Abstract
Capnocytophaga canimorsus can be a virulent pathogen, whereas C. cynodegmi is of low virulence. Heterogeneity within these species, their frequency in dogs, and pathogenicity factors are largely unknown. Strains from blood cultures from patients presumptively identified as C. canimorsus (n = 25) and as C. cynodegmi by rrs analysis (n = 4), blood cultures from dogs (n = 8), blood cultures from cats (n = 2), and cultures from swabs from dog mouths (n = 53) were analyzed. PCR-restriction fragment length polymorphism (PCR-RFLP), a species-specific PCR on rpoB, and rrs sequencing were used. All 29 strains from human blood cultures could be grouped into three PCR-RFLP types. One included the C. canimorsus type strain, and the other types were closely related. Two canine strains were C. canimorsus and grouped into the least common RLFP pattern group. Five were C. cynodegmi and clustered with the reference strain. One canine and both feline strains were distinct. Four human strains that presumptively had been identified as C. cynodegmi by RNA gene sequence analysis clustered with the C. canimorsus strains by both PCR-RFLP and the sequence-specific PCR of the rpoB gene. C. canimorsus DNA was present in 73% (range, 61 to 85%) of dogs' mouths, and C. cynodegmi DNA was present in 96% (range, 94 to 100%) of dogs' mouths. As defined by rpoB PCR-RFLP and by PCRs using specific primers, all strains from human blood were C. canimorsus. The sequencing of rrs genes suggested the presence of different gene copies in a few strains, indicating that the method is less appropriate for species identification. Both species are present in the majority of dogs. Additional Capnocytophaga species occur in dogs' and cats' mouths.
Capnocytophaga canimorsus, originally described as DF-2, can infect humans after bites from dogs and occasionally from cats. The usual clinical presentation of infection with this bacterium is fulminant sepsis, which has a high mortality. Infection with C. canimorsus also can lead to endocarditis, meningitis, eye infections, and mycotic aneurysms. Although high-risk groups such as patients after splenectomy or with alcoholism have been defined, the infection also can occur in healthy persons (14, 19).
A second Capnocytophaga species, Capnocytophaga cynodegmi, also has been cultured from the oral flora of dogs (3). This species, originally referred to as DF-2-like, has been cultured only infrequently from patients with infections. In the initial report characterizing this species, six strains originating from wound or eye infections are reported (3). Since then, only one patient with a wound infection (9) and two cases of bacteremia (11, 20) with this species have been described. In the latter two cases, only biochemical determination was performed; one of the two strains was identified only on the basis of a positive nitrate test, which is not a common feature of this species. Therefore, C. cynodegmi appears to be an extremely rare pathogen with low virulence.
In the original publication in which both species were described, discrimination was based mainly on DNA-DNA hybridization experiments, since strains of both species had biochemical features in common (3). Although nowadays sequencing the 16S rRNA gene is a tool that is frequently used to identify bacterial species, especially those species that are difficult to identify by conventional methods, a homology of more than 97% between C. canimorsus and C. cynodegmi is within the range of one species. The rpoB gene is used frequently for differentiating species that are closely related on the basis of the 16S rRNA gene (5, 12, 13, 18) and has a homology of only 83% in both species; therefore, it is a more suitable target for their genetic characterization. An alternative molecular method to sequencing for distinguishing between and within species is PCR-restriction fragment length polymorphism (PCR-RFLP). This technique has been applied successfully to identify other Capnocytophaga species originating from the human mouth (4).
It is unclear why C. canimorsus infections are so infrequent. One possibility is that the bacterium occurs in low frequency in dogs. C. canimorsus has been described routinely as a commensal microorganism in dog mouths. This is based on three different studies, which report a frequency of this bacterium of 8% (1), 24% (23), and 25% (2). Bailie et al. (1) published long before C. cynodegmi was identified and cannot have been able to discriminate between the species. In their description of the methods to identify C. canimorsus, Blanche et al. (2) refer to a publication that also did not differentiate between C. canimorsus and C. cynodegmi. Therefore, in both studies the occurrence of C. canimorsus may have been overestimated. In addition, in neither of these two studies was a selective medium was used, and this may have resulted in an underestimation of the colonization rate of dogs by C. canimorsus. In contrast, Westwell et al. (23) used a selective medium to isolate Capnocytophaga species strains and discriminated between DF-2 and DF-2-like strains. DF-2-like strains were identified in 11% of the dogs. The authors did not report whether they also looked for double infections by screening multiple colonies from every dog. In a very recent paper, Capnocytophaga species strains, characterized as C. canimorsus by rrs sequence analysis, were cultured from 61% of 103 dogs (15). However, rrs sequence analysis might not be the most appropriate way to distinguish between C. canimorsus and C. cynodegmi.
Another explanation is a difference in virulence among C. canimorsus strains. It has been described that C. canimorsus produces a toxin affecting macrophages that is not present in C. cynodegmi (7). One possibility is that not all C. canimorsus strains produce this toxin. It also could be that strains from dogs are genetically more diverse, and that only a subpopulation of these strains is pathogenic for humans.
The aim of the present study was to characterize oxidase-positive Capnocytophaga species strains in more detail, to develop species-specific real-time PCRs for C. canimorsus and C. cynodegmi, and to assess the presence of both pathogens in the oral flora of dogs with this technique without culturing.
MATERIALS AND METHODS
Bacterial isolates.
Reference strains for C. canimorsus (ATCC 35979 T) and C. cynodegmi (ATCC 49045) were obtained from the ATCC. Reference strains for C. gingivalis (DSM 3290 T), C. sputigena (DSM 7273 T), C. haemolytica (DSM 11385 T), C. granulosa (DSM 11449 T), and C. ochracea (DSM 7271 T) were obtained from the German Collection of Microorganisms and Cell Cultures. Twenty-eight other oxidase-positive Capnocytophaga species strains cultured from the blood of human patients were kindly provided by different clinical laboratories from The Netherlands. Four of these strains (98-196, 02-195, 01-351, and 02-39) had been identified presumptively as C. cynodegmi by the partial sequencing of the rrs gene, whereas the other 24 strains had been identified as C. canimorsus by this method (Table 1). Sixteen bacterial and yeast strains from other species (Haemophilus influenzae, Staphylococcus aureus, Neisseria meningitidis, Pseudomonas aeruginosa, Salmonella enteritidis, Salmonella enterica serovar Typhimurium, Bordetella pertussis, Acinetobacter baumannii, Moraxella catarrhalis, Streptococcus bovis, Streptococcus salivarius, Chlamydia pneumoniae, Bacillus cereus, Serratia marcescens, Escherichia coli, and Candida albicans) were obtained from our collections for specificity testing.
TABLE 1.
Characteristics of Capnocytophaga species strains used in this study
| Strain | Origin | Typing bya:
|
|||||
|---|---|---|---|---|---|---|---|
|
rpoB RFLP
|
Sequence analysis (16S rRNA; nt 38-492)
|
rpoB PCR | |||||
| Pattern
|
Type | Type (according nomenclature of Fig. 2) | Accession no. | ||||
| StyI | MseI | ||||||
| ATCC 49045 | Patient, wound (reference strain) | 4 | 4 | Ccyn1 | CcynA | AY643076.1 | C. cynodegmi |
| ATCC 35979 | Patient, blood (reference strain) | 2 | 2 | Ccan2 | CcanA | AY643075.1 | C. canimorsus |
| 03-349 | Patient, blood | 1 | 1 | Ccan1 | CcanA | FJ669146 | C. canimorsus |
| 03-308 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 03-620 | Patient, blood | 1 | 1 | Ccan1 | CcanB | FJ669152 | C. canimorsus |
| 03-612 | Patient, blood | 2 | 2 | Ccan2 | CcanB | FJ669151 | C. canimorsus |
| 05-80 | Patient, blood | 3 | 3 | Ccan3 | CcanA | EU124407 | C. canimorsus |
| 05-29 | Patient, blood | 1 | 1 | Ccan1 | CcanB | FJ669153 | C. canimorsus |
| 97-16 | Patient, blood | 1 | 1 | Ccan1 | CcanA | FJ669154 | C. canimorsus |
| 601231 | Patient, blood | NA | NA | ND | ND | C. canimorsus | |
| 602032 | Patient, blood | NA | NA | ND | ND | C. canimorsus | |
| 601232 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 601233 | Patient, blood | 2 | 2 | Ccan2 | ND | ND | C. canimorsus |
| 601234 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 601251 | Patient, blood | 2 | 2 | Ccan2 | CcanC | EU124411 | C. canimorsus |
| 602031 | Patient, blood | 1 | 1 | Ccan1 | CcanB2 | EU124410 | C. canimorsus |
| 602035 | Patient, blood | 2 | 2 | Ccan2 | ND | ND | C. canimorsus |
| 602091 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 602241 | Patient, blood | 2 | 2 | Ccan2 | ND | ND | C. canimorsus |
| 603292 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 603231 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 603232 | Patient, blood | 2 | 2 | Ccan2 | ND | ND | C. canimorsus |
| 603233 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 603234 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 603235 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 604051 | Patient, blood | 1 | 1 | Ccan1 | ND | ND | C. canimorsus |
| 98-196 | Patient, blood | 1 | 1 | Ccan1 | CcynA4 | FJ669146 | C. canimorsus |
| 02-195 | Patient, blood | 1 | 1 | Ccan1 | CcynA3 | FJ669149 | C. canimorsus |
| 01-351 | Patient, blood | 1 | 1 | Ccan1 | CcynA1 | FJ669147 | C. canimorsus |
| 02-39 | Patient, blood | 1 | 1 | Ccan1 | CcynA2 | FJ669148 | C. canimorsus |
| LUMC-HC 1 | Dog mouth | 4 | 5 | Ccyn2 | CcynA7 | EU124420 | C. cynodegmi |
| LUMC-HC 2 | Dog mouth | 4 | 5 | Ccyn2 | CcynA6 | EU124413 | C. cynodegmi |
| LUMC-HC 3 | Dog mouth | 2 | 3 | Ccan4 | CcanA | EU124409 | C. canimorsus |
| LUMC-HC 8 | Dog mouth | 4 | 5 | Ccyn2 | CcynA9 | EU124418 | C. cynodegmi |
| LUMC-HCJ15 | Dog mouth | 3 | 3 | Ccan3 | CcanA | EU124408 | C. canimorsus |
| LUMC-HJB21 | Dog mouth | 4 | 5 | Ccyn2 | ND | C. cynodegmi | |
| LUMC-DogNido1 | Dog mouth | 6 | 7 | C_dog | Cnew | EU124412 | Negative |
| LUMC-DogNido3 | Dog mouth | 4 | 5 | Ccyn2 | CcynA6 | EU124414 | C. cynodegmi |
| LUMC-HA1 | Dog mouth | 4 | 4 | Ccyn1 | ND | ND | C. cynodegmi |
| LUMC-HA2 | Dog mouth | 4 | 4 | Ccyn1 | CcynA5 | EU124415 | C. cynodegmi |
| LUMC-HA3 | Dog mouth | 4 | 4 | Ccyn1 | CcynA8 | EU124419 | C. cynodegmi |
| LUMC-CatG1 | Cat mouth | 6 | 6 | C_cat | Ccyncat | EU124417 | Negative |
| LUMC-CatE l1 | Cat mouth | 6 | 6 | C_cat | Ccyncat | EU124416 | Negative |
NA, not amplifiable; ND, not done.
Additionally, Capnocytophaga species strains were cultured from swabs from the gums of 14 dogs and 2 cats. Swabs were stored at 4°C in Stuart medium up to 2 days before inoculation, with the exception of three swabs that were stored for 7 to 9 days. Swabs were inoculated onto culture plates containing 5% sheep blood agar and gentamicin (BioMérieux) for 1 week at 37°C in the presence of 5% CO2. Oxidase-positive strains that were gram-negative rods by Gram staining were further identified by API Rapid ID32 A, (BioMérieux, Lyon, France). By this approach, 11 strains were obtained from dogs and 2 from cats.
Canine samples for PCR and culture.
Fifty-three dogs attending a veterinary clinic for various reasons had their mouths swabbed with informed consent from the owners. Cotton-tip swabs were rubbed on the gums or on the tongue and suspended in phosphate-buffered saline and stored at −20°C until processed for PCR. The age and sex of 43 of these dogs were reported; their mean age was 6 years (range, 2 months to 15 years). Twenty-seven dogs (63%) were male.
Culture.
To obtain strains and to compare culture and PCR results, cultures were performed initially from specimens from eight dogs; cultures were performed on the same day on sheep blood agar plates (BioMérieux) containing gentamicin. Colonies that were oxidase and catalase positive and showed gram-negative rods on Gram stain were further analyzed by both rpoB PCRs as described below.
Since the initial yield was only four strains (three C. cynodegmi and one C. canimorsus strain), samples from five additional dogs were obtained and cultured in the same way. This resulted in an additional three C. cynodegmi strains and one strain resembling a Capnocytophaga species, although it was oxidase negative. An additional attempt to obtain more C. canimorsus strains was performed by culturing additional samples from nine dogs on sheep blood agar plates with gentamicin and performing the C. canimorsus PCR as described below on the mixture of colonies that appeared on the plates. These samples had been stored for 2 to 9 days in Stuart transport medium. A positive signal was obtained from only three culture plates; after subculture, a C. canimorsus strain was grown from only one dog, which later appeared to be the same dog as the one from which we had cultured a C. canimorsus strain earlier.
DNA extraction.
DNA was extracted from all strains and all canine specimens with the use of the QIAamp DNA mini kit (Qiagen, Hilden, Germany). For comparison, the MagNa Pure LC instrument also was used to extract DNA from the first 38 canine specimens. The efficiency of the extraction of nucleic acids from capnocytophagae by these two methods was comparable (data not shown).
Development of PCR-RFLP on rpoB for characterization of strains.
For PCR-RFLP, the chosen rpoB primers were based on sequences published in the NCBI database: for C. canimorsus CIP103936 T and C. cynodegmi CIP103937 T, the accession numbers were AY643069 and AY643070, respectively. The primers 5′-CAGTTTATGGATCAAACCAATCC-3′ and 5′-CCATCTCTCCRAAACGCTGAC-3 were expected to amplify a 2,190-bp fragment. Sequences were chosen using Beacon Designer software (Premier Biosoft International, Palo Alto, CA). PCR was performed for 40 cycles at an annealing temperature of 58°C. Ten microliters of the amplicon subsequently was digested with StyI (10 U/μl) for 3 h or MseI (5 U/μl) for 1 h. Restriction fragments were analyzed on an standard 1.5% agarose gel or a 4.0% NuSieve GTG agarose (Invitrogen) gel, respectively.
Development of species-specific rpoB PCRs.
Two different species-specific PCRs for C. canimorsus and C. cynodegmi were designed based on the above-mentioned NCBI rpoB gene sequences, with specific primers for C. canimorsus (5′-TTTCAGCTTCATTAATTCCTTTCC-3′ and 5′-GCCTGACGCATCATATTCG-3′) and C. cynodegmi (5′-GAATTTCGGCTTCATTGATTCC-3′ and 5′-CGCATCATATTTGACCCCATC-3′). The 6-carboxyfluorescein-490-labeled probes were 5′-CGATGATGCGAACCGTGCGTTGAC-3′ for C. canimorsus and 5′-CTTGGAACACGATGATGCGAACCG-3′ for C. cynodegmi. PCR amplification was performed in 50 cycles with an annealing temperature of 62°C in the presence of 3.5 mM MgCl2 for the PCR for C. cynodegmi and 4.5 mM MgCl2 for the PCR for C. canimorsus. In previous experiments, a range of MgCl2 concentrations was used to optimize sensitivity and specificity for both PCRs, and these MgCl2 concentrations resulted in optimal sensitivity and specificity for both species.
Nucleotide sequencing of the 16S rRNA of cultured strains.
The partial nucleotide sequencing of nucleotide (nt) 38 to 492 of the 16S rRNA was performed by PCR amplification using primers 5′-TAACACATGCAAGTCGAGGG-3′ and 5′-ATCTCCGTATTACGGCGGC-3′ and subsequent sequence analysis on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Inc.). The entire rrs gene of a number of strains, including those strains previously identified as C. cynodegmi strains from patients, was sequenced using the forward primers 16S8F (5′-AGA GTT TGA TCM TGG YTC AG-3′), 16S339F (5′-CTC CTA CGG GAG GCA GCA G-3′), 16S518F (5′-CCA GCA GCC GCG GTA ATA-3′), 16S785F (5′-GGA TTA GAT ACC CTG GTA G-3′), and 16S912F (5′-CAA AKGA ATT GAC GGG GGC-3′). The reverse primers were 16S575R (5′-CTT TAC GCC CAR TRA WTC CG-3′), 16S802R (5′-CTA CCA GGG TAT CTA ATC C-3′), 16S930R (5′-GCC CCC GTC AAT TCM TTT G-3′), 16S1115R (5′-AGG GTT GCG CTC GT RCG-3′), and 16S1512R (5′-ACG GHT ACC TTG TTA CGA CTT-3′).
RESULTS
Characterization of oxidase-positive Capnocytophaga species strains by PCR-RFLP.
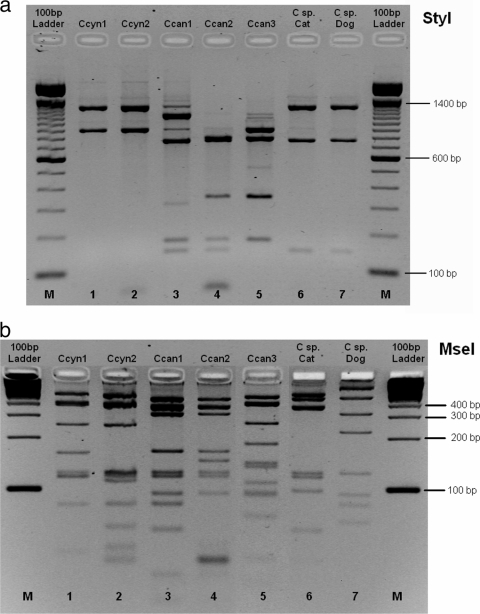
The amplification of the rpoB gene was successful for 40/43 Capnocytophaga species strains included in the study (Table 1), as well as for both feline isolates (LUMC-CatG1 and LUMC-CatE1) and the oxidase-negative strain (LUMC-DogNido1). The reference strains from C. ochracea, C. haemolytica, C. sputigena, C. granulosa, and C. gingivalis gave no amplification product with the primers for the rpoB gene. Subsequent digestion with the restriction enzymes StyI and MseI resulted in different RFLP types, as shown in Fig. 1. For StyI digestion, one of the patterns was in agreement with the pattern predicted by the in silico digestion of the amplification product of strain 24231, described as C. canimorsus in GenBank (accession no. AY643071), and consisted of fragments of 1,127, 756, 169, and 136 bp (StyI type Ccan1) (Fig. 1a, lane 4). The second pattern of strain ATCC 35979 (Fig. 1a, lane 5) consisted of fragments of 788, 756, 339, 169, and 136 bp (StyI type Ccan2) and was in accordance with the sequence of this strain (AY643069). We also found a third pattern in which, compared to the sequence of type 2, one StyI restriction site apparently was not present, resulting in pattern 3 (892, 788, 339, and 169 bp; StyI type Ccan3) (Fig. 1a, lane 6). We also could distinguish three different MseI patterns, of which patterns Ccan1 and Ccan2 corresponded to predicted patterns based on the in silico digestion of the published sequences, and the third pattern showed a number of differences (Fig. 1b).
FIG. 1.
StyI and MseI restriction patterns of the PCR amplification product of the rpoB gene (see Materials and Methods). DNA fragments were separated on a 1.5% standard agarose gel after StyI digestion (a) or on a 4% NuSieve agarose gel after MseI digestion (b). Lanes M, 100-bp marker; lanes 1, C. cynodegmi ATCC 49045; lanes 2, C. cynodegmi LUMC-Ccyn01; lanes 3, C. canimorsus 601232; lanes 4, C. canimorsus ATCC 35979; lanes 5, C. canimorsus LUMC-HCJ15; lanes 6, Capnocytophaga sp. strain LUMC-CatG1; lanes 7, Capnocytophaga sp. strain LUMC-DogNido1.
Fifteen human C. canimorsus strains fell into type 1 (Table 2). Interestingly, the four strains 98-196, 02-195, 01-351, and 02-39, presumptively identified as C. cynodegmi based on the partial sequencing of the rrs gene, also clustered in this PCR-RFLP group. Eight C. canimorsus isolates belonged to type 2. Only two C. canimorsus isolates from patients, as well as one of the dog isolates, belonged to type 3. The second dog isolate had StyI pattern type 2 and MseI pattern type 3 and is classified as type 4 in Table 2.
TABLE 2.
Distribution of human and canine Capnocytophaga strains and types as defined by StyI and MseI digestion patterns
| Species | Type | Origin | n | Digestion pattern (no.)
|
|
|---|---|---|---|---|---|
| StyI | MseI | ||||
| C. canimorsus | 1 | Patienta | 19 | 1 | 1 |
| 2 | Patient | 8 | 2 | 2 | |
| 3 | Patient | 1 | 3 | 3 | |
| 3 | Dog | 1 | 3 | 3 | |
| C. canimorsus | 4 | Dog | 1 | 2 | 3 |
| C. cynodegmi | 1 | Patientb | 1 | 4 | 4 |
| 1 | Dog | 3 | 4 | 4 | |
| C. cynodegmi | 2 | Dog | 5 | 4 | 5 |
| Putative new species | 1 | Cat | 2 | 6 | 6 |
| Putative new species | 2 | Dog | 1 | 6 | 7 |
Including the strains 98-196, 02-195, 01-351, and 02-19, which were identified presumptively as C. cynodegmi earlier (see the text).
Strain ATCC 49045.
C. cynodegmi strains could be grouped in two different types, differing only in their MseI digestion patterns: type 4, which fit the pattern of strain ATCC 49045 (Fig. 1, lanes 1) and in accordance with the prediction on the basis of sequence AY643070 (strain CIP103937 T = ATCC 49044 T), and type 5, showing one or several additional restriction sites in a 157-bp fragment (Fig. 1, lanes 2). Both types showed the same StyI pattern as that predicted (1,296 and 892 bp). The reference strain, which is a strain from a wound of a patient after a dog bite, clustered with one of these types. The two cat Capnocytophaga species strains showed identical PCR-RFLP patterns that differed from the sequences of the other strains. Compared to the sequence of C. cynodegmi, one additional StyI restriction site is present, resulting in fragments of 1,296, 756, and 136 bp (type 6). The strain Nido1, belonging to a newly discovered canine Capnocytophaga species, had an StyI digestion pattern that was identical to that of C. cynodegmi, but it showed a difference after MseI digestion (Table 2) .
rrs gene analysis of Capnocytophaga species strains.
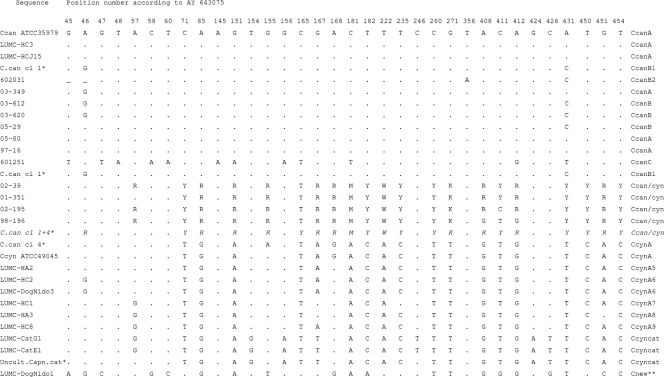
The partial sequencing of the rrs gene (nt 38 to 492) was performed on nine C. canimorsus strains cultured from human patients, including strains for each C. canimorsus type as described above, on the ambiguous four isolates 98-196, 02-195, 01-351, and 02-39 and on all 10 cultured Capnocytophaga species strains from dogs' and cats' mouths. Algorithms for similarity and phylogenetic trees treat ambiguous nucleotides as different features, resulting in different clusters in cases of differences between multiple copies, therefore the nucleotides that differ between strains are shown in Fig. 2. The sequence of the reference strain was designated type CcanA. Eight out of nine strains from patients as well as both strains typed as C. canimorsus from dogs had, at most, two nucleotide substitutions compared to the sequence of the reference strain (types B and B2). One strain showed 11 substitutions and was designated CcanC. The reference C. cynodegmi strain ATCC 49045 showed 20 nucleotide substitutions compared to the sequence of the C. canimorsus reference strain. The sequences of the six canine C. cynodegmi strains were highly related to that of the reference strain, showing only two to five nucleotide substitutions. Four strains from patients (98-196, 02-195, 01-351, and 02-39) showed a mixture of 2 nt at 21 positions (Fig. 2). At these positions, both a nucleotide corresponding to the reference sequence of C. canimorsus and a nucleotide corresponding to the reference sequence of C. cynodegmi were found. Interestingly, two sequences from GenBank, described as C. canimorsus 24231 clone 1 and clone 4, have been published, of which clone 1 is almost identical to the C. canimorsus reference sequence, and clone 4 is almost identical to the C. cynodegmi reference sequence. A mixture of these two sequences would result in almost the same sequence as the one we found. Since the sequence analysis of the 16S rRNA gene includes all copies of this gene on the chromosome, base pair differences in the various copies may result in ambiguous nucleotides in the consensus sequence. The two feline strains also were related to the C. cynodegmi sequence, but they showed four additional nucleotide differences and their sequences were almost identical to sequence AF426105, which originated from an uncultivable strain from a cat with sinusitis and rhinitis (8). These strains probably represent a new Capnocytophaga species, of which the pathogenicity is unknown. Finally, one strain, DogNido1, from a dog, had more than 40 nucleotide differences and was more similar to C. haemolytica than to other species. It should be noted that this strain was oxidase negative and probably represents a newly identified commensal canine Capnocytophaga species.
FIG. 2.
rrs sequence comparison (nt 38 to 492) of reference sequences of C. canimorsus (ATCC 35979 T, AY643075), C. cynodegmi (ATCC49044 T, AY643076), and a number of Capnocytophaga species strains used in the present study. Nucleotides differing from the reference sequence are shown. A dot indicates identity to the sequence of C. canimorsus, and a dash indicates a deletion. Sequences AY643077 (C. canimorsus 24231 clone 1), AY643078 (C. canimorsus 24231 clone 4), and AF426105 (an uncultivable Capnocytophaga species strain from cat) (marked with asterisks) are shown for comparison. Strain LUMC-DogNido1 (marked with a double asterisk) has 27 additional nucleotide changes from the sequence of the reference strain ATCC 35979, which are not shown in the figure. Nucleotide positions are according to the sequence of accession number AY643075.
In addition, the complete sequence analysis of the rrs gene was performed for six human C. canimorsus strains as well as for the ambiguous strains 01-351, 02-39, 02-195, and 98-196 (Fig. 3). Homology for this region of the gene was even greater than the homology in the 5′ region of the gene. There was only one major difference in nt 1075 to 1112, in which all sequenced strains except strain 01-351 differed from the reference sequences of C. canimorsus and C. cynodegmi, which are identical in this region. The nine strains had 12 mutations and 2 deletions compared to the reference sequences. The same mutations were found in clones 1 and 4 of the above-mentioned C. canimorsus strain 24231 (AY643077 and AY643078, respectively). Nine of the 10 ambiguous nucleotides in this part of the sequence of the remaining strain 01-351 are a consensus of strain 24231 and of the type strains of C. canimorsus and C. cynodegmi.
FIG. 3.
rrs sequence comparison (nt 493 to 1350) of selected Capnocytophaga species strains. Only nucleotides differing from the reference sequence are shown. For further information, see the legend to Fig. 2.
We used the Ribosomal Database Project II-release10 tools Seqmatch and Tree Builder (http://rdp.cme.msu.edu/) to generate a phylogenetic tree (see the supplemental material). Whereas C. cynodegmi and C. canimorsus grouped into two different branches, a number of strains grouped between those branches as an independent group that could not be assigned to either species, although by rpoB RFLP and specific real-time PCR these strains clearly were C. canimorsus.
Development of specific real-time PCRs for C. cynodegmi and C. canimorsus.
Real-time PCRs to detect C. canimorsus and C. cynodegmi were developed using the rpoB gene as the target gene. Amplification efficiency was optimal at an MgCl2 concentration of 4.5 mM for the C. canimorsus PCR and 3.5 mM for the C. cynodegmi PCR. Optimal specificity was reached at 62°C (data not shown). At this temperature, 13 fg C. cynodegmi DNA still was detectable, whereas 70 pg C. canimorsus DNA gave no signal at all. The C. canimorsus PCR still was positive at an input of 7 fg. At an input of 130 pg, C. cynodegmi DNA resulted in a very weakly positive signal in the PCR, but a smaller amount gave no signal.
DNA of C. ochracea, C. sputigena, C. gingivalis, C. haemolytica, and C. granulosa, as well as DNA from 16 different bacterial and yeast species (see Materials and Methods), gave no reaction in PCRs for both C. cynodegmi and C. canimorsus.
We tested DNA from 25 C. canimorsus isolates from patients, from the 2 C. canimorsus strains from dogs, and from the 4 strains 98-196, 02-39, 02-195, and 01-351. All 31 strains gave a positive reaction in the C. canimorsus PCR only. Eight C. cynodegmi strains cultured from dogs all reacted in the C. cynodegmi PCR. The two strains from cats and the strain Nido1 did not react in either PCR.
Frequency of C. canimorsus and C. cynodegmi in dog mouths as assessed by real-time PCR.
C. canimorsus DNA was detected in 39 out of 53 dogs (73%; 95% confidence interval [CI], 61 to 85%) in either a gum or a tongue swab, whereas C. cynodegmi DNA was detected in 51 out of 53 dogs (96%; 95% CI, 94 to 100%) (Table 3) . Negative controls containing buffer only, which were tested regularly among the canine samples, reacted uniformly negatively in both PCRs.
TABLE 3.
Occurrence of C. canimorsus and C. cynodegmi DNA in dogs, as assessed by species-specific PCRa
| Specimen type |
C. canimorsus
|
C. cynodegmi
|
||||
|---|---|---|---|---|---|---|
| + | +/− | − | + | +/− | − | |
| Tongue | 38 | 1 | 14 | 49 | 4 | |
| Gums | 34 | 2 | 17 | 49 | 4 | |
| Any | 39 | 2 | 12 | 51 | 2 | |
+, positive; +/−, weakly positive; −, negative.
Specimens from the tongue were slightly more frequently positive for C. canimorsus DNA. Quantitative results showed no differences between average cycle threshold (CT) values of positive tongue and gum samples or between average CT values in the PCRs for C. canimorsus and C. cynodegmi. Both dogs that showed no detectable C. cynodegmi DNA also showed no detectable C. canimorsus DNA. No relationship between C. canimorsus positivity and breed, age, gender, or clinical status could be detected. For instance, most of the infection-negative dogs were young (1 year or younger), but some very young dogs also appeared to be positive. On the other hand, some of the older dogs appeared to be negative in the PCR. Also, the condition of the gums was not related to the presence or absence of C. canimorsus as detected by PCR.
C. canimorsus was grown from only 1 (5%) dog of the 21 dogs from which samples were cultured. C. cynodegmi was cultured from 6/14 dogs that were screened for the presence of this species. In the corresponding samples, DNA from C. canimorsus and C. cynodegmi, respectively, also was detectable.
DISCUSSION
Whereas biochemical methods still are useful to identify rapidly growing strains in the clinical microbiology laboratory, molecular methods are becoming more common to identify slowly growing strains. In addition, molecular methods are of use in identifying the strains in the case of inconclusive phenotypical data. We used rrs gene sequencing, the amplification of a specific target in the rpoB gene, and the PCR-RFLP of the rpoB gene as tools to characterize oxidase-positive Capnocytophaga species strains of human, canine, and feline origin.
The C. canimorsus strains from patients used in our study already had been characterized by rrs gene analysis and were found to be highly related to the type strain ATCC 35979. The rrs genes of the type strains of C. canimorsus and C. cynodegmi differ in only 19 nucleotides, resulting in a homology of 98.6%.
The four strains with heterogeneous sequences in the first 500 nt of the rrs genes had the same genetic distance to both C. cynodegmi and C. canimorsus. Ambiguous nucleotides in 16S rRNA gene analysis can be explained by the existence of multiple different gene copies in a strain. The rrs sequences of clones 1 and 4 from strain 24231 (AY643077 and AY643078) support this, since the consensus sequence of both clones results in the same ambiguous nucleotides as those of the four patient strains (Fig. 2). Sequencing the entire gene introduced more copy variation. The ambiguous nucleotides of strain 03-351 (Fig. 3) are in favor of the multicopy theory.
The sequences of the single-copy genes rpoB (AY643071) and gyrA (AY643074) of strain 24231 have a similarity of 98 to 99% to the C. canimorsus type strain. Therefore, we conclude that the rpoB gene is a better tool than the analysis of the rrs gene to distinguish between C. canimorsus and C. cynodegmi. Using the rpoB-based techniques, we conclude that the strains 01-351, 02-39, 02-195, and 98-196 belong to the species C. canimorsus.
C. canimorsus is a serious pathogen causing sepsis, meningitis, and occasionally other infections, such as keratitis and endocarditis, in healthy and immunocompromised patients. Infection usually is associated with contact with dogs. A few cases of infections have been described after contact with cats or without any animal contact. Whereas dog bites are very common, infections with C. canimorsus are rare. One explanation is that the prevalence of C. canimorsus in dogs is low. Our PCR study, however, showed that this is not the case, and that 74% of routinely tested dogs had C. canimorsus DNA in their mouths. C. cynodegmi DNA was found even more frequently, being present in 96% of the dogs. The higher frequency of C. cynodegmi is in contrast with the results of Westwell et al., who found C. canimorsus in 24% and C. cynodegmi in 11% of dogs after the culturing and biochemical identification of isolates. For culturing, we used sheep-blood agar plates containing gentamicin, whereas Westwell et al. used horse blood agar plates supplemented with cysteine, kanamycin, and vancomycin. It is possible that this accounts for the discrepancy. We were much less successful with the culture of C. canimorsus: we could culture a strain from only 2 out of 22 dog samples, and it turned out that both of these samples originated from the same dog, so our culture rate was only 1/21 (5%). We looked only for C. cynodegmi strains in the first 14 samples from different dogs and cultured this species from six dogs (43%). Our combined sensitivity for both species therefore can be calculated as 7/14 (50%). Our finding that C. cynodegmi was more frequently cultured from dogs than C. canimorsus is in agreement with our PCR findings. A much higher sensitivity for PCR compared to that of culture also has been shown for other slowly growing microorganisms, such as Bordetella pertussis (6). Apparently, many strains do not grow so well on the routinely used culture media. Unfortunately, a gold standard is lacking. Using our specific real-time PCRs, a panel of strains, including all so-far-described Capnocytophaga species, were not reactive, and two newly described Capnocytophaga species described in the present paper did not react in our PCRs. Therefore, we are confident that specific C. canimorsus and C. cynodegmi DNA was amplified from the samples. Also, the frequency of other difficult-to-culture pathogens in dogs, such as Mycoplasma (10), Ehrlichia, and Babesia (16), also have been approximated by PCR.
In a recent paper, the presence of C. canimorsus in dogs by culture was estimated at 60% (15). However, these authors used rrs gene sequencing and subsequent cluster analysis to discriminate between C. canimorsus and C. cynodegmi. The analysis of their 16S rRNA sequences (GQ167551 to GQ167622) shows that the 22 strains of a C. canimorsus subcluster had a maximal homology of only 95% with the type strains of C. canimorsus and C. cynodegmi. As indicated above, the identity of the other C. cynodegmi and C. canimorsus strains should be confirmed by other methods. The authors suggested that they could differentiate C. canimorsus and C. cynodegmi on colony morphology; this is not supported by the original colony description (3) and is not our experience. Mally et al. (15) found that the combined yield for both species, after correction for the subcluster, is 41%, which is comparable to our combined yield of 50%.
Whereas we could confirm that C. canimorsus is present in the mouths of most dogs, we could not confirm that these dogs harbored the same genotype. With PCR-RFLP, we could distinguish three different PCR-RFLP types. Although our two dog strains were in the least common group, more dog strains need to be cultured and typed to see whether all C. canimorsus-positive dogs can infect patients with this pathogen. A toxin produced by some strains could be a virulence factor (7); however, we could not confirm its presence (data not shown), and in a recent publication a toxin was not found (21). Recently, some potential virulence mechanisms for C. canimorsus have been revealed. It was shown that human macrophages incubated with C. canimorsus failed to produce tumor necrosis factor alpha, interleukin-1α (IL-1α), IL-6, Il-8, and gamma interferon; that TLR-4 could not be activated by the reference strain; and that even tumor necrosis factor alpha release by another pathogen, Yersinia enterocolitica, was impaired by live C. canimorsus (21). The resistance to phagocytosis of C. canimorsus by macrophages and the blocking of the ability of macrophages to kill other bacteria also were demonstrated by the same group of investigators (17), as well as its resistance to killing by complement and polymorphonuclear leukocytes (22). However, C. canimorsus sepsis is characterized by extensive signs of inflammation, and alternative virulence mechanisms should be present. It would be highly interesting to look at these features in a number of genetically well-characterized C. canimorsus strains obtained from dogs.
In addition to C. canimorsus and C. cynodegmi, we also obtained evidence for the existence of other, so-far-uncultured oxidase-positive Capnocytophaga spp. in dogs' and cats' mouths. The virulence potential of such species still is unknown. A case report describing a cat with sinusitis and rhinitis (8) showed the presence of DNA in a supernatant on a cell culture vial that was identical to that of strains LUMC-CatG1 and LUMC-CatE1; the authors, however, were not able to culture the strain. It is not clear whether this strain was a pathogen. The catalase-negative, oxidase-positive strain LUMC-DogNido1 is a novel type for which pathogenic properties still are unknown.
The rarity of C. canimorsus infections in humans in spite of their abundance in dog mouths as well as the almost complete absence of infections with other canine Capnocytophaga species remain unaccounted for. The further analysis of strains and patients should reveal specific host and bacterial factors contributing to severe sepsis in selected cases.
Supplementary Material
Acknowledgments
We thank P. Van Keulen (Amphia Hospital, Breda, The Netherlands), A. Van Griethuijsen (Rijnstate Hospital, Arnhem, The Netherlands), D. Potters (Viecurie Hospital, Venlo, The Netherlands), R. Schouten (Gelderse Vallei Hospital, Ede, The Netherlands), J. Weel (Laboratory for Public Health Friesland, Leeuwarden, The Netherlands), E. Mooi (Hospital Groene Hart, Gouda, The Netherlands), and F. Heilman (Gelre Hospital, Apeldoorn, The Netherlands) for providing us with strains.
We thank Maaike van de Beld and Wim Sandman for their technical assistance.
Footnotes
Published ahead of print on 29 July 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bailie, W. E., E. C. Stowe, and A. M. Schmitt. 1978. Aerobic bacterial flora of oral and nasal fluids of canines with reference to bacteria associated with bites. J. Clin. Microbiol. 7:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanche, P., E. Bloch, and D. Sicard. 1998. Capnocytophaga canimorsus in the oral flora of dogs and cats. J. Infect. 36:134. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, D. J., D. G. Hollis, G. R. Fanning, and R. E. Weaver. 1989. Capnocytophaga canimorsus sp. nov. (formerly CDC group DF-2), a cause of septicemia following dog bite, and C. cynodegmi sp. nov., a cause of localized wound infection following dog bite. J. Clin. Microbiol. 27:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciantar, M., H. N. Newman, M. Wilson, and D. A. Spratt. 2005. Molecular identification of Capnocytophaga spp. via 16S rRNA PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 43:1894-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahllöf, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dragsted, D. M., B. Dohn, J. Madsen, and J. S. Jensen. 2004. Comparison of culture and PCR for detection of Bordetella pertussis and Bordetella parapertussis under routine laboratory conditions. J. Med. Microbiol. 53:749-754. [DOI] [PubMed] [Google Scholar]
- 7.Fischer, L. J., R. S. Weyant, E. H. White, and F. D. Quinn. 1995. Intracellular multiplication and toxic destruction of cultured macrophages by Capnocytophaga canimorsus. Infect. Immun. 63:3484-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey, E., B. Pressler, J. Guy, C. Pitulle, and E. Breitschwerdt. 2003. Capnocytophaga sp. isolated from a cat with chronic sinusitis and rhinitis. J. Clin. Microbiol. 41:5321-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerster, J. C., and J. Dudler. 2004. Cellulitis caused by Capnocytophaga cynodegmi associated with etanercept treatment in a patient with rheumatoid arthritis. Clin. Rheumatol. 23:570-571. [DOI] [PubMed] [Google Scholar]
- 10.Kenny, M. J., S. E. Shaw, F. Beugnet, and S. Tasker. 2004. Demonstration of two distinct hemotropic mycoplasmas in French dogs. J. Clin. Microbiol. 42:5397-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khawari, A. A., J. W. Myers, D. A. Ferguson, Jr., and J. P. Moorman. 2005. Sepsis and meningitis due to Capnocytophaga cynodegmi after splenectomy. Clin. Infect. Dis. 40:1709-1710. [DOI] [PubMed] [Google Scholar]
- 12.Kim, B. J., S. K. Hong, K. H. Lee, Y. J. Yun, E. C. Kim, Y. G. Park, G. H. Bai, and Y. H. Kook. 2004. Differential identification of Mycobacterium tuberculosis complex and nontuberculous mycobacteria by duplex PCR assay using the RNA polymerase gene (rpoB). J. Clin. Microbiol. 42:1308-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko, K. S., T. Kuwahara, L. Haehwa, Y. J. Yoon, B. J. Kim, K. H. Lee, Y. Ohnishi, and Y. H. Kook. 2007. RNA polymerase beta-subunit gene (rpoB) sequence analysis for the identification of Bacteroides spp. Clin. Microbiol. Infect. 13:48-54. [DOI] [PubMed] [Google Scholar]
- 14.Lion, C., F. Escande, and J. C. Burdin. 1996. Capnocytophaga canimorsus infections in human: review of the literature and cases report. Eur. J. Epidemiol. 12:521-533. [DOI] [PubMed] [Google Scholar]
- 15.Mally, M., C. Paroz, H. Shin, S. Meyer, L. V. Soussoula, U. Schmiediger, C. Saillen-Paroz, and G. R. Cornelis. 2009. Prevalence of Capnocytophaga canimorsus in dogs and occurrence of potential virulence factors. Microbes Infect. 11:509-514. [DOI] [PubMed] [Google Scholar]
- 16.Matjila, P. T., A. L. Leisewitz, F. Jongejan, and B. L. Penzhorn. 2008. Molecular detection of tick-borne protozoal and ehrlichial infections in domestic dogs in South Africa. Vet. Parasitol. 155:152-157. [DOI] [PubMed] [Google Scholar]
- 17.Meyer, S., H. Shin, and G. R. Cornelis. 2008. Capnocytophaga canimorsus resists phagocytosis by macrophages and blocks the ability of macrophages to kill other bacteria. Immunobiology 213:805-814. [DOI] [PubMed] [Google Scholar]
- 18.Mollet, C., M. Drancourt, and D. Raoult. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005-1011. [DOI] [PubMed] [Google Scholar]
- 19.Pers, C., B. Gahrn-Hansen, and W. Frederiksen. 1996. Capnocytophaga canimorsus septicemia in Denmark, 1982-1995: review of 39 cases. Clin. Infect. Dis. 23:71-75. [DOI] [PubMed] [Google Scholar]
- 20.Sarma, P. S., and S. Mohanty. 2001. Capnocytophaga cynodegmi cellulitis, bacteremia, and pneumonitis in a diabetic man. J. Clin. Microbiol. 39:2028-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin, H., M. Mally, M. Kuhn, C. Paroz, and G. R. Cornelis. 2007. Escape from immune surveillance by Capnocytophaga canimorsus. J. Infect. Dis. 195:375-386. [DOI] [PubMed] [Google Scholar]
- 22.Shin, H., M. Mally, S. Meyer, C. Fiechter, C. Paroz, U. Zaehringer, and G. R. Cornelis. 2009. Resistance of Capnocytophaga canimorsus to killing by human complement and polymorphonuclear leukocytes. Infect. Immun. 77:2262-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westwell, A. J., K. Kerr, M. B. Spencer, and D. N. Hutchinson. 1989. DF-2 infection. BMJ 298:116-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.