Abstract
Double-strand breaks (DSBs) are harmful DNA lesions that can generate chromosomal rearrangements or chromosome losses if not properly repaired. Despite their association with a number of genetic diseases and cancer, the mechanisms by which DSBs cause rearrangements remain unknown. Using a newly developed experimental assay for the analysis of translocations occurring between two chromosomes in Saccharomyces cerevisiae, we found that a single DSB located on one chromosome uses a short homologous sequence found in a third chromosome as a bridge to complete DSB repair, leading to chromosomal translocations. Such translocations are dramatically reduced when the short homologous sequence on the third chromosome is deleted. Translocations rely on homologous recombination (HR) proteins, such as Rad51, Rad52, and Rad59, as well as on the break-induced replication-specific protein Pol32 and on Srs2, but not on Ku70. Our results indicate that a single chromosomal DSB efficiently searches for short homologous sequences throughout the genome for its repair, leading to triparental translocations between heterologous chromosomes. Given the abundance of repetitive DNA in eukaryotic genomes, the results of this study open the possibility that HR rather than nonhomologous end joining may be a major source of chromosomal translocations.
Genomic instability can be a source of cell death and cancer (3, 34). It is usually manifested as mutations and chromosomal rearrangements, including translocations, deletions, inversions, and duplications, often leading to gene fusions that may play a key role in the initial steps of tumorigenesis and subsequent cancer development (41, 42, 44). It is widely assumed that all rearrangement events are initiated by DNA double-strand breaks (DSBs) (1, 27, 49). In somatic cells, DSBs generally occur during DNA replication or by the action of environmental agents, such as genotoxic chemicals or ionizing radiation. Nevertheless, the mechanisms responsible for chromosomal rearrangements are not completely understood.
Eukaryotic cells have evolved two main pathways to repair DSBs: nonhomologous end joining (NHEJ) and homologous recombination (HR). NHEJ involves processing of the two break ends so that either addition or deletion of nucleotides can occur prior to ligation and, consequently, mutations may be introduced. The involvement of NHEJ in the generation of chromosomal rearrangements has been extensively studied (9, 32, 62), and a number of NHEJ-mediated chromosomal rearrangements associated with cancer have been reported (4, 35). HR requires significant amounts of homology between the broken DNA ends and the intact DNA sequence to be used for repair. For this reason, HR is typically error free. However, HR can also be a source of chromosomal rearrangements when occurring between DNA repeats located in different chromosomes (33, 54).
HR may occur via different mechanisms, depending on the nature of the DNA ends, the location of the homologous partners, and the length of homology (reviewed in reference 46). After a chromosome break, cells carry out a genome-wide search for homologous sequences that are used as template for the repair of the broken chromosome. When both ends of the DSB (two-ended DSB) are homologous to sequences present in an intact chromosome, HR may proceed by DSB repair (DSBR) or synthesis-dependent strand annealing (SDSA) mechanisms (reviewed in reference 21). When only one end of the DSB (one-ended DSB) is homologous to sequences elsewhere in the genome, break-induced replication (BIR) becomes the alternative HR repair mechanism, as shown for yeast (39). Whereas HR repair of two-ended DSBs is completed with the capture of the second end, repair of one-ended DSBs relies on DNA synthesis primed by the invading end all the way to the end of the chromosome or until it encounters a barrier (43).
In yeast, both DSBR/SDSA and BIR require the DSB repair genes of the RAD52 epistasis group, including the RAD51 strand exchange factor and the RAD52 single-stranded DNA binding protein (reviewed in reference 30). However, RAD51 may be dispensable in BIR occurring between particular substrates, such as inverted repeats (5, 12, 58). BIR requires additional specific replication proteins, such as Pol32, one of the subunits of DNA polymerase δ that is dispensable for replication and DSB repair mechanisms other than BIR (36).
To gain further insight into the mechanisms of chromosomal translocations, we devised an intron-based chromosomal translocation assay in Saccharomyces cerevisiae, in which a DSB is generated in a single chromosome by the HO endonuclease. We show that DSB-induced translocations occur via triparental recombination events. A short homologous sequence in a third chromosome serves as a bridge template for HR events occurring between two nonhomologous chromosomes. The triparental HR events that occur in our assays give rise mainly to reciprocal translocations that require Rad52, Rad51, and importantly, Pol32. Rad59, as well as the nonreplicative DNA helicase Srs2, are also required, although to a lesser extent, whereas Ku70 or Mus81 endonuclease play no role. We conclude that BIR-mediated triparental recombination could be a major mechanism for chromosomal translocations in eukaryotic cells.
MATERIALS AND METHODS
Yeast strains and plasmids.
The yeast strains used in this study are listed in Table S1 in the supplemental material. All strains were isogenic to strain W303. rad52Δ, rad51Δ, and srs2Δ single-deletion mutants were obtained with 60-mer oligonucleotides via the PCR-based gene replacement method (61). All deletions were confirmed by PCR and Southern blot analysis. pol32Δ, rad59Δ, rad1Δ, ku70Δ, and mus81Δ mutant strains were obtained by genetic crosses. The ACT1 intron was deleted as previously described (23) to obtain the ACT1Δi strain.
The recombination system was constructed by chromosome integration of plasmids specifically constructed for this study. For this, the LEU2 gene under the control of the GAL1 promoter from plasmid p314GLB (47) was split by digestion with EcoRV to generate two LEU2 fragments, leu2Δ5′ (750 nucleotides [nt]) and leu2Δ3′ (350 nt plus the transcriptional termination sequence from the CYC1 gene). The intron of the yeast ACT1 gene (ACT1-i, 308 bp) was amplified by PCR by using oligonucleotides ACT-s and ACT-as (see Table S2 in the supplemental material) and Pfu polymerase (Promega). The blunt-ended PCR fragment containing the ACT1 intron was cloned into the GLB plasmid to generate plasmid GLB-ACT-i. The desired orientation of the cloned ACT1-i was confirmed by DNA sequencing. Plasmid GLB-ACT-i was digested with XhoI, which had a unique restriction site in the intron (located at nucleotide position 62 from the 5′ donor splice site); blunt ended; and finally ligated to the URA3 cassette (plasmid GLB-ACT-i-U). To place these constructs in the yeast chromosomes, the fragment GAL1p::leu2Δ3′::ACT1-iΔ3′::URA3 was amplified by PCR using oligonucleotides IIIint-s and IIIint-as (see Table S2 in the supplemental material) and was integrated next to the SUP61 locus, at chromosome III, in the yeast strain MKOS3C (see Table S1 in the supplemental material), by transformation. The recovered recombinants were tested for both the correct integration and the desired orientation of the DNA fragment by Southern analysis.
On the other hand, a hygromycine (HYG) cassette, followed by the 117-bp region surrounding the HO cleavage site present at the MATa locus, was cloned at the same blunt ended-XhoI restriction site of plasmid GLB-ACT-i (plasmid GLB-HYG::HO-ACT-i). The desired orientation of the HYG-HO cassette was confirmed by DNA sequencing. The fragment HYG-HOcs::ACT1-iΔ5′::leu2Δ5′ was amplified by PCR with oligonucleotides XVint-s and XVint-as (see Table S2 in the supplemental material) and was integrated into chromosome XV at the HIS3 locus in the WS strain (see Table S1 in the supplemental material). Diploid strains with the complete experimental system were obtained by crossing the derivative haploid strains MKOS3C and WS carrying the recombination systems, and the corresponding haploid strains with the complete experimental system were obtained by sporulation and tetrad dissection.
Determination of recombination frequencies.
Yeast colonies were grown on yeast extract-peptone-adenine-dextrose (YPAD) plates. For recombination analyses, independent colonies were picked from plates and grown overnight in liquid minimal medium (synthetic complete [SC] medium-3% glycerol-2% lactate). HO endonuclease expression was induced by the addition of 2% galactose. After 24 h of incubation in the presence of galactose, appropriate dilutions were plated on SC-galactose to determine the total cell number and on SC-galactose without leucine to determine the number of recombinants. Recombination frequencies were obtained as the median value for at least six independent colonies for each test performed for each strain. Statistical analysis was performed using the χ2 test.
Kinetic analysis of BIR intermediates by PCR.
After galactose induction, yeast cells were taken at different times and genomic DNAs were extracted according to standard methods. Analysis of the DNA intermediates produced by the first HR step was done by PCR using oligonucleotides p1 and p2. PCR detection of the DNA intermediates produced by the second HR step was performed with oligonucleotides p3 and p4. Control PCR products were detected by using oligonucleotides p1 and p4. Whereas PCR analysis of the DNA strand invasion at the HMR locus was done with oligonucleotides p3-1 and p15-1, DNA strand invasion at MAT was monitored by using the oligonucleotides p3-2 and p15-2. PCR detection of DNA translocations between chromosomes III and XV was performed with oligonucleotides p3-3 and p15-3. Oligonucleotide sequences are listed in Table S2 in the supplemental material. PCR amplifications were performed with a standard profile of 95°C for 30 s, 51°C for 30 s, and 72°C for 60 s during 35 cycles. For MAT DNA strand invasion reactions, PCR extension time was increased up to 4 min. PCR primers were used at 1 μM, deoxynucleoside triphosphate mix was used at 200 μM, and GoTaq DNA polymerase (Promega) was used at 5 U/ml under otherwise standard conditions. PCR products were subjected to gel electrophoresis in 0.8% agarose and stained with ethidium bromide.
Pulsed-field gel electrophoresis (PFGE) and Southern analysis.
Recombinant yeast colonies were grown in 10 ml of YPAD medium overnight at 30°C. Chromosome plugs were made according to the manufacturer's instructions (Bio-Rad). Agarose gels (0.9%) were run in a Bio-Rad CHEF MapperXA apparatus for 16 h at 6 V/cm with a switch time of 70 s and for an additional 12 h at 6 V/cm with a switch time of 120 s. Then, gels were stained with ethidium bromide. After denaturation, agarose gels were transferred to Hybond-N membranes (GE Healthcare) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Southern hybridizations and radiolabeled DNA probes were done according to the standard procedures. The different DNA probes used in this work were amplified by PCR using the oligonucleotides listed in Table S2 in the supplemental material and were specific to the following open reading frames: probe 1, YCR001w; probe 2, MAT; probe 3, YCR061w; probe 4, YCR089w; probe 5, yCR105w; probe LEU, related to chromosome III; probe 6, YOR164w; and probe 7, YOR238w, related to chromosome XV.
RESULTS
A new assay for DSB-induced chromosomal translocations.
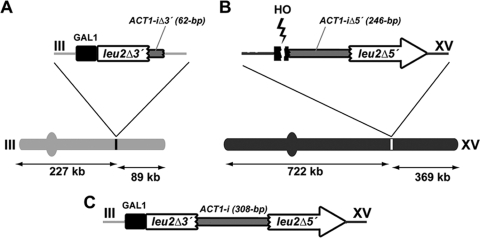
We devised an intron-based assay for the selection of chromosomal rearrangements induced by an HO-mediated DSB located at one specific site on chromosome XV of the yeast S. cerevisiae. As illustrated in Fig. 1, the system is based on the two nonhomologous halves of the LEU2 gene integrated in chromosomes III and XV (see Materials and Methods). The 5′ half (leu2Δ3′), followed by the 5′-end 62 bp of the ACT1 intron, was placed under the control of the galactose-inducible GAL1 promoter (GAL1p) and integrated at the right arm of chromosome III at the 227-kb position (Fig. 1A). The 3′ half (leu2Δ5′) was located downstream from the remaining 246 bp of the ACT1 intron (ACT1-i), integrated at the right arm of chromosome XV at the 722-kb position (Fig. 1B). Upstream of this ACT1-i-leu2Δ5′ fragment, the 117-bp HO endonuclease recognition site was inserted. These genetically engineered chromosomes III and XV were created in both diploid (α/a-inc) and haploid (a-inc) cells containing the HO endonuclease gene under the GAL1 promoter, integrated at the ADE3 locus of chromosome VII, and most of the endogenous LEU2 gene deleted from its original locus on chromosome III. As expected, DSBs were observed at the inserted HO site on chromosome XV 30 min after HO induction, while no breaks were detected at MATa-inc even 24 h later (data not shown). A DSB at the HO site of chromosome XV could lead to Leu-positive (Leu+) recombinants by translocation of the leu2Δ5′ fragment from chromosome XV close to the leu2Δ3′ on chromosome III, with a functional intron in between (Fig. 1C).
FIG. 1.
Schematic representation of intron-based assay for chromosomal translocations in yeast. (A) A fragment of the LEU2 gene (leu2Δ3′) under the control of the GAL1 promoter, plus the first 62 bp of the yeast ACT1 intron (ACT1-iΔ3′), was integrated in the right arm of chromosome III. (B) The 117-bp HO endonuclease cleavage sequence was cloned into the right arm of chromosome XV. The downstream 246 bp of the ACT1 intron (ACT1-iΔ5′) and the 3′ half of the LEU2 gene sequence (leu2Δ5′) were integrated next to the HO cut site. The sizes of the chromosomal fragments with respect to the integrated sequences are shown in both cases. There was no sequence homology between the two DNA fragments integrated in chromosomes III and XV. The endogenous LEU2 gene was deleted from the ClaI site to the EcoRV site. (C) A chromosomal translocation between these two DNA fragments generates a galactose-inducible full-length LEU2 gene harboring the yeast ACT1 intron sequence.
After 24 h of induction with galactose, Leu+ recombinants were obtained in haploid cells at a frequency of 10.2 × 10−6 ± 3.6 × 10−6 (mean ± standard deviation) (Fig. 2B). With shorter induction times, we obtained similar frequencies of Leu+ recombinants (24 × 10−6 ± 8.9 × 10−6 after 4 h and 10.4 × 10−6 ± 2.1 × 10−6 after 10 h), which indicated that the time range of HO induction used does not significantly affect the Leu+ recombinant frequency. In diploid cells, the frequency of Leu+ recombinants was 15 times lower than in haploids (Fig. 2B), likely due to the presence of the additional nonrecombinant homologous chromosome in diploids, which provided a competing partner for the repair of the HO-induced DSB via an HR event not resulting in Leu+ recombinants. However, other possible explanations cannot be discarded.
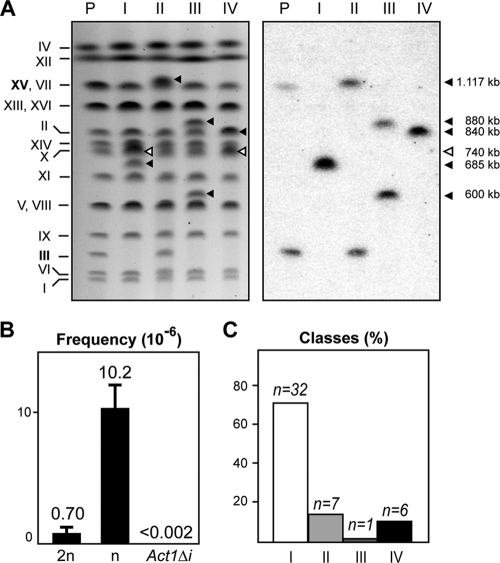
FIG. 2.
Molecular analysis of chromosomal translocations resulting from the repair of a single HO-induced DSB. (A) Molecular karyotype of Leu+ recombinants analyzed by PFGE. Agarose gels were stained with ethidium bromide (left) and analyzed by Southern blotting using a radiolabeled LEU2 probe (right). Electrophoretic mobilities of all nonrecombining chromosomes are indicated on the left. The LEU2 signal in parental cells corresponds to the two nonhomologous halves of LEU2 integrated in chromosomes III and XV. Chromosomes XV and VII have the same electrophoretic mobility in the experimental conditions used here. The translocated chromosomes containing LEU2 are marked with black triangles, and those without LEU2 sequences are marked with open triangles. The sizes of the translocated chromosomes were estimated by adjusting their position in the gel to a regression curve made with the known sizes of unrearranged chromosomes and their electrophoretic positions. P, parental strain; I, class I recombinant; II, class II recombinant; III, class III recombinant; IV, class IV recombinant. (B) Frequencies of Leu+ recombinants after HO-induced DSBs in both diploid (2n) and haploid (n) wild-type cells and in ACT1Δi cells, in which the intron of the endogenous ACT1 locus was deleted. Mean values and standard deviations are shown. (C) Distribution of the different classes of Leu+ recombinants in haploid wild-type cells. The percentage of each class is plotted in the histogram. The absolute number of recombinants of each class is indicated on top of each bar.
These results indicated that chromosomal rearrangements occur efficiently between chromosomes XV and III as a consequence of the single HO-induced DSB.
Different types of chromosomal translocations are induced by a single HO break.
Chromosome rearrangements leading to the Leu+ recombinants were analyzed by PFGE in a number of independent events. Translocations were visualized by ethidium bromide staining and Southern analysis using a LEU2-specific probe, and the size of the new, rearranged chromosomes was estimated (Fig. 2A). On the basis of their molecular karyotypes, haploid Leu+ recombinants were grouped in four main classes, the most frequently represented recombinant class being class I (70%), followed by classes II (15%), IV (13%), and III (2%) (Fig. 2C). Since the same recombinant types were recovered in diploid cells after DSB induction (see Figure S1 in the supplemental material), we decided to pursue the molecular characterization of the chromosomal rearrangements in the haploid strain only. It is worth noticing, however, that the distribution of recombinant classes was different in diploids than in haploids (see Figure S1 in the supplemental material), likely as a consequence of the presence of a competing homologous chromosome for the repair of the single HO-induced DSB.
Class I recombinants were characterized by the appearance of two novel chromosomes (685 and 740 kb long) concomitant with the loss of parental chromosomes III and XV (Fig. 2A). Such chromosome losses were more evident for chromosome III than for chromosome XV, since the electrophoretic mobility of the latter was coincident with that of the nonrecombinant chromosome VII (Fig. 2A). The LEU2 signal was clearly detected in the smaller of the two novel chromosomes (Fig. 2A). Class II recombinants showed a molecular karyotype similar to that of parental cells, in which the LEU2 signal associated with both chromosome III and XV (Fig. 2A). Chromosome XV in this recombinant class appeared to have a slightly lower electrophoretic mobility (Fig. 2A), presumably due to the acquisition of unknown genetic information through some chromosomal exchange. Class III recombinants were characterized by the appearance of two novel rearranged chromosomes (600 and 880 kb long), which was accompanied by the loss of parental chromosomes III and XV (Fig. 2A). The LEU2 signal was specifically associated with the rearranged chromosomes. Finally, class IV was also characterized by two novel recombinant chromosomes (740 and 840 kb long). The smallest one was similar to that found in class I recombinants, and the largest one was only visible by hybridization with the LEU2 probe, because its electrophoretic mobility was coincident with that of the nonrecombinant chromosome II (Fig. 2A).
Mapping the chromosomal translocation breakpoints.
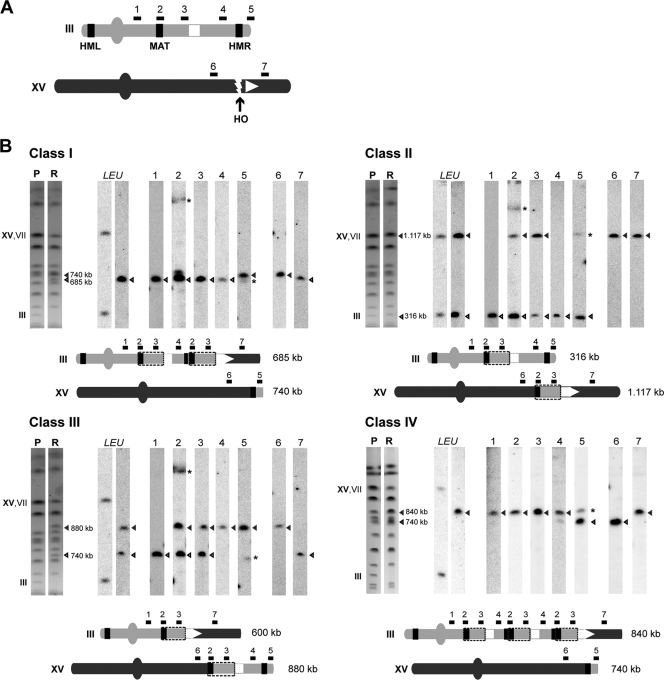
To map the breakpoints of the chromosomal translocations, we analyzed PFGE gels by Southern blotting using radiolabeled DNA probes from different chromosomal regions (see Materials and Methods), including the right arm of chromosome III and two locations flanking the HO site at the right arm of chromosome XV (Fig. 3A). The results obtained are summarized in Fig. 3, where the deduced genetic maps of the translocated chromosomes are shown. In class I recombinants, whereas probes 1, 2, 3, 4, LEU, and 7 hybridized with the smallest novel chromosome (685 kb long), probes 5 and 6 were associated with the larger one (740 kb) (Fig. 3B). These data suggested the occurrence of a reciprocal translocation involving both chromosome III and chromosome XV at the site of the HO break (Fig. 3B). In class II recombinants, it is noteworthy that probes 2 and 3 hybridized with both chromosome XV and III (Fig. 3B), indicating the existence of a duplicated chromosome III fragment, of at least 27 kb, inserted into chromosome XV during the DSB repair process. A similar conclusion could be extracted from the analysis of class III recombinants (Fig. 3B), since the same two chromosome III probes hybridized with the two new-sized chromosomes. Moreover, in class III recombinants, probe 6, which represents the fragment of chromosome XV proximal to the centromere, hybridized with the largest rearranged chromosome (880 kb long), whereas probe 7, representing the fragment of chromosome XV distal to the centromere, hybridized with the smallest (600 kb) chromosome (Fig. 3B). These results suggested that the duplication was accompanied by a reciprocal translocation involving both chromosomes III and XV. Class IV recombinants were similar to class I recombinants since they shared one of the rearranged chromosomes, which hybridized with probes 5 and 6. The largest rearranged chromosome was associated with all the remaining probes tested (Fig. 3B), this chromosome being almost 100 kb larger than that in the class I recombinants.
FIG. 3.
Molecular analysis of chromosomal translocation breakpoints. (A) The different DNA probes used, specific to the right arm of either chromosome III (1, 2, 3, 4, and 5) or chromosome XV (6 and 7), are indicated by black lines at the top of the schematic representation of each chromosome. The two nonhomologous halves of LEU2, integrated at chromosome III and XV, are represented as a white square and a white triangle, respectively. The HO cleavage site on chromosome XV and the HML, MAT, and HMR loci on chromosome III are indicated. (B) Mapping the chromosomal translocation breakpoints. Southern analysis was carried out with the PFGE gels for the four different classes (I to IV) of Leu+ recombinants. Ethidium bromide-stained gels and the results of Southern analysis, using LEU2 as a probe (LEU), of parental (P) and recombinant (R) strains are shown on the left of each panel. Recombinants were analyzed by Southern blotting with all other radiolabeled probes (1 to 7). Asterisks indicate nonspecific cross-hybridizations. The electrophoretic positions of unrearranged chromosomes III, VII, and XV are indicated. The new, rearranged chromosomes appearing on each gel are marked with triangles, and their estimated sizes are indicated. A schematic representation of translocations is shown at the bottom of each panel. The recombinant LEU2 gene harboring the complete yeast ACT1 intron at the translocation breakpoint is shown as a white arrowhead. Chromosomal fragments marked with boxes represent chromosomal duplications.
To confirm the genetic map of the new, translocated chromosomes, we further analyzed all breakpoints by PCR using specific primers that anneal at the different fragments of chromosomes III and XV involved in the rearrangements (see Figure S2 in the supplemental material). Intriguingly, these results suggested that whereas class I and IV recombinants are originated by a common recombination event, class II and III recombinants are generated by another common event.
Importantly, as deduced from the genetic maps of the translocated chromosomes, the rearrangements generated in class I and IV recombinants appeared to involve intrachromosomal duplications, which were characterized by the juxtaposition of the probe 2-containing region immediately downstream from the probe 4-containing region (Fig. 3). A detailed analysis of these chromosomal junctions by Southern blotting confirmed this hypothesis and demonstrated that such intrachromosomal duplications are specific to class I and IV recombinants (see Figure S3 in the supplemental material).
On the basis of the previous results, we expected that the HMR locus should be specifically necessary for the origin of recombinant classes I and IV, because the breakpoints of such recombinant classes mapped at the HMR sequence. To assay this possibility, we generated a yeast strain in which the endogenous HMR locus was deleted. The frequency of HO-induced Leu+ recombinants in hmrΔ cells decreased fivefold with respect to the wild-type levels (see Figure S4 in the supplemental material). The decrease was in agreement with the predicted lack of class I and IV recombinants, which constituted up to 80% of total recombinants in wild-type cells (see Fig. 2). As expected, PFGE analysis of the molecular karyotype of the Leu+ recombinants recovered from hmrΔ cells revealed that no class I and IV recombinants but only class II recombinants were obtained (see Figure S4 in the supplemental material). Altogether, these results fully confirmed the genetic maps established for each class of translocated chromosome.
The endogenous ACT1 intron is essential for the DSB-induced translocations.
To decipher the mechanism involved in the repair of the HO-mediated DSBs that give rise to a functional LEU2 gene, we also analyzed the breakpoints of the Leu+ recombinants by PCR using primers flanking the intron sequences. DNA sequencing of these breakpoints demonstrated that all Leu+ recombinants analyzed contained a full ACT1 intron sequence (data not shown). This result suggested that the repair of the HO-induced DSB on chromosome XV relied on the endogenous ACT1 intron of chromosome VI, which served as a bridge for a recombination reaction occurring between the two leu2 halves on chromosomes XV and III (Fig. 4A).
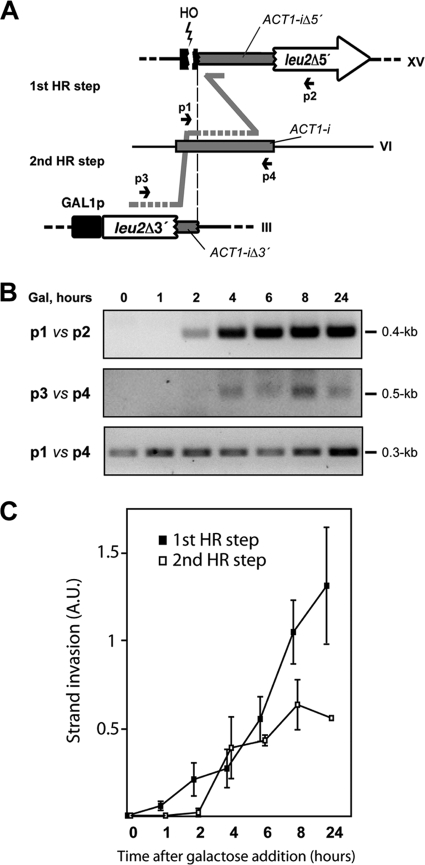
FIG. 4.
Molecular analysis of triparental recombination intermediates by PCR. (A) Schematic representation of the HR reactions analyzed. There was no sequence homology between the two DNA fragments integrated in chromosomes III and XV. The different chromosomes have been aligned according to the homology of the ACT1-i (a vertical dashed line is used as a guide). (B) PCR detection of the two consecutive HR steps leading to a full-length LEU2. Genomic DNA samples were extracted at intervals after induction of HO cleavage with galactose (Gal). For the first HR step, two specific oligonucleotides annealing at chromosome XV (p2) and chromosome VI (p1) were used. The PCR yielded a specific 0.4-kb amplimer visible 2 h after induction of HO cleavage by the addition of galactose (top panel). For the second HR step, which occurs between chromosomes VI and III, oligonucleotides p3 and p4 were used. The PCR rendered a 0.5-kb amplimer that was visible 4 h after DSB induction (middle panel). A third PCR amplification using oligonucleotides p1 and p4, done as a control, generated a 0.3-kb product (bottom panel). (C) Recombination intermediates corresponding to the two consecutive HR steps during triparental recombination were amplified as described for panel B using genomic DNA samples extracted from at least three independent kinetics assays. PCR products were quantified, and values were normalized to those of the control product generated with oligonucleotides p1 and p4. Mean values and standard deviations are shown. A.U., arbitrary units.
If this hypothesis was correct, we should detect the recombination intermediates by PCR using specific oligonucleotides annealing at the appropriate sides of the ACT1 intron on chromosomes XV and VI (Fig. 4A). As can be seen by the results in Fig. 4B, recombination intermediates covering chromosomes XV and VI were detected after 2 h of DSB induction. These recombination intermediates would include the 3′ end of the ACT1 intron, which is also present at chromosome III (see scheme in Fig. 4A). The only way this event can be completed, leading to a translocation containing a full ACT1 intron, is via a second recombination event with the 5′ half of the ACT1 intron placed on chromosome III. This second recombination step was also monitored by PCR using specific oligonucleotides annealing at chromosomes VI and III, and the strand invasion intermediate was detected after 4 h of DSB induction (Fig. 4B). Therefore, it seems that a triparental recombination event is responsible for the translocations detected. As one-ended DSBs are repaired via break-induced replication (BIR) (28), it is likely that the PCR fragments derive from BIR intermediates. Indeed, the molecular nature of the intermediates detected is consistent with the idea that triparental translocations would be occurring by BIR (52) and is in agreement with the recently reported unstable nature of the BIR intermediates formed from one-ended DSBs (54). Interestingly, after genomic DNA samples were treated with the S1 single-stranded DNA nuclease, PCR detection of the first HR step intermediates decreased dramatically (data not shown). This suggests that a pool of single-stranded DNA recombination intermediates are generated after HO cleavage in the first invasion step, consistent with a BIR reaction.
To confirm the role of the endogenous ACT1 intron as a bridge template during DNA recombination that leads to triparental translocations, we generated an identical yeast strain in which the endogenous ACT1 intron was deleted from the ACT1 locus, and we analyzed the Leu+ recombination frequency. In the absence of intron at the ACT1 locus, the frequency of Leu+ recombinants dropped by 4 orders of magnitude (Fig. 2B). This result confirmed the essential role of the endogenous ACT1 intron in the generation of the observed Leu+ recombinants and that triparental recombination via BIR plays a crucial role in repairing induced DSBs through the generation of chromosomal translocations.
The second end of the HO-induced DSB is also processed via BIR.
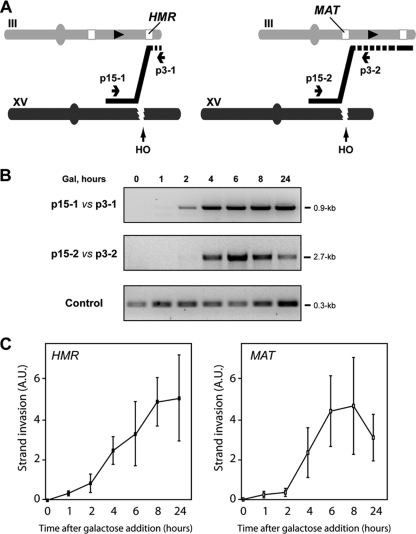
We have seen that from the two DNA ends generated by the HO break at chromosome XV, the centromere-distal end was able to promote a triparental reaction that generated a Leu+ translocation via the ACT1 intron. The question remaining is the fate of the second, centromere-proximal end of the induced DSB. Since the 117-bp HO cut site corresponds to the Y/Z sequences of both the MATa and HMR loci on chromosome III, the centromere-proximal end of the DSB generated at chromosome XV (the one that does not contain any ACT1 intron sequence) has 75 bp of homology with such Y/Z sequences of both the MATa and HMR locus (Fig. 5). Consequently, the centromere-proximal end of the HO-induced break has the potential to initiate a second one-ended BIR reaction in these loci at chromosome III, priming DNA synthesis outwards to the telomere and leading to an additional translocation. To confirm this hypothesis, we used specific oligonucleotides annealing to the appropriate regions of both chromosome XV and chromosome III around the HO sequences for PCR analyses. As shown in Fig. 5, recombination intermediates were detected at both the MAT locus and HMR locus by PCR. Strand invasion at the HMR locus that occurred after DSB induction generated a recombination intermediate that covers the 20-kb region toward the right telomere (Fig. 5A). This reaction was the same for recombinants of classes I and IV, as shown by PCR analysis of the chromosomal junctions (see Figure S2 in the supplemental material). The recombination intermediate at the MAT locus was also detected after DSB induction and would be responsible for the translocation, including 116 kb of the farthest right end of chromosome III toward the telomere (Fig. 5A). This intermediate was the same for the recombinants of classes II and III, as shown by PCR analysis of the chromosomal junctions (see Figure S2 in the supplemental material).
FIG. 5.
Repair of the centromere-proximal HO-mediated DSB by BIR with both HMR and MAT loci. (A) BIR can be initiated by the centromere-proximal induced-DSB end at the HMR/MAT locus. Strand invasion at both the HMR locus and the MAT locus, represented as a black line, was detected by PCR with oligonucleotides p15-1 and p3-1 (left) and p15-2 and p3-2 (right), respectively. The location of the GAL1-leu2Δ3′ fragment in chromosome III is represented as a black arrowhead. Both the HMR locus and the MAT locus are represented as white boxes in chromosome III. DNA synthesis is indicated with a stretch of dashed black line. (B) PCR detection of the exchange at the HMR locus gave rise to a 0.9-kb product that was visible 2 h after HO induction (top panel). PCR detection of the exchange at the MAT locus generated a 2.7-kb product that was visible 4 h after HO induction (middle panel). A third PCR amplification was done as a control, using oligonucleotides p1 and p4 (see Fig. 4), which generated a 0.3-kb product (bottom panel). In all cases, genomic DNA samples were extracted at different intervals after HO induction. Gal, galactose. (C) Quantification of strand invasion at HMR (left) and MAT (right) loci. Recombination intermediates were amplified as described for panel B using genomic DNA extracted from at least three independent kinetics assays. PCR products were quantified, and values were normalized to those for the control product generated with oligonucleotides p1 and p4. Mean values and standard deviations are shown. A.U., arbitrary units.
These results demonstrate that the centromere-proximal end of the DSB generated after HO induction on chromosome XV is also efficiently repaired by an independent recombination event, consistent with the molecular nature of the reciprocal translocations detected by PFGE and with the idea that these occur by independent BIR events.
HR functions are required for triparental translocations.
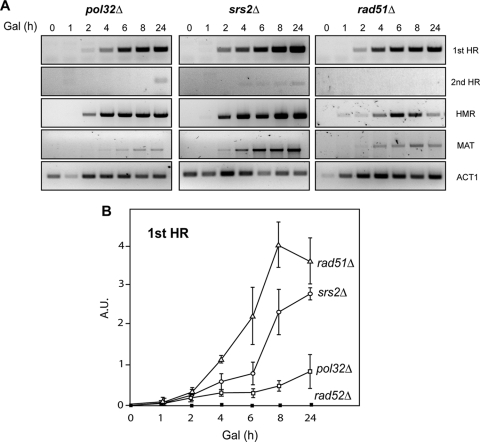
To test whether triparental translocations were dependent on HR functions, we first analyzed the effects of rad52Δ and rad51Δ mutations on HO-induced chromosomal translocations. These mutations abolish the strand annealing and reciprocal exchange steps, respectively, of HR in yeast. No Leu+ recombinants were recovered after HO-mediated DSB induction in rad52Δ mutants (see below). Furthermore, we were unable to detect by PCR the intermediate of the first recombination step in this mutant (Fig. 6B). Therefore, DSB-induced triparental translocations are absolutely dependent on Rad52. No Leu+ recombinants were recovered after DSB induction in the rad51Δ mutants, either (Fig. 7A). In this case, however, the first reaction intermediate between chromosomes XV and VI could be detected by PCR, whereas the second recombination intermediate, occurring between chromosomes VI and III, was undetectable (Fig. 6). This result implies that the first HR step could also occur in the absence of Rad51, as shown in specific BIR events between inverted repeats (5, 37, 48), whereas the second BIR reaction, between chromosomes VI and III, would occur at a much lower frequency. Indeed, PCR analysis of the HMR-MAT strand invasion steps in rad51Δ mutants revealed recombination intermediates in all cases (Fig. 6A). This result would be consistent with a low frequency of effective BIR reactions that would lead to a detectable intermediate in the first invasion event but not in the second, presumably as a consequence of the need to switch templates.
FIG. 6.
Molecular analysis of the different BIR-mediated DNA synthesis steps during triparental recombination in mutant strains. (A) PCR analysis of the different recombination intermediates in mutant strains. Genomic DNA samples from pol32Δ, srs2Δ, and rad51Δ strains were extracted at intervals after the induction of HO cleavage with galactose. PCR detection of the different recombination intermediates (corresponding to the first HR, second HR, MAT and HMR strand invasion, and DNA synthesis steps, as well as the control ACT1 amplimers) was done as described in the Fig. 4 and 5 legends. (B) PCR analysis of the first-step recombination intermediates in mutant strains. Recombination intermediates were amplified using genomic DNA samples extracted from three independent kinetics assays of pol32Δ, srs2Δ, rad51Δ, and rad52Δ strains. PCR products corresponding to the first HR step were quantified, and values were normalized to those for the control product generated with oligonucleotides p1 and p4 (see Fig. 4). Mean values and standard deviations are shown. Gal, galactose; A.U., arbitrary units.
FIG. 7.
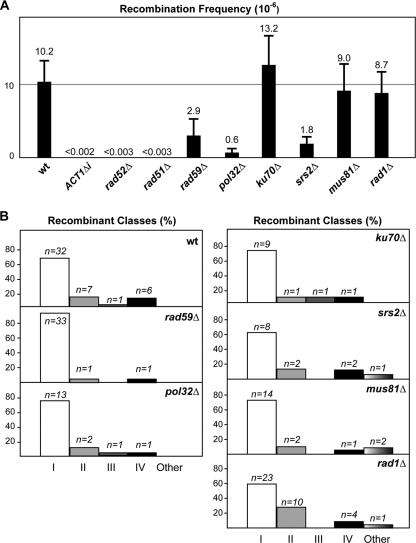
Frequencies of Leu+ recombinants and distribution of recombinant classes after HO-induced DSBs in wild-type and mutant strains. (A) Frequencies of Leu+ recombinants. (B) Distribution of the different classes of recombinants. The distribution is shown as the percentages of the total number of recombinants analyzed in a histogram. The total number of recombinant events recovered for each class (n) is shown with the corresponding histogram bar.
As Rad59, a homolog of Rad52 that also promotes strand annealing, has been shown to be critical in Rad51-independent HR, including RAD51-independent RAD52-dependent BIR between inverted repeats (5), we decided to test whether Rad59 was also required for triparental translocations. The Leu+ recombinant frequency in rad59Δ mutants decreased 3.5-fold with respect to that of wild-type cells (Fig. 7A). Moreover, the distribution of the Leu+ recombinant classes showed a bias toward class I recombinants compared to the distribution in wild-type cells (χ2 = 6.18, P < 0.05) (Fig. 7B). Therefore, viable triparental translocations depend completely on the Rad51 and Rad52 functions and, significantly, on Rad59, as expected for HR mitotic recombination. In agreement with these results, NHEJ does not participate in the triparental translocations observed, since deletion of Ku70 had no effect on either the frequency or distribution of recombinant classes (Fig. 7).
Requirement of POL32 for triparental translocations.
It has recently been reported that the nonessential subunit of replicative DNA polymerase δ, Pol32, is specifically required for BIR in yeast (36). Therefore, after confirming that HR was responsible for triparental translocations, we evaluated the dependence of the HO-induced translocations on Pol32 as a more direct way of confirming that triparental translocations occurred via BIR. As can be seen by the results in Fig. 7A, the recombination frequency in pol32Δ mutants decreased 16-fold with respect to that of the wild type. This indicates a relevant role for Pol32 in the triparental recombination events, even though they were not fully abolished since some Leu+ recombinants were recovered. Consistently, whereas the first HR invasion step was easily detected by PCR in pol32Δ mutants, the second HR step occurred at a lower efficiency in pol32Δ mutants and was detected much later (Fig. 6A). When we analyzed the molecular karyotype of the Leu+ recombinants recovered from pol32Δ cells, no differences were detected in the distribution of translocation classes with respect to their distribution in the wild-type recombinants (Fig. 7B). These results suggested that Pol32 plays an important role in triparental translocations, even though a less-efficient Pol32-independent mechanism can also repair HO-induced DSBs by triparental translocations. The results also indicate that BIR accompanied by template switching is a major source for triparental translocations.
A role for nonreplicative helicase Srs2 but not for Rad1 and Mus81 endonucleases in BIR-mediated triparental translocations.
Nonreplicative helicases play important roles in the maintenance of genome integrity (25, 51). The Srs2 helicase seems to have a complex role in HR (25). It appears that it prevents potentially lethal recombination intermediates by displacing Rad51 nucleofilaments from single-stranded DNA (29, 60) and crossovers by unwinding the invading strand from the donor DNA (16). We wondered whether Srs2 helicase contributed to the chromosomal translocations generated in our assay. As shown by the results in Fig. 7A, the frequency of DSB-induced Leu+ recombinants in srs2Δ mutants was approximately sixfold lower than for wild-type cells, the distribution of the recombinant classes being similar (Fig. 7B). However, early HR intermediates, as detected by PCR, accumulated in srs2Δ mutants at levels similar to the levels in the wild type (Fig. 6), consistent with the idea that the function of Srs2 in recombination is only relevant after strand invasion. These data suggested that Srs2 has a significant role in the outcome of the triparental recombination reaction.
Finally, two different and homologous endonuclease complexes, Rad1/Rad10 and Mus81/Mms4, have been involved in the cleavage of the different types of DNA branched substrates generated during DNA repair (6), Mus81 being a eukaryotic candidate for replication-mediated processing of cruciform structures (50, 56). To assess whether such endonucleases could have a role during triparental translocations that could imply the formation of cruciform intermediates or branched structures, we tested the effect of both rad1Δ and mus81Δ mutations on Leu+ translocations. As shown in Fig. 7A, neither rad1Δ nor mus81Δ mutants were affected in the frequency of Leu+ recombinants with respect to their frequency in the wild-type strain. These data suggest that neither endonuclease has a general role during the triparental recombination process.
DISCUSSION
Using a newly developed experimental assay for chromosomal rearrangements in S. cerevisiae, we found that a single DSB located on chromosome XV uses a short homologous sequence found in chromosome VI as a bridge to complete DSB repair, leading to translocations between chromosomes XV and III. Such translocations are dramatically reduced when the short homologous sequence on chromosome VI is deleted. Translocations rely on HR proteins, such as Rad51, Rad52, and Rad59, as well as on the BIR-specific protein Pol32 and, to a lesser extent, on the DNA helicase Srs2, but not on Ku70 and Mus81. Our results indicate that a single chromosomal DSB efficiently searches for short homologous sequences throughout the genome for its repair. This leads to triparental translocations between heterologous chromosomes via an HR mechanism that primarily involves two rounds of BIR. Therefore, in the presence of short repeated sequences, translocations generated by one DSB occur preferentially via BIR-mediated triparental recombination rather than by NHEJ. Given the abundance of repetitive DNA in eukaryotic genomes, the results of this study open the possibility that HR may be a major source of chromosomal translocations associated with genetic diseases and cancer.
Despite the absence of direct homology between the two chromosomal fragments involved in the translocations between chromosomes XV and III selected in our assay, a homologous partner in a third chromosome is used to repair the DSB via HR. The existence in chromosome VI of a 246-bp sequence homologous to the centromere-distal end of the induced DSB (the downstream half of the ACT1 intron) provides a bridge to complete a triparental recombination event. Triparental translocations require at least two rounds of strand invasion and DNA synthesis. The molecular nature of these translocation events suggests that BIR initiated in one chromosome may be unstable and suffer dissociation and template switching, as recently reported with a minichromosome assay in which only one end of a DSB was homologous with a parental chromosome (54).
As previously shown with other assays, one single DSB can lead to translocations (8, 17, 18, 20, 38, 45). However, in contrast to previous systems studied in which translocations can be generated by an efficient ectopic crossover/gene conversion mechanism (17, 19, 20), the very low frequency and the nature of the translocation events analyzed here suggests that our system detects a different type of events. Not only do they involve three different chromosomes and two intermolecular HR steps, but they occur primarily by BIR.
The frequencies of these events in our diploid cells are 1.5- to 15-fold lower than was reported for a similar assay involving one single recombination step (45). Whether this difference is due to the possibility that the two subsequent recombination steps observed in our strain might not be independent from each other, to a position effect, to the chromatin structure or transcriptional state of the donor sequence, to strain background, etc, is uncertain.
Differential role of HR genes and mechanistic insights into multiparental BIR-mediated translocations.
Both Rad52 and Rad51 are strictly required for the completion of the BIR-mediated triparental translocation events, as determined by PCR assays and genetic analysis. Nevertheless, although no intermediates were observed for either the first or second HR steps in rad52Δ mutants, only the intermediates of the second exchange reaction were not observed in rad51Δ mutants. It is unclear how a DNA strand invasion can occur in the absence of Rad51 in the first HR step in our assays. The suggested requirements for Rad51 in HR seem to depend on the chromatin context (55), on the length of the recombining sequences (24), or on topological constraints (37). It has been proposed that recombination between inverted repeats can occur by Rad51-independent BIR events (5, 12, 58). It is also possible that relaxation of the DNA surrounding the DSB site after cleavage spreads to the repeat used as a donor, facilitating the opening of the DNA strands and the invasion catalyzed by Rad52 and Rad59 in the absence of Rad51 (2). The existence of a Rad59-dependent single-strand-annealing mechanism associated with BIR in the absence of Rad51 (24, 26) would indeed be consistent with the significant reduction of translocations in rad59Δ cells in our assays.
Nevertheless, an important difference between the first HR event between chromosomes XV and VI and the second exchange between chromosomes VI and III may be that the former is originated after one induced DSB, whereas the latter would be initiated from an ongoing recombination intermediate on chromosome VI. If BIR generates unstable intermediates, as previously proposed (54), dissociation of the first HR intermediate and subsequent template switching would trigger a second HR step with chromosome III which might not be initiated by a DSB. The question raised would be whether HR initiated by template switching or by an unstable intermediate different from a DSB is more dependent on Rad51.
The lack of detection of translocations in rad51Δ cells, as well as the strong reduction of the translocations in pol32Δ mutants, indicates that Rad51-dependent BIR has, indeed, a major role in the completion of the chromosomal translocations observed here, since Pol32 has been reported to be important for BIR (36). Nevertheless, we observed that the triparental translocations are not completely abolished in pol32Δ mutants, the type of translocations generated being the same as in wild-type cells. This indicates that Pol32 only affects the efficiency rather than the outcome of the BIR event. These results suggest the existence of a Pol32-independent pathway to generate translocations, which is consistent with the detection by PCR of recombinant intermediates in the first exchange reaction in pol32Δ mutants, as well as in the second exchange reaction after longer times of HO induction. This would be in agreement with recent data suggesting that the pol32Δ mutation does not eliminate DNA synthesis from one-ended strand invasion intermediates, although its frequency is strongly reduced (53). Furthermore, the absence of Pol32 leads to unusual repair outcomes, such as half-crossovers (14, 53).
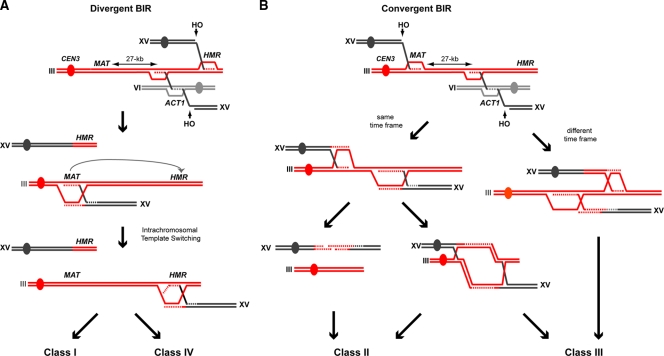
One important feature of our assays, in contrast to other, previously described BIR assays (8, 13, 54), is that both ends of the induced DSB must be repaired to guarantee the viability of the haploid cells. Whereas the centromere-distal end is repaired by BIR-mediated triparental recombination, the centromere-proximal end is repaired primarily by BIR via the HMR (class I and IV recombinants) or the MAT (class II and III recombinants) homologous sequences (Fig. 8). Since both ends of the broken chromosome need to be rescued, the frequency of Leu+ recombinants measures the repair of both ends and not just that of the end involved in template switching. Given this limitation, we cannot attribute the decrease in translocation frequencies observed in the mutants to failures in the rescue of either one or the other end of the DSB.
FIG. 8.
Model of triparental BIR-dependent translocations. After induction of HO-mediated DSBs, all Leu+ recombinants repaired the centromere-distal DSB end at chromosome XV by a triparental BIR reaction that uses the homology with the endogenous ACT1 intron at chromosome VI as a bridge template. The centromere-proximal DSB end on chromosome XV has 75 nt of homology with both HMR and MATa sequences where it can initiate a second BIR reaction required for the complete repair of the DSB. (A) DSB repair of both centromere-proximal and -distal ends by divergent BIR reactions. The HO-mediated centromere-proximal DSB end (top) can be repaired via BIR with the HMR locus, synthesizing a stretch of DNA of about 20 kb up to the right telomere of chromosome III. In order to avoid the loss of essential genes located between the two different BIR invasion sites, the triparental BIR reaction that moves toward the chromosome III centromere (CEN3) must switch the template from the MAT/Y sequence to the homologous HMR/Y sequence (gray arrow). Then, the BIR reaction can continue DNA synthesis back toward the CEN3. Depending on the number of intrachromosomal template-switching rounds, class I (one round) and IV (two rounds) recombinants could be generated. This multiple BIR reaction would be finally resolved before reaching CEN3, leading to the Leu+ chromosome III-XV translocation genetically detected in these studies. (B) DSB repair of both the centromere-proximal and -distal end by convergent BIR reactions. The HO-mediated centromere-proximal DSB end can be repaired via BIR at the MAT locus. In this case, two opposite BIR reactions would be generated, which might occur with either similar or different time frames. If each BIR reaction occurs in a different time frame (right), the reaction initiating at the MAT locus would reach the telomere while the reaction traveling toward the centromere would be finally solved as described for panel A. This scenario would generate class III recombinants. If the two opposite BIR reactions take place in the same time frame (left), both recently synthesized DNA molecules can either reanneal in a SDSA-like reaction (class II recombinants) or lead to double-Holliday junction structures to be resolved (class II and III recombinants, depending on whether the two Holliday junctions are resolved in the same orientation—that is, both horizontal or both vertical—or in different orientations—that is, one horizontal and the other vertical—respectively).
Interestingly, triparental recombination initiated at the centromere-distal DSB end occurs inwards toward the centromere of chromosome III (Fig. 8). In all recombinant classes except class II, DNA synthesis can continue after template switching, along chromosome III until it reaches the centromere of chromosome III. There, the HR event would be resolved, since the BIR reaction is not able to proceed through centromeres (43), providing an explanation for the absence of a parental nonrecombinant chromosome III in the Leu+ translocants.
In our assays, whereas the centromere-distal end is repaired by BIR-mediated triparental recombination, the centromere-proximal end is repaired primarily by BIR via the HMR (class I and IV recombinants) or the MAT (class II and III recombinants) homologous sequences (Fig. 8). Since the chromosome III region between MAT and HMR contains essential genes, cells in which the centromere XV-proximal end of the DSB invades HMR (class I and IV recombinants) need to maintain such an essential region for viability (Fig. 8A). Our data suggest that the triparental reaction occurring toward the chromosome III centromere can switch between the MAT/Y and HMR/Y template sequences, creating duplications of the internal regions (Fig. 3 and 8A) and consistent with the observation that deletion of the HMR locus completely abolished class I and IV recombinants.
Interestingly, in class II recombinants, there are two BIR-independent events, both initiated on each end of the induced DSB (Fig. 8B). In this case, two convergent BIR reactions would encounter each other and cDNA strands would be synthesized which could theoretically anneal with each other after dissociation, in a SDSA-like mechanism. In this sense, it is known that HR is favored in diploid versus haploid yeast (57). Consistent with this, class II recombinants are overrepresented in diploids with respect to their occurrence in haploids in our experimental system. On the other hand, the frequency of class II recombinants was strongly reduced in rad59Δ mutants, which suggests that, indeed, the annealing of the two recently synthesized DNA strands becomes necessary, consistent with Rad59 being absolutely required for the annealing of homologous sequences, especially when they are short (55). Related to this, it has recently been reported that Rad59 is also required for chromosomal translocations generated by simultaneous HO-mediated DSB (45).
Finally, since Srs2 helicase favors SDSA reactions (16), a reduction in the frequency of recombinants in srs2Δ mutants should be expected in our experimental assays. Consistently, the frequency of the translocations drops fivefold in srs2Δ cells versus the levels in the wild type. One possibility could be that the effect of the absence of Srs2 in the triparental recombination events could be related to the function of Srs2 in the recovery from checkpoint-mediated arrest after a DSB (59), and in agreement with this possibility, the detection of HR intermediates in srs2Δ mutants was similar to that in the wild type.
Biological relevance of BIR-mediated translocations.
Our results support a model in which BIR is the natural consequence of DNA strand invasion during any type of HR repair event when the second end of the break is not captured and does not generate a Rad51 nucleofilament that could block DNA synthesis. In the case of class II recombinants, as each asymmetrical end invades sites separated by 27 kb, DNA synthesis initiated at each end is able to go all the way until the two strands encounter each other or until they find the invading point of the other end, where the cruciform structure or the Rad51 nucleofilament would impede the progress of DNA synthesis (2). This is the first evidence that HR DSB repair can be seen as two independent events that encounter each other in the same invaded template, implying that the coordinated invasion of the two ends of the break is not necessarily required.
Finally, our results suggest that template switching events occurring during BIR (54) can be very efficient, generating multiple recombination events. It is noteworthy that template switching might have an important role in the genesis and/or development of some human genomic disorders that are characterized by the presence of genomic rearrangements, such as duplications and translocations (7, 11, 22, 31). From our data, it is tempting to speculate that BIR-mediated triparental HR events can be a powerful mechanism generating chromosomal translocations, which can be a source of tumorigenesis. In this sense, an involvement of BIR-mediated DSB repair pathways in the generation of oncogenic translocations and gene amplifications affecting the immunoglobulin H and c-myc loci in B cells has been suggested (15). The relevance of HR in the development of B cells has become more evident with the publication of several recent works (10, 40). Interestingly, a prevalent use of nonconservative HR pathways, such as BIR, in B cells has recently been suggested, implying that they may have a role in the diversification of immunoglobulin genes (40). This same type of event may cause the genesis of lymphomas by the generation of chromosomal rearrangements when short homologous sequences on different chromosomes are used. The demonstration that one single DSB in the genome can efficiently lead to chromosomal translocations between nonhomologous chromosomes via multiparental HR events provides a new and possibly major mechanism for the generation of translocations, opening new perspectives for the origin of chromosomal rearrangements in eukaryotic cells.
Supplementary Material
Acknowledgments
This work was funded by grants from the Spanish Ministry of Research and Innovation (BFU2006-05260 and CDS2007-0015) and Junta de Andalucia (CVI-624 and BIO-102). J.F.R. and B.G.-G. were the recipients of postdoctoral and predoctoral grants, respectively, from the Spanish Ministry of Science and Education.
We thank H. Klein, J. Haber, and A. Gabriel for providing yeast strains and plasmids and B. Pardo for helpful discussions and critical reading of the manuscript.
Footnotes
Published ahead of print on 3 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agarwal, S., A. A. Tafel, and R. Kanaar. 2006. DNA double-strand break repair and chromosome translocations. DNA Repair (Amsterdam) 5:1075-1081. [DOI] [PubMed] [Google Scholar]
- 2.Aguilera, A. 2001. Double-strand break repair: are Rad51/RecA-DNA joints barriers to DNA replication? Trends Genet. 17:318-321. [DOI] [PubMed] [Google Scholar]
- 3.Aguilera, A., and B. Gomez-Gonzalez. 2008. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9:204-217. [DOI] [PubMed] [Google Scholar]
- 4.Aplan, P. D. 2006. Causes of oncogenic chromosomal translocation. Trends Genet. 22:46-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai, Y., and L. S. Symington. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025-2037. [DOI] [PubMed] [Google Scholar]
- 6.Bastin-Shanower, S. A., W. M. Fricke, J. R. Mullen, and S. J. Brill. 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23:3487-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauters, M., H. Van Esch, M. J. Friez, O. Boespflug-Tanguy, M. Zenker, A. M. Vianna-Morgante, C. Rosenberg, J. Ignatius, M. Raynaud, K. Hollanders, K. Govaerts, K. Vandenreijt, F. Niel, P. Blanc, R. E. Stevenson, J. P. Fryns, P. Marynen, C. E. Schwartz, and G. Froyen. 2008. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res. 18:847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosco, G., and J. E. Haber. 1998. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150:1037-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burma, S., B. P. Chen, and D. J. Chen. 2006. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amsterdam) 5:1042-1048. [DOI] [PubMed] [Google Scholar]
- 10.Caddle, L. B., M. G. Hasham, W. H. Schott, B. J. Shirley, and K. D. Mills. 2008. Homologous recombination is necessary for normal lymphocyte development. Mol. Cell. Biol. 28:2295-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho, C. M., F. Zhang, P. Liu, A. Patel, T. Sahoo, C. A. Bacino, C. Shaw, S. Peacock, A. Pursley, Y. J. Tavyev, M. B. Ramocki, M. Nawara, E. Obersztyn, A. M. Vianna-Morgante, P. Stankiewicz, H. Y. Zoghbi, S. W. Cheung, and J. R. Lupski. 2009. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum. Mol. Genet. 18:2188-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortes-Ledesma, F., C. Tous, and A. Aguilera. 2007. Different genetic requirements for repair of replication-born double-strand breaks by sister-chromatid recombination and break-induced replication. Nucleic Acids Res. 35:6560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, A. P., and L. S. Symington. 2004. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 24:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deem, A., K. Barker, K. Vanhulle, B. Downing, A. Vayl, and A. Malkova. 2008. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179:1845-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Difilippantonio, M. J., S. Petersen, H. T. Chen, R. Johnson, M. Jasin, R. Kanaar, T. Ried, and A. Nussenzweig. 2002. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196:469-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupaigne, P., C. Le Breton, F. Fabre, S. Gangloff, E. Le Cam, and X. Veaute. 2008. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol. Cell 29:243-254. [DOI] [PubMed] [Google Scholar]
- 17.Fasullo, M., T. Bennett, P. AhChing, and J. Koudelik. 1998. The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed translocations. Mol. Cell. Biol. 18:1190-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasullo, M., T. Bennett, and P. Dave. 1999. Expression of Saccharomyces cerevisiae MATa and MAT alpha enhances the HO endonuclease-stimulation of chromosomal rearrangements directed by his3 recombinational substrates. Mutat. Res. 433:33-44. [DOI] [PubMed] [Google Scholar]
- 19.Fasullo, M., P. Giallanza, Z. Dong, C. Cera, and T. Bennett. 2001. Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics 158:959-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fasullo, M., C. St. Amour, and L. Zeng. 2005. Enhanced stimulation of chromosomal translocations and sister chromatid exchanges by either HO-induced double-strand breaks or ionizing radiation in Saccharomyces cerevisiae yku70 mutants. Mutat. Res. 578:158-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haber, J. E., G. Ira, A. Malkova, and N. Sugawara. 2004. Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday's model. Philos. Trans. R. Soc. Lond. B 359:79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastings, P. J., G. Ira, and J. R. Lupski. 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5:e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haviv-Chesner, A., Y. Kobayashi, A. Gabriel, and M. Kupiec. 2007. Capture of linear fragments at a double-strand break in yeast. Nucleic Acids Res. 35:5192-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang, L. E., and L. S. Symington. 2000. Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 20:9162-9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khanna, K. K., and S. P. Jackson. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:247-254. [DOI] [PubMed] [Google Scholar]
- 28.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 30.Krogh, B. O., and L. S. Symington. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38:233-271. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. A., C. M. Carvalho, and J. R. Lupski. 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131:1235-1247. [DOI] [PubMed] [Google Scholar]
- 32.Lee, K., Y. Zhang, and S. E. Lee. 2008. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature 454:543-546. [DOI] [PubMed] [Google Scholar]
- 33.Lemoine, F. J., N. P. Degtyareva, K. Lobachev, and T. D. Petes. 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell 120:587-598. [DOI] [PubMed] [Google Scholar]
- 34.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 35.Lieber, M. R., K. Yu, and S. C. Raghavan. 2006. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amsterdam) 5:1234-1245. [DOI] [PubMed] [Google Scholar]
- 36.Lydeard, J. R., S. Jain, M. Yamaguchi, and J. E. Haber. 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448:820-823. [DOI] [PubMed] [Google Scholar]
- 37.Malagon, F., and A. Aguilera. 2001. Yeast spt6-140 mutation, affecting chromatin and transcription, preferentially increases recombination in which Rad51p-mediated strand exchange is dispensable. Genetics 158:597-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malkova, A., M. L. Naylor, M. Yamaguchi, G. Ira, and J. E. Haber. 2005. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell. Biol. 25:933-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McEachern, M. J., and J. E. Haber. 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75:111-135. [DOI] [PubMed] [Google Scholar]
- 40.Mierau, M., G. A. Drexler, A. Kutzera, K. Braunschmidt, J. Ellwart, F. Eckardt-Schupp, E. Fritz, J. Bachl, and B. Jungnickel. 2008. Non-conservative homologous recombination in human B lymphocytes is promoted by activation-induced cytidine deaminase and transcription. Nucleic Acids Res. 36:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mills, K. D., D. O. Ferguson, and F. W. Alt. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194:77-95. [DOI] [PubMed] [Google Scholar]
- 42.Mitelman, F., B. Johansson, and F. Mertens. 2007. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 7:233-245. [DOI] [PubMed] [Google Scholar]
- 43.Morrow, D. M., C. Connelly, and P. Hieter. 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nickoloff, J. A., L. P. De Haro, J. Wray, and R. Hromas. 2008. Mechanisms of leukemia translocations. Curr. Opin. Hematol. 15:338-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pannunzio, N. R., G. M. Manthey, and A. M. Bailis. 2008. RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae. DNA Repair (Amsterdam) 7:788-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piruat, J. I., and A. Aguilera. 1998. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 17:4859-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rattray, A. J., and L. S. Symington. 1994. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics 138:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson, C., and M. Jasin. 2000. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405:697-700. [DOI] [PubMed] [Google Scholar]
- 50.Roseaulin, L., Y. Yamada, Y. Tsutsui, P. Russell, H. Iwasaki, and B. Arcangioli. 2008. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 27:1378-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt, K. H., and R. D. Kolodner. 2006. Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc. Natl. Acad. Sci. USA 103:18196-18201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt, K. H., J. Wu, and R. D. Kolodner. 2006. Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom's syndrome protein. Mol. Cell. Biol. 26:5406-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, C. E., A. F. Lam, and L. S. Symington. 2009. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase δ complex. Mol. Cell. Biol. 29:1432-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, C. E., B. Llorente, and L. S. Symington. 2007. Template switching during break-induced replication. Nature 447:102-105. [DOI] [PubMed] [Google Scholar]
- 55.Sugawara, N., E. L. Ivanov, J. Fishman-Lobell, B. L. Ray, X. Wu, and J. E. Haber. 1995. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 373:84-86. [DOI] [PubMed] [Google Scholar]
- 56.Taylor, E. R., and C. H. McGowan. 2008. Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc. Natl. Acad. Sci. USA 105:3757-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valencia-Burton, M., M. Oki, J. Johnson, T. A. Seier, R. Kamakaka, and J. E. Haber. 2006. Different mating-type-regulated genes affect the DNA repair defects of Saccharomyces RAD51, RAD52 and RAD55 mutants. Genetics 174:41-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.VanHulle, K., F. J. Lemoine, V. Narayanan, B. Downing, K. Hull, C. McCullough, M. Bellinger, K. Lobachev, T. D. Petes, and A. Malkova. 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol. Cell. Biol. 27:2601-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaze, M. B., A. Pellicioli, S. E. Lee, G. Ira, G. Liberi, A. Arbel-Eden, M. Foiani, and J. E. Haber. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10:373-385. [DOI] [PubMed] [Google Scholar]
- 60.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 61.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 62.Yu, X., and A. Gabriel. 2004. Reciprocal translocations in Saccharomyces cerevisiae formed by nonhomologous end joining. Genetics 166:741-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.