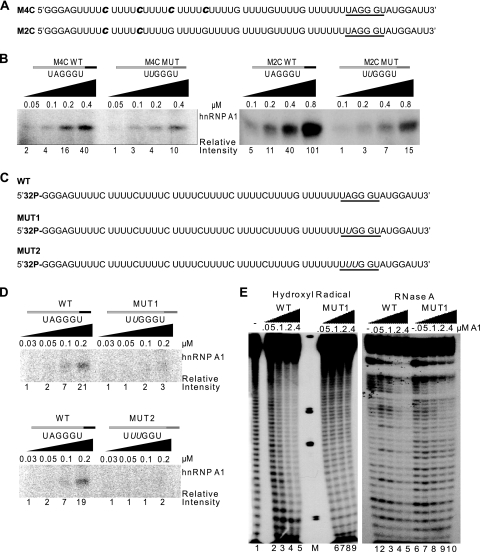
The pre-mRNA we used to test for hnRNP A1 oligomerization by UV cross-linking has six labeled C nucleotides (Fig. 1A). To determine more precisely how far hnRNP A1 spreads from the site of initial binding, we generated transcripts for cross-linking with fewer labeled Cs placed at different positions to see if we could still detect label transfer, reflecting cooperative binding (Fig. 6A). When the first two labeled Cs upstream of the high-affinity binding site were replaced with unlabeled Gs, we still detected label transfer to hnRNP A1 (Fig. 6B, left panel; see Fig. S6A in the supplemental material), indicating that cooperative binding extends beyond ∼20 nt. Similarly, when the next two labeled C nucleotides were also replaced by unlabeled Gs, we continued to detect a signal (Fig. 6B, right panel; see Fig. S6B in the supplemental material), indicating cooperative binding beyond ∼30 nt. Finally, we prepared a substrate by 32P 5′-end-labeling an otherwise unlabeled RNA transcript (Fig. 6C), and again, we detected cooperative binding by comparing the WT with the mutant (MUT1 or MUT2) transcripts (Fig. 6D, top and bottom panels; see Fig. S6C and S6D in the supplemental material). As expected, the signals became progressively weaker as transcripts with fewer labeled nucleotides were analyzed. We conclude that multiple molecules of hnRNP A1 bind consecutively along the RNA, all the way to its 5′ end.
FIG. 6.
Mapping the extent of hnRNP A1 cooperative spreading along the RNA. (A) Sequences of synthetic RNA transcripts for UV cross-linking experiments. The radiolabeled cytidines are indicated in bold italic. (B) UV cross-linking of the 16 to 32 nM RNA transcripts from panel A and the corresponding ESSm controls, in the presence of increasing recombinant hnRNP A1 (0.05 to 0.8 μM). Left panel, cross-linked products after RNase digestion of the top transcript (16 nM) in panel A and its ESSm counterpart. Right panel, Idem for the bottom transcript (32 nM) in panel A and its ESSm counterpart. Band intensities were measured on a phosphorimager, and normalized values are shown below the gel. (C) Sequences of 5′-end-labeled synthetic RNA transcripts for UV cross-linking and footprinting experiments. The underlined hexanucleotide is a high-affinity hnRNP A1 binding site: UAGGGU is the WT version, UUGGGU is the inactive mutant version 1 (MUT1), and UUUGGU is the inactive mutant version 2 (MUT2), with the mutated nucleotide shown in italic. (D) UV cross-linking with 8 nM WT and MUT1 and MUT2 RNAs from panel C in the presence of increasing concentrations of recombinant hnRNP A1 (0.03 to 0.2 μM). Detection and quantitation of the cross-linked products were as for Fig. 1B. (E) Footprinting assays with the first two RNA transcripts from panel C (WT and MUT1). The left panel shows a hydroxyl radical footprinting assay, and the right panel shows an RNase A footprinting assay; both were carried out with 16 nM 5′-end-labeled RNA in the presence of increasing recombinant hnRNP A1 (0 to 0.4 μM). M, molecular weight markers.
To address the cooperative spreading of hnRNP A1 using a different technique, we carried out hydroxyl radical footprinting using the first two 32P-5′-end-labeled RNAs in Fig. 6C. Figure 6E, left panel, shows the hydroxyl radical footprinting results. With increasing recombinant hnRNP A1, the region protected by hnRNP A1 increased (cf. WT in lanes 2 to 5 with MUT in lanes 6 to 9). Figure 6E, right panel, shows RNase A footprinting, which gives consistent results (cf. WT in lanes 2 to 5 with MUT in lanes 7 to 10). In addition, we obtained consistent results by RNase A footprinting using 3′-end-labeled RNA (see Fig. S7 in the supplemental material). Both chemical and RNase footprinting methods show that the entire length of the WT RNA is protected by cooperative binding of hnRNP A1. These results indicate that cooperative binding of hnRNP A1 to RNA resembles tightly packed “beads on a string” and does not require RNA secondary structure. As shown above, such structures, when present, can actually be unwound by hnRNP A1.
“Cross talk” between hnRNP A1 molecules bound at distant sites.
Finally, we investigated whether distant high-affinity hnRNP A1 binding sites can influence how hnRNP A1 binds to each site and subsequently spreads. Relevant to this, hnRNP A1 was reported to dimerize upon binding to distant sites, resulting in looping out of the RNA between the two sites (47). We generated five RNA constructs (Fig. 7A and C). First, we placed two identical ESSs at different positions along the RNA constructs (Fig. 7A): the first construct, XT1, has the two ESSs juxtaposed, separated by only 2 nt, and placed at the 3′ end of the RNA; the second construct, XT2, has one ESS at the 3′ end and the other in the middle of the RNA; the third construct, XT3, has one ESS at the 3′ end and the other at the 5′ end of the RNA; the fourth construct, XT3m1, has a mutant ESS (ESSm) at the 5′ end of the RNA (Fig. 7C); and the fifth construct, XT3m2, has a mutant ESS (ESSm) at the 3′ end (Fig. 7C). UV cross-linking of each of these constructs in the presence of increasing amounts of recombinant hnRNP A1 was compared with that of an RNA with a single ESS at the 3′ end (WT) (Fig. 1A).
FIG. 7.
“Cross talk” between hnRNP A1 molecules bound at distant sites. (A) Sequences of synthetic RNA transcripts for UV cross-linking experiments. The radiolabeled cytidines are indicated in bold italic. The underlined hexanucleotide is a high-affinity hnRNP A1 binding site (ESS). (B) UV cross-linking with 8 nM RNA transcripts from panel A and WT control from Fig. 1A, in the presence of increasing recombinant hnRNP A1 (0.03 to 0.2 μM). Detection and quantitation of the cross-linked products were as for Fig. 1B. The position of the ESS in each of the RNAs is indicated by a dark line. (C) Sequences of synthetic RNA transcripts for UV cross-linking experiments. The radiolabeled cytidines are indicated in bold italic. The underlined hexanucleotide is a high-affinity hnRNP A1 binding site: UAGGGU is the WT version, and UUGGGU is the inactive, mutant version, with the mutated nucleotide shown in italic. (D) UV cross-linking with 16 nM RNA transcripts from panel C and the XT3 control from panel A, in the presence of increasing recombinant hnRNP A1 (0.05 to 0.4 μM). Detection and quantitation of the cross-linked products were as for Fig. 1B. The position of the ESS in each of the RNAs is indicated by a dark line; a mutant ESS is indicated by an asterisk.
When the two binding sites were separated by only 2 nt, there was no apparent cross talk between the two sites, i.e., no additive or synergistic effect compared to the control WT RNA (Fig. 7B, left panel, cf. WT with XT1; see Fig. S8A in the supplemental material). When the distance separating the two high-affinity binding sites was greater, the signal relative to the WT RNA increased (Fig. 7B, middle panel, cf. WT with XT2; see Fig. S8B in the supplemental material), indicating an additive effect, or cross talk, between the two sites. With the ESS at both ends of the RNA, the signal increased even further (Fig. 7B, right panel, cf. WT with XT3; see Fig. S7C in the supplemental material). When the ESS at either end of the RNA construct was inactivated by a point mutation (Fig. 7C), cooperative-binding-dependent cross talk was lost (Fig. 7D, cf. XT3 with XT3m1 [left panel] [see Fig. S7D in the supplemental material] and also XT3 with XT3m2 [right panel] [see Fig. 7E in the supplemental material]).
We note that the type of cross talk shown in this experiment is cooperative-binding-dependent and differs from the mechanism proposed by Nasim et al. (47): if the RNA between the two distant high-affinity sites is looped out and not bound by hnRNP A1, label transfer would not occur, because all the radiolabeled nucleotides are present along the RNA sequences between the two ESSs at the ends. On the other hand, looping may be the preferred configuration with other substrates, and it is also possible that bridging of two distant sites by hnRNP A1 dimerization results in looping of the intervening RNA and its coating by additional molecules of hnRNP A1.
DISCUSSION
We have demonstrated that RNA secondary structure is not required for hnRNP A1 cooperative binding to RNA, even though hnRNP A1 can bind to RNAs that are highly structured in solution, such as the HIV-1 tat pre-mRNA (14, 38). We found that hnRNP A1 can unwind RNA secondary structure in a cooperative-binding-dependent manner. This result is consistent with hnRNP A1's established properties as a single-stranded RNA/DNA binding protein that can coat the entire length of a polynucleotide (9, 12, 16). In vivo, this type of binding could play a multitude of roles in cotranscriptional and posttranscriptional RNA processing, including splice site recognition, alternative splicing regulation, mRNA susceptibility to RNases, nuclear export of mature mRNA, etc., as well as in telomere length regulation (34, 64).
We showed that displacement of GST-MS2 protein bound to a hairpin and unwinding of this hairpin structure by hnRNP A1 require cooperative binding. Thus, UP1 had little or no activity in the GST-MS2 displacement and hairpin-unwinding assays. This is consistent with UP1 lacking the C-terminal glycine-rich domain, which is necessary for cooperative binding and splicing silencing (16, 44, 67). In addition, when we prevented the initial binding of hnRNP A1 by introducing a point mutation in the high-affinity binding site, the protein could no longer displace bound GST-MS2 protein or unwind a hairpin.
We further showed that a 6-nt hnRNP A1 SELEX winner sequence (UAGGGU) has ESS activity and that a single point mutation in this sequence is enough to disrupt hnRNP A1 cooperative binding and splicing silencing.
Cooperative binding by hnRNP A1 was shown to spread from the 3′ end of an HIV-1 RNA toward the 5′ end of the exon and to inhibit splicing by blocking an SC35-dependent ESE (67). However, it was not known whether cooperative binding of hnRNP A1 can also proceed in a 5′-to-3′ direction and likewise inhibit splicing. Here, we observed that 5′-to-3′ cooperative spreading does occur but appears to be considerably weaker than 3′-to-5′ spreading. We generated β-globin minigene derivatives with two exons of the same length (101 nt) and with the identical 6-nt ESS at the 5′ end of exon 1 in one construct and at the 3′ end of exon 2 in the other construct. Using these pre-mRNAs, we observed strong inhibition of splicing in vitro for the pre-mRNA with the ESS at the 3′ end of exon 2, whereas splicing of the pre-mRNA with the ESS at the 5′ end of exon 1 was unaffected (data not shown). However, when we reduced the size of exon 1 with the ESS at the 5′ end to 64 nt, splicing was strongly inhibited. This inhibition of splicing can be attributed to hnRNP A1 cooperative binding, as strong splicing inhibition depended on addition of recombinant hnRNP A1.
Figure 8 shows our model for hnRNP A1 cooperative binding. hnRNP A1 can displace a protein bound to a secondary structure that interrupts the path of hnRNP A1 spreading. Moreover, hnRNP A1 unwinds the structure to then spread further and displace bound SC35 from an ESE.
FIG. 8.
Model of hnRNP A1 cooperative binding. 1, hnRNP A1 binds to the ESS, MS2 coat protein binds to the MS2 hairpin, and SC35 binds to the ESE. 2 and 3, hnRNP A1 cooperative spreading displaces bound MS2 coat protein. 4 and 5, hnRNP A1 unwinds MS2 hairpin and continues cooperative spreading. 6 and 7, hnRNP A1 cooperative spreading displaces bound SC35 from the ESE and inhibits splicing.
A form of cross talk or communication between two hnRNP A1 molecules bound at distant sites has been described (47). This cross talk allows the skipping of an exon between the two flanking intronic binding sites, through protein-protein interaction between hnRNP A1 molecules bound at these sites causing the exon to loop out. A similar looping out may also occur within a long intron, thereby increasing the efficiency of splicing between two distant splice sites (47). Here we also investigated if there is cross talk between two molecules of hnRNP A1 bound at nonadjacent high-affinity sites. Our results are consistent with a kind of cross talk that does not necessarily involve looping out of the RNA. We found that when two high-affinity hnRNP A1 binding sites are juxtaposed, the extent of hnRNP A1 cooperative spreading toward the 5′ end of the RNA is similar to that observed with a single site. In contrast, as the distance between the two sites increases, the extent of cooperative binding increases, and it is maximal when the two high-affinity sites are placed at both ends of the RNA. In the context of our experiments, looping out of naked RNA between the two high-affinity binding sites would not have resulted in label transfer to hnRNP A1 after RNase digestion, as all the labeled nucleotides were placed between the two high-affinity sites. Thus, with these particular substrates, either there is no looping or the RNA in the loop is also bound by hnRNP A1 molecules.
We termed the kind of interaction between two hnRNP A1 sites observed here cooperative-binding-dependent cross talk. With the two hnRNP A1 binding sites placed at the 5′ and 3′ ends of the RNA, hnRNP A1 binding initially at the 5′ site would spread toward the 3′ end and, simultaneously, hnRNP A1 binding initially at the 3′ end would spread toward the 5′ end. Convergent spreading would increase the rate at which the gap between the two binding sites is filled with hnRNP A1 molecules, compared to a single initial binding site. The looping model (47) and the cross talk model reported here may each apply in different situations, although what pre-mRNA contexts or cellular conditions determine one or the other mode of binding remains unknown.
The results presented here indicate that hnRNP A1 can unwind an RNA hairpin, even when the hairpin is protected by a tightly bound protein. However, it is possible that more extensive secondary/tertiary structures and/or very tightly bound proteins could be more effective at blocking hnRNP A1 propagation than the MS2 hairpin with or without bound MS2 coat protein.
In short, we have described the features of hnRNP A1 cooperative binding. This cooperative binding, as shown in the model in Fig. 8, unwinds RNA secondary structure and preferentially spreads in a 3′-to-5′ direction to displace SR proteins bound at an ESE, thereby inhibiting splicing. 5′-to-3′ cooperative spreading of hnRNP A1 appears to be less robust, but within certain distance constraints, it may also be sufficient to unwind RNA secondary structure, displace bound SR proteins, and/or displace U1 snRNP from a 5′ splice site to inhibit splicing.
Supplementary Material
Acknowledgments
We thank Michele Hastings, Zuo Zhang, and Qingshuo Zhang for generous gifts of recombinant proteins and Mads Jensen, Olga Anczuków-Camarda, Isabel Aznarez, and Yimin Hua for helpful comments on the manuscript.
This work was supported by grant CA13106 from the NCI.
Footnotes
Published ahead of print on 10 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdul-Manan, N., and K. R. Williams. 1996. hnRNP A1 binds promiscuously to oligoribonucleotides: utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 24:4063-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt, B. A., Z. H. Si, and C. M. Stoltzfus. 1995. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 15:4606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birney, E., S. Kumar, and A. R. Krainer. 1993. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21:5803-5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchette, M., and B. Chabot. 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 18:1939-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burd, C. G., and G. Dreyfuss. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13:1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cáceres, J. F., and A. R. Krainer. 1993. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 12:4715-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caputi, M., and A. M. Zahler. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartegni, L., M. Maconi, E. Morandi, F. Cobianchi, S. Riva, and G. Biamonti. 1996. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 259:337-348. [DOI] [PubMed] [Google Scholar]
- 10.Chew, S. L., H. X. Liu, A. Mayeda, and A. R. Krainer. 1999. Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 96:10655-10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke, P. A. 1999. RNA footprinting and modification interference analysis. Methods Mol. Biol. 118:73-91. [DOI] [PubMed] [Google Scholar]
- 12.Cobianchi, F., R. L. Karpel, K. R. Williams, V. Notario, and S. H. Wilson. 1988. Mammalian heterogeneous nuclear ribonucleoprotein complex protein A1. Large-scale overproduction in Escherichia coli and cooperative binding to single-stranded nucleic acids. J. Biol. Chem. 263:1063-1071. [PubMed] [Google Scholar]
- 13.Cullen, B. R. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28:419-424. [DOI] [PubMed] [Google Scholar]
- 14.Damgaard, C. K., T. O. Tange, and J. Kjems. 2002. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8:1401-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Gatto-Konczak, F., M. Olive, M. C. Gesnel, and R. Breathnach. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding, J., M. K. Hayashi, Y. Zhang, L. Manche, A. R. Krainer, and R. M. Xu. 1999. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 13:1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyfuss, G., M. J. Matunis, S. Piñol-Roma, and C. G. Burd. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289-321. [DOI] [PubMed] [Google Scholar]
- 18.Eperon, I. C., O. V. Makarova, A. Mayeda, S. H. Munroe, J. F. Cáceres, D. G. Hayward, and A. R. Krainer. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 20:8303-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge, H., P. Zuo, and J. L. Manley. 1991. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell 66:373-382. [DOI] [PubMed] [Google Scholar]
- 20.Ghigna, C., M. Moroni, C. Porta, S. Riva, and G. Biamonti. 1998. Altered expression of heterogenous nuclear ribonucleoproteins and SR factors in human colon adenocarcinomas. Cancer Res. 58:5818-5824. [PubMed] [Google Scholar]
- 21.Graveley, B. R. 2005. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 123:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graveley, B. R., and T. Maniatis. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1:765-771. [DOI] [PubMed] [Google Scholar]
- 23.Hanamura, A., J. F. Cáceres, A. Mayeda, B. R. Franza, Jr., and A. R. Krainer. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4:430-444. [PMC free article] [PubMed] [Google Scholar]
- 24.Herrick, G., and B. Alberts. 1976. Purification and physical characterization of nucleic acid helix-unwinding proteins from calf thymus. J. Biol. Chem. 251:2124-2132. [PubMed] [Google Scholar]
- 25.Hua, Y., T. A. Vickers, H. L. Okunola, C. F. Bennett, and A. R. Krainer. 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 82:834-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Y., and J. A. Steitz. 2005. SRprises along a messenger's journey. Mol. Cell 17:613-615. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Kan, P. M. Loerch, C. D. Armour, R. Santos, E. E. Schadt, R. Stoughton, and D. D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141-2144. [DOI] [PubMed] [Google Scholar]
- 28.Kan, Z., D. States, and W. Gish. 2002. Selecting for functional alternative splices in ESTs. Genome Res. 12:1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karni, R., E. de Stanchina, S. W. Lowe, R. Sinha, D. Mu, and A. R. Krainer. 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashima, T., N. Rao, and J. L. Manley. 2007. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 104:3426-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krainer, A. R., G. C. Conway, and D. Kozak. 1990. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 4:1158-1171. [DOI] [PubMed] [Google Scholar]
- 32.Krainer, A. R., T. Maniatis, B. Ruskin, and M. R. Green. 1984. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36:993-1005. [DOI] [PubMed] [Google Scholar]
- 33.Krainer, A. R., A. Mayeda, D. Kozak, and G. Binns. 1991. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66:383-394. [DOI] [PubMed] [Google Scholar]
- 34.LaBranche, H., S. Dupuis, Y. Ben-David, M. R. Bani, R. J. Wellinger, and B. Chabot. 1998. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19:199-202. [DOI] [PubMed] [Google Scholar]
- 35.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 36.LeCuyer, K. A., L. S. Behlen, and O. C. Uhlenbeck. 1995. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry 34:10600-10606. [DOI] [PubMed] [Google Scholar]
- 37.Lin, S., and X. D. Fu. 2007. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 623:107-122. [DOI] [PubMed] [Google Scholar]
- 38.Marchand, V., A. Mereau, S. Jacquenet, D. Thomas, A. Mougin, R. Gattoni, J. Stévenin, and C. Branlant. 2002. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 323:629-652. [DOI] [PubMed] [Google Scholar]
- 39.Maris, C., C. Dominguez, and F. H. Allain. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272:2118-2131. [DOI] [PubMed] [Google Scholar]
- 40.Mayeda, A., D. M. Helfman, and A. R. Krainer. 1993. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol. Cell. Biol. 13:2993-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayeda, A., and A. R. Krainer. 1999. Mammalian in vitro splicing assays. Methods Mol. Biol. 118:315-321. [DOI] [PubMed] [Google Scholar]
- 42.Mayeda, A., and A. R. Krainer. 1999. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 118:309-314. [DOI] [PubMed] [Google Scholar]
- 43.Mayeda, A., and A. R. Krainer. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68:365-375. [DOI] [PubMed] [Google Scholar]
- 44.Mayeda, A., S. H. Munroe, J. F. Cáceres, and A. R. Krainer. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 13:5483-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milligan, J. F., and O. C. Uhlenbeck. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180:51-62. [DOI] [PubMed] [Google Scholar]
- 46.Nadler, S. G., B. M. Merrill, W. J. Roberts, K. M. Keating, M. J. Lisbin, S. F. Barnett, S. H. Wilson, and K. R. Williams. 1991. Interactions of the A1 heterogeneous nuclear ribonucleoprotein and its proteolytic derivative, UP1, with RNA and DNA: evidence for multiple RNA binding domains and salt-dependent binding mode transitions. Biochemistry 30:2968-2976. [DOI] [PubMed] [Google Scholar]
- 47.Nasim, F. U., S. Hutchison, M. Cordeau, and B. Chabot. 2002. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA 8:1078-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okazaki, Y., M. Furuno, T. Kasukawa, J. Adachi, H. Bono, S. Kondo, I. Nikaido, N. Osato, R. Saito, et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420:563-573. [DOI] [PubMed] [Google Scholar]
- 49.Paradis, C., P. Cloutier, L. Shkreta, J. Toutant, K. Klarskov, and B. Chabot. 2007. hnRNP I/PTB can antagonize the splicing repressor activity of SRp30c. RNA 13:1287-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrotti, D., and P. Neviani. 2007. From mRNA metabolism to cancer therapy: chronic myelogenous leukemia shows the way. Clin. Cancer Res. 13:1638-1642. [DOI] [PubMed] [Google Scholar]
- 51.Pontius, B. W., and P. Berg. 1992. Rapid assembly and disassembly of complementary DNA strands through an equilibrium intermediate state mediated by A1 hnRNP protein. J. Biol. Chem. 267:13815-13818. [PubMed] [Google Scholar]
- 52.Pontius, B. W., and P. Berg. 1990. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: implications for a mechanism for rapid molecular assembly. Proc. Natl. Acad. Sci. USA 87:8403-8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romaniuk, P. J., and O. C. Uhlenbeck. 1983. Joining of RNA molecules with RNA ligase. Methods Enzymol. 100:52-59. [DOI] [PubMed] [Google Scholar]
- 54.Schmucker, D., J. C. Clemens, H. Shu, C. A. Worby, J. Xiao, M. Muda, J. E. Dixon, and S. L. Zipursky. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101:671-684. [DOI] [PubMed] [Google Scholar]
- 55.Shamoo, Y., U. Krueger, L. M. Rice, K. R. Williams, and T. A. Steitz. 1997. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution. Nat. Struct. Biol. 4:215-222. [DOI] [PubMed] [Google Scholar]
- 56.Shaw, S. D., S. Chakrabarti, G. Ghosh, and A. R. Krainer. 2007. Deletion of the N-terminus of SF2/ASF permits RS-domain-independent pre-mRNA splicing. PLoS One 2:e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, D. J., C. C. Query, and M. M. Konarska. 2008. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol. Cell 30:657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staknis, D., and R. Reed. 1994. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol. Cell. Biol. 14:7670-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stoltzfus, C. M., and J. M. Madsen. 2006. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV Res. 4:43-55. [DOI] [PubMed] [Google Scholar]
- 60.Tange, T. O., and J. Kjems. 2001. SF2/ASF binds to a splicing enhancer in the third HIV-1 tat exon and stimulates U2AF binding independently of the RS domain. J. Mol. Biol. 312:649-662. [DOI] [PubMed] [Google Scholar]
- 61.Vitali, J., J. Ding, J. Jiang, Y. Zhang, A. R. Krainer, and R. M. Xu. 2002. Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1. Nucleic Acids Res. 30:1531-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, Z., and C. B. Burge. 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14:802-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu, R. M., L. Jokhan, X. Cheng, A. Mayeda, and A. R. Krainer. 1997. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure 5:559-570. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, Q. S., L. Manche, R. M. Xu, and A. R. Krainer. 2006. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA 12:1116-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Z., and A. R. Krainer. 2007. Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components. Proc. Natl. Acad. Sci. USA 104:11574-11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu, J., and A. R. Krainer. 2000. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 14:3166-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]
- 68.Zuo, P., and J. L. Manley. 1993. Functional domains of the human splicing factor ASF/SF2. EMBO J. 12:4727-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.