Abstract
Whole-genome sequencing of human adenovirus type 11 (HAdV-11) strain QS, isolated in China, was conducted, and its sequence was compared with the sequences of strains within the species of HAdVs. The HAdV-11 QS genome contains 34,755 nucleotides. Similar to the other HAdV subgenus B sequences, the HAdV-11 QS genome coded 37 functional proteins and could be divided into four early, two intermediate, and five late transcription regions. The amino acid sequences of the fiber and the hypervariable regions (HVRs) within the hexon gene of HAdV-11 QS were identical to the corresponding sequences of the HAdV-11a strain; further analyses that compared those amino acid sequences with the amino acid sequences of the HAdV species subgenus B:2 strains revealed that the highest degree of homology (>99.2%) existed between HAdV-11 QS and the prototypical HAdV-14 strain, except for a few coding sequences of HVRs within the hexon gene, DNA polymerase, pVI, and pre-terminal protein. This indicate that HAdV-11 strain QS, isolated in China, is a recombinant adenovirus of HAdV-14, and the recombination analyses also confirmed this finding. It is difficult to clarify the time and manner of the recombination, and further investigations are required to determine whether the emergence of recombination between HAdV-11a and HAdV-14 might increase virulence, thereby posing a new global challenge with regard to acute respiratory diseases in the near future.
Adenoviruses (AdVs) have thus far been classified into five genera, namely, Atadenovirus, Aviadenoviridae, Mastadenoviridae, and Siadenoviridae (2) and unclassified genus Adenoviridae. Human AdVs (HAdVs) belonging to the Mastadenoviridae genus are composed of seven subgenera (subgenera A to G), including 52 serotypes, on the basis of their hemagglutination, oncogenicity in rodents, restriction fragment analyses, tropism, genome homology (3, 13), and serum neutralization ability. Generally, AdV infections are subclinical; however, AdVs are associated with a broad spectrum of illnesses, including conjunctivitis, a febrile upper respiratory tract illness, pneumonia, and gastrointestinal diseases (34). HAdV type 11 (HAdV-11), which belongs to subgenus B:2, can further be divided into at least two genomic types, designated HAdV-11p and HAdV-11a, on the basis of the similarities in the fragment comigration patterns during restriction genome typing (20). HAdV-11p is generally the causative agent of kidney and urinary tract infections, while HAdV-11a is associated with respiratory tract infections. HAdV-14, another component of subgenus B:2, has been recognized as a pathogen that causes acute respiratory illnesses (24, 34), and these subgenus B:2 viruses have generally been recognized to be mild or rare pathogens. Until recently, HAdV-11a has been found in China, Spain, Turkey, and Latin America (7, 14, 20), while diseases associated with HAdV-14 have also been reported from many other regions, such as Taiwan (6) and the United States (24). However, inconsistent with the findings of previous reports, AdVs appear to have been exhibiting a tendency to cause more severe clinical outcomes, including death (21, 24).
In 2006, an outbreak of HAdV-associated diseases occurred in Qishan County of Shaanxi Province, China. One of the patients died of acute respiratory disease-induced multiple-organ failure during the outbreak. On the basis of the epidemiological data and the results of multiple laboratory assays (36), HAdV-11a was identified as the pathogen responsible for that outbreak. Interestingly, we found that there was the possibility of intraspecies recombination between HAdV-11a and HAdV-14 on the basis of whole-hexon-gene sequence analysis (36). Our proposition differed from the findings in previous reports that HAdV-11 is very closely related to serotypes 34 and 35 (20, 31). In order to gain a better understanding of this phenomenon, we determined and analyzed the whole-genome sequence of HAdV-11 strain QS isolated from that outbreak.
MATERIALS AND METHODS
Virus and preparation of viral DNA.
The viral strain designated HAdV-11 QS in this study was isolated from hydrothorax fluid samples from the patient who died during the AdV outbreak in Qishan County, Shaanxi Province, China, in 2006. The virus was cultured and propagated in a HEp-2 cell line, which was maintained in minimal essential medium supplemented with 2% fetal bovine serum. Viral DNA was extracted from the infected cultures by using a QIAamp DNA mini kit (Qiagen, Valencia, CA), according to the manufacturer's instructions. The DNA was solubilized with an elution buffer and was stored at −70°C until further use.
PCR strategy and methodology.
Appropriate primers were designed according to the HAdV sequences available from GenBank. For genome sequencing, a standard PCR with 61 primer pairs was conducted to amplify the overlapping fragments encompassing the entire genome of HAdV-11 QS, except its 5′ and 3′ termini. For the terminal genome sequences, a method similar to that which uses a 5′/3′ rapid amplification of cDNA ends kit was followed. First, the covalent junction between the purified DNA template and the terminal protein (TP) was broken by the addition of 0.4 N NaOH. The mixture was then incubated at room temperature for 1 h and further purified by chromatography. Subsequently, in the presence of T4 DNA ligase (Promega), the blunt-ended DNA templates without TP were linked by using a DNA double-strand linker containing two synthesized single strands (5′-CGGTCGTGAGTGCTTATAG-3′ and 5′-PCTATAAGCACTCACCGTC-3′). Lastly, heminested PCR was used for amplification of the terminal DNA sequences to obtain the sequencing templates. In addition to the common primer (5′-CGGTCGTGAGTGCTTATAG-3′), external primers 5′-CAGTCCACGGAACTCAAAT-3′ and 5′-CTGCACCATTCCCAGTA-3′ and internal primers 5′-TCTTCTCGCTGGCACTCA-3′ and 5′-AGCCATGGCTTACCAGAC-3′ were used to amplify the 5′- and 3′-terminal ends, respectively.
DNA sequencing and analyses.
The amplified fragments were purified by agarose gel electrophoresis by using a QIAquick gel extraction kit (Qiagen, KK, Japan). The sequencing reactions were performed bidirectionally with the appropriate primers and cycle sequencing kits (ABI Prism BigDye Terminator, version 3.1; PE Applied Biosystems) and were then resolved with a model 3100 genetic analyzer (Applied Biosystems).
The DNA sequences were assembled with the Sequencher program (version 4.0.5; Gene Codes Corporation). We divided the whole AdV genome into 1-kb-long nonoverlapping segments by the method of Lauer et al. (19) and systematically queried the sequence against the sequences in the nonredundant NCBI database using the BLASTX program. We searched the database using the default parameters with the BLOSUM62 matrix and gap penalties of 11 (existence) and 1 (extension). The DNA and protein sequence alignments were created by using BioEdit sequence alignment editor software (version 5.0.9; Tom Hall, North Carolina State University) and an online gene-wise program (http://www.ebi.ac.uk/Wise2/advanced.html). Phylogenetic tree analyses and constructions were conducted by using the Mega program (version 4.0; Sudhir Kumar, Arizona State University). The splice position and functional genome were predicted by using an online splice program (http://www.fruitfly.org/seq_tools/splice. html) and the online GEN SCAN program (http://genes.mit.edu/GENSCAN.html). Recombination analyses among HAdV strains 11p,14, 34, 35, and QS were performed with SimPlot software (Johns Hopkins University School of Medicine, Baltimore, MD).
Nucleotide sequence accession numbers.
The nucleotide sequence of the whole genome of HAdV strain QS, which was determined in this study, has been deposited in the GenBank nucleotide sequence database under accession number FJ643676.
RESULTS
The whole genome of HAdV strain QS comprised 34,755 nucleotides (nt); and the plus strand had a base composition of 26.1% A, 24.4% C, 24.4% G, and 25.1% T. Similar to the sequences of other HAdV subgenus B strains, the HAdV-11 QS genome also coded 37 functional proteins, which could be divided into four early, two intermediate, and five late transcription products. The detailed genome organization, putative splice sites, and polyadenylation signals are described in Table 1.
TABLE 1.
Genome organization of HAdV-11 QSa
| Name of region | Nucleotide locality | Gene product | No. of amino acid residues | Transcription start site | ATG site | Stop site | Splice donor site | Splice acceptor site | Poly(A) signal |
|---|---|---|---|---|---|---|---|---|---|
| ITR | 1-137 | ||||||||
| EIA | 495-1505 | 6.9 kDa | 58 | 495 | 587 | 1355 | 658 | 1251 | 1505 |
| 25.1 kDa | 231 | 495 | 587 | 1459 | 1073 | 1251 | 1505 | ||
| 28.8 kDa | 262 | 495 | 587 | 1459 | 1166 | 1251 | 1505 | ||
| E1B | 1562∼3927 | 20.8 kDa | 180 | 1562 | 1616 | 2158 | N | N | 3927 |
| 54.7 kDa | 494 | 1562 | 1921 | 3405 | N | N | 3927 | ||
| pIX | 14.2 kDa | 139 | ND | 3488 | 3907 | N | N | 3927 | |
| E2A | 23376c-21784c | DBP | 518 | 26844c | 23376c | 21820c | ND | ND | 21784c |
| E2B | 10360c-3975c | pTP | 638 | 10360c | 8444c | ND | ND | 3975c | |
| DNA polymerase | 1122 | 8441c | 5073c | ND | ND | 3975c | |||
| Iva2 | 50.9 kDa | 448 | ND | 5594c | 3970c | 5582c | 5303c | 3975c | |
| E3 | 26841-30583 | 12.2 kDa | 105 | 26841 | 27159 | 27476 | 26869 | 26909 | 30583 |
| 14.3 kDa | 131 | 26841 | 27430 | 27825 | 26869 | 27354 | 30583 | ||
| 18.5 kDa | 166 | 26841 | 27810 | 28310 | 26869 | 27785 | 30583 | ||
| 20.3 kDa | 181 | 26841 | 28330 | 28875 | 26869 | 28328 | 30583 | ||
| 20.2 kDa | 183 | 26841 | 28893 | 29444 | 26869 | 28890 | 30583 | ||
| 10.3 kDa | 91 | 26841 | 29488 | 29763 | 26869 | 29471 | 30583 | ||
| 15.2 kDa | 134 | 26841 | 29768 | 30172 | 26869 | 29678 | 30583 | ||
| 15.3 kDa | 135 | 26841 | 30165 | 30572 | 26869 | 30142 | 30583 | ||
| E4 | 34455c-31777c | ORF6 | 34454c | 32935c | 32036c | 34381c | 33105c | 31776c | |
| ORF4 | 34454c | 33206c | 32838c | 34381c | 33300c | 31776c | |||
| ORF3 | 34454c | 33568c | 33215c | 34381c | 33597c | 31776c | |||
| ORF2 | 34454c | 33954c | 33565c | 34381c | 33972c | 31776c | |||
| ORF1 | 34454c | 34376c | 33999c | N | N | 31776c | |||
| L1 | 10652-13607 | 43.9 kDa | 386 | 5894 | 10652 | 11812 | 9586 | 10651 | 13607 |
| pIIIa | 587 | 5894 | 11838 | 13601 | 9586 | 11837 | 13607 | ||
| L2 | 13682-17317 | Penton | 557 | 5894 | 13682 | 15355 | 9586 | 15058 | 17317 |
| pVII | 192 | 5894 | 15360 | 15938 | 9586 | 15212 | 17317 | ||
| pV | 351 | 5894 | 15981 | 17036 | 9586 | 15972 | 17317 | ||
| pX | 47 | 5894 | 17152 | 17295 | 9586 | 16759 | 17317 | ||
| L3 | 17377-21766 | pVI | 246 | 5894 | 17377 | 18117 | 9586 | 17062 | 21766 |
| Hexon | 946 | 5894 | 18233 | 21073 | 9586 | 17976 | 21766 | ||
| 23K protein | 209 | 5894 | 21113 | 21742 | 9586 | 20521 | 21766 | ||
| L4 | 23407-27465 | 100K protein | 812 | 5894 | 23407 | 25845 | 9586 | 22677 | 27465 |
| 21.6 kDa | 191 | 5894 | 25577 | 26152 | 9586 | 25575 | 27465 | ||
| 14.6 kDa | 130 | 5894 | 26034 | 26426 | 9586 | 25575 | 27465 | ||
| pVIII | 227 | 5894 | 26476 | 27159 | 9586 | 26326 | 27465 | ||
| L5 | 30755-31755 | Fiber | 325 | 5894 | 30775 | 31752 | 9586 | 30774 | 31755 |
| ITR | 34755c-34619c |
N, no predicted splicing site; ND, data not determined. The beginning of each region was defined by the location of the TATA box or the ATG of the first ORF, while the end of each region was defined by the first poly(A) signal nucleotide.
Genome structure analyses of HAdV-11 QS.
The inverted terminal repeats (ITRs) were 137 bp in length and existed at the two ends of the HAdV-11 strain QS genome, and their function was probably associated with the initiation of DNA replication by a strand displacement mechanism (4). The extreme terminal sequence of the ITRs (CATCATCAAT) was relatively conserved and was the same motif as that found in subgenera B (HAdV-11p, HAdV-14, HAdV-34, and HAdV-35), C (HAdV-1 and HAdV-5), D (HAdV-17, HAdV-26, and HAdV-46), E (HAdV-4), and F (HAdV-40). Between bases 9 and 18 of HAdV-11 QS there was another conserved motif (ATAATATACC) which was present in almost all HAdVs and which was reported to be directly involved in interactions with a complex of pre-TP (pTP) and DNA polymerase within the origin of DNA replication (32).
Early gene regions.
It is recognized that the ends of the HAdV genome, including the E1 and E4 regions, are transcribed first, followed by transcription of the delayed early units (IX, Iva2, and E2 late) and the major late transcriptional units. The early transcription units of the HAdV genomes contained five components (E1A, E1B, E2, E3, and E4) (4). Compared with the sequence of HAdV-11p (GenBank accession number AY163756), the core transcription promoters of the early transcription units (E1A, E1B, E2, E3, and E4) of HAdV-11 QS were predicted to be the TATTTATA (nt 495 to 502), TATATA (nt 1562 to 1567), TATATTAT (nt 26844c to 26837c, where c indicates the complementary strand), TATAAAAA (nt 26841 to 26847), and TATATATA (nt 34454c to 34447c) motifs, respectively. On the basis of the differences in the polyadenylation signals, the E1 transcription unit could further be differentiated into two parts, namely, E1A and E1B. The proteins encoded by E1A probably have an important role in transcriptional activation and stimulating the host cell into the S phase, while those of E1B might inhibit the apoptosis mediated by p53 (4). Similarly, the E2 region could be also divided into E2A and E2B, which are probably crucial for AdV DNA replication (4) and which are located in the complementary strand. In all, three nonstructural proteins were encoded by in the E2 region: DNA-binding protein (DBP), pTP, and DNA polymerase. In addition to the TATA box, there was an SP1 transcription factor-binding site (GGGCGG; nt 23505c to 23500c) that encoded the DBP. The DBP amino acid sequence alignments among HAdV subgenera A to G showed that the DBP of HAdV-11 strain QS contained four conserved regions (CR1 to CR4) and two zinc-binding sites, one of which was HGCNDYEGKLKCLH and the other of which was four discontinuous cysteines. The E3 open reading frames (ORFs) of HAdV-11 QS encoded a total of eight predicted proteins (12.2, 14.3, 18.5, 20.3, 20.2, 10.3, 15.2, and 15.3 kDa) that were probably involved in evasion from the host immune response but that were not indispensable for viral growth and propagation (27). The E3 region is therefore a potential insertion site for gene therapy. In addition, a 9-kDa protein that was present only in subgenus B:1 (31) had not been found within the E3 region of HAdV-11 QS, and on the basis of that finding, HAdV-11 QS could further be confirmed to belong to subgenus B:2. The E4 region encoded five predicted proteins (14.2, 14.3, 13.6, 14.2, and 34.6 kDa) that probably possess a range of functions, such as the modulation of DNA replication, mRNA transcription, transport, translation, and cell apoptosis (4). Each of the splice acceptor and donor sites in the early transcription genes were determined by using Splice Site Finder software (available online at http://www.fruitfly.org/seq_tools/splice.html) and are described in detail in Table 1.
Intermediate gene regions.
Although IX and IVa2 were apparently included in the E1B and E2B gene regions, they could be classified as intermediate genes according to the different transcription stages to which they belong (23, 26, 31). The IX region (nt 3488 to 3907) encoded a 14.2-kDa protein (pIX), and the IVa2 region (nt 5594c to 3970c) encoded a 50.9-kDa protein (pIVa2). Inconsistent with the findings for the early genes, there was no predicted TATA box for pIX and pIVa2 in HAdV-11 strain QS. In addition to the common role of pIX and pIVa2 in enhancing the activity of the major late promoter (MLP), pIX functioned as one of the minor capsid proteins and pIVa2 mediated serotype-specific binding with the genome packaging signal (4, 35). The amino acid sequence alignments of pIVa2 of HAdV-2, HAdV-3, HAdV-5, and HAdV-7 with pIVa2 of HAdV-11 QS revealed that pIVa2 of HAdV-11 QS had maximum similarity with pIVa2 of HAdV-3 (93.5%), followed by that with pIVa2 of HAdV-7 (92.6%), HAdV-2 (79.9%), and HAdV-5 (79.9%). This finding therefore suggests that the IVa2 region is also a potential target site for the reconstruction of chimeric AdV vectors.
Late gene regions.
The late genes of HAdV-11 strain QS shared a common transcription promoter, namely, MLP (nt 5894 to 5900) (4), but utilized multiple poly(A) signals, owing to which five subunits (subunits L1 to L5) could be recognized. In addition, there were some other conserved control elements in HAdV-11 QS, such as an inverted CAAT box (nt 5849c to 5845c), an upstream stimulatory factor-binding site (nt 5865 to 5873), an initiator (INR) element (nt 5923 to 5934), the DE1 region (nt 6010 to 6020), and the DE2 region (nt 6025 to 6042). By examining the alignments of the MLPs of HAdV-3, HAdV-5, and HAdV-14 and the MLP of HAdV-11 QS, we observed that there was only one variation among these important control elements, i.e., a variation in the sixth base of the INR element, which indicated the importance of MLP in transcription regulation and control. Following the MLP, the tripartite leader (TPL) could be found in all late RNA species from L1 to L5. Three regions of strain QS, i.e., the first (nt 5925 to 5965), second (nt 6984 to 7056), and third (nt 9499 to 9585) leader sequences, constituted the TPL.
The late genes of HAdV-11 strain QS encoded a total of 14 predicted proteins, and these probably play an important role in ensuring the structural integrity of the virion (4). The L1 region had two predicted proteins, i.e., a 43.9-kDa protein and pIIIa, which shared a common poly(A) signal at nt 13607. The former was the analog of the L1 protein with a molecular weight of 52,000/55,000 (L1-52/55K protein), which is involved in virion assembly and in the DNA encapsidation process (10, 11). pIIIa, which extended from the exterior to the interior surface of the capsid, presumably performs a rivet-like function to stabilize the interfaces between the two facets adjacent to the capsid (28).
Within the L2 region, four predicted ORFs encoded pIII (62.5 kDa), pVII (21.3kDa), pV (40.1kDa), and pX (5.2kDa); and their common poly(A) signal was situated at nt 17317. pIII, also termed the penton base protein, was one of the major capsid proteins and measured 557 residues in length; it was presumed to mediate virus internalization by interacting with the ανβ3 and ανβ5 integrins through the RGD motif or with the α4β1 integrin through the LDV motif (18, 33). Multiple pIII amino acid sequence alignments among subgenera A to G showed that HAdV-11 QS had a penton base, i.e., a fiber-interacting domain (ESRLSNLLGIRKK) (5), and had the closest similarity with subgroup B, particularly serotype 14 (99.4%), followed by subgroups E (81.9%), D (75.6%), G (71.9%), A (71.6%), F (71.2%), and C (67.6%), in that order. The L2-encoded proteins (pVII, pV, and pX) together constituted the virion core and were in contact with the viral DNA through arginine-rich regions (4). The putative proteins pVII, pV, and pX were 192, 351, and 47 residues in length, respectively. pVII probably performs a histone-like function to condense the viral DNA (4); pV functions as a bridge between pVI and pIII to stabilize the virion (4); pX, also known as mu, is associated with AdV DNA condensation and charge neutralization (16).
Three L3-encoded proteins, namely, pVI (26.6 kDa), hexon (106.9 kDa), and the 23K protease (23.7 kDa), shared a common poly(A) signal at nt 21766. pVI was 246 residues in length, and it putatively transports hexon molecules into the nucleus and participates in the disruption of the endosomal membrane during infection with the virus (4, 15). The hexon, also known as pII, was 946 residues in length and is the major structural component of the capsid. In all, seven hypervariable regions (HVRs) of the hexon were regarded as type-specific epitopes of the AdV (29). On the basis of this finding, we confirmed that HAdV-11 QS belonged to HAdV-11a because it shared maximum homology with another strain of HAdV-11a (GenBank accession number AY972815) (36). The 23K protein analog of HAdV-11 QS was 209 residues in length and is presumably required for the cleavage of the viral protein precursors during virus maturation and assembly (4).
Four predicted proteins (91, 21.6, 14.6, and 25 kDa) identified in the L4 region used a common putative poly(A) signal at nt 27465. The 91-kDa protein (the analog of L4-100K) might play a role in the hexon trimer assembly and may be necessary for the efficient initiation of late viral protein synthesis (4, 12). Compared to the other B subgenera, the L4 region in this B subgenus contained two partially overlapping ORFs which encoded two proteins (21.6 and 14.6 kDa) whose functions remained to be clarified. The predicted pVIII (25 kDa) was 227 residues in length and interacted with three other proteins (pIIIa, pVI, and pIX) to stabilize the virion capsid (4).
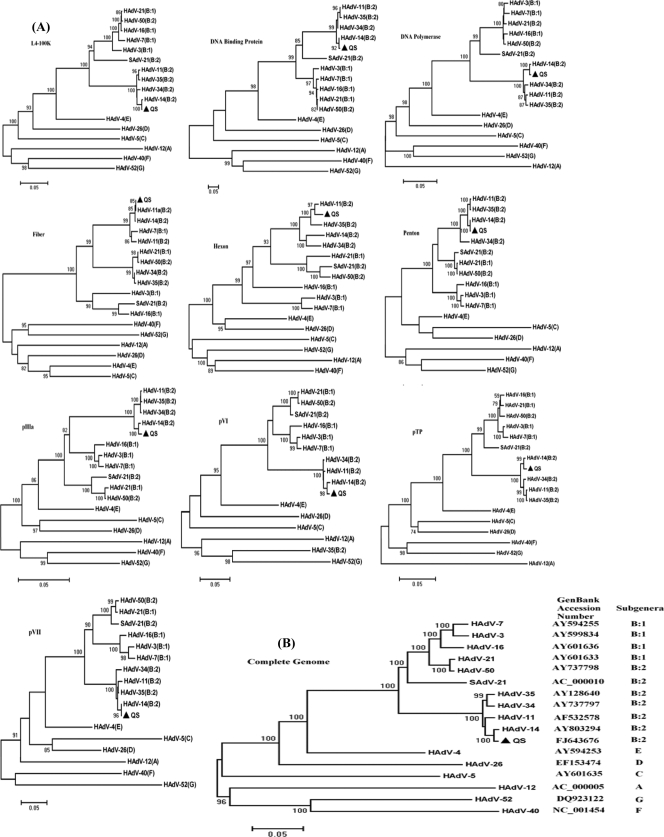
The L5 region (nt 30775 to 31755) encoded a 35.3-kDa homolog (325 residues in length) of the fiber protein whose poly(A) signal was located at nt 31755. The fiber could structurally be divided into three parts, including an N-terminal tail, a central shaft with repeated motifs, and a C-terminal globular knob (4, 9). Generally, the knob is responsible for binding to the cellular receptors, such as the coxsackievirus-AdV receptor. However, it has been reported that the HAdV-11 knob may also interact with the membrane cofactor protein, also known as CD 46 (30), which enables HAdV-11 to be an alternative vector for gene therapy in place of HAdV-2 or HAdV-5. The N-terminal end of the fiber of HAdV-11 strain QS contains a hydrophobic motif (FNPVYPY) which has been reported to mediate the interaction between the penton base and the fiber through hydrogen bonds and salt bridges (37). Comparison of the amino acid sequence of the fiber with the amino acid sequences of the fibers of other B subgenera revealed that the variation was mainly present in the regions of the shaft and the knob. Further phylogenetic analyses suggested that the amino acid sequence of the subgenus B fiber could be classified into a large cluster and that HAdV-11 QS had a closer relationship with HAdV-14 (99%), HAdV-11p (92.3%), and HAdV-7 (90.7%). Comparisons of the nucleotide sequences of the fiber region also yielded a similar result. It was notable that the fiber of HAdV-11 QS showed maximum homology with that of another available HAdV-11a strain (GenBank accession number L08232) but exhibited greater dissimilarities with the prototype of HAdV-11 than with the prototype of HAdV-14 (Fig. 1A). Although simian adenovirus (SAdV) is a different species, the fiber of SAdV type 21 (SAdV-21) showed 55.4% amino acid sequence homology with that of HAdV-11 QS, which conspicuously exceeded the homology with other HAdV subgenera (homologies of subgenera A, C, D, E, G, and F, 15.2%, 16.6%, 24.1%, 22.3%, 21.3%, and 19.5%, respectively). At the same time, comparisons of the corresponding nucleotide sequences also revealed the similar relationships of the various subgenera and SAdV-21, such that they could be classified into subgroups: one subgroup consisting of the whole B subgenera and SAdV-21; one subgroup consisting of subgenera F and G; and one subgroup consisting of subgenera A, C, D and E, respectively (Fig. 1A).
FIG. 1.
(A) Phylogenetic analyses of a few main functional genes of HAdV-11 strain QS (L4-100K, DBP, DNA polymerase, fiber, hexon, penton, pIIIa, pVI, pTP, and pVII). The corresponding subgenus types are indicated within parentheses after the respective serotypes. The GenBank accession numbers of the available fiber of HAdV-11a is L08232. (B) Phylogenetic analysis of the whole-genome sequence of HAdV. The GenBank accession numbers of the following serotypes are given in parentheses: HAdV-3 (AY599834), HAdV-4 (AY594253), HAdV-5 (AY601635), HAdV-7 (AY594255), HAdV-11 (AF532578), HAdV-12 (AC_000005), HAdV-14 (AY803294), HAdV-16 (AY601636), HAdV-21 (AY601633), HAdV-26 (EF153474), HAdV-34 (AY737797), HAdV-35 (AY128640), HAdV-40 (NC_001454), HAdV-50 (AY737798), HAdV-52 (DQ923122), and SAd-21(AC_000010). A, HAdV-A; B:1, HAdV-B1; B:2, HAdV-B2; C, HAdV-C; D, HAdV-D; E, HAdV-E; F, HAdV-F; G, HAdV-G. The relative phylogenetic distances were measured from the lowest scales of each phylogenetic tree (0.05).
Whole-genome phylogenetic analyses among HAdV subgenera A to G revealed that HAdV-11 strain QS could be clustered into a subgroup along with HAdV-11, HAdV-14, HAdV-34, and HAdV-35; and there was maximum homology between HAdV-11 QS and HAdV-14 (Fig. 1B).
DISCUSSION
Recombination not only is a well-known feature of AdV genetics but also is an important driving force for virus evolution. Generally, AdV recombination occurs much more frequently within a species than among species (22, 34) and has been suggested to occur in a few local regions, such as the fiber gene and nonstructural regions (22, 31). AdVs have been reported to be continuously detectable in the tonsils or other adenoid tissues after acute infection (1, 8, 34), which enables the formation of new recombinants during coinfection with different AdV serotypes (22). In this study, phylogenetic analyses of the coding regions of the nonstructural proteins (L4-100K, DBP, and DNA polymerase), major capsid proteins (fiber, hexon, and penton base protein), minor capsid proteins (pIIIa and pVI), core proteins (pTP and pVII), and even the complete genome of the prototype strains of subgenera A to G revealed that, except for the hexon and fiber genes, HAdV-11 strain QS had the highest degree of homology with the prototype HAdV-14 strain (strain de Wit, GenBank accession number AY803294) (Fig. 1). The corresponding proteins also showed a similar trend, including even up to 100% similarity at local amino acid sequences, such as L1-52/55K, pVII, pV, and pIVa2 (Table 2); however, at the hexon gene, there was an apparent difference between HAdV-11 QS and HAdV-14. With regard to the fiber gene, although HAdV-11 QS shared maximum homology with HAdV-11a (GenBank accession number L08232), comparisons among the prototypes showed that HAdV-11 QS had a closer phylogenetic relationship with the prototype HAdV-14 strain than with the prototype HAdV-11 strain (Fig. 1A). This finding is different from the finding presented previously that HAdV-11 is very closely related to serotypes 34 and 35, which was determined on the basis of the differences in restriction fragment migration patterns (20). This phenomenon indicates that recombination probably occurred between HAdV-11 QS (HAdV-11a) and HAdV-14. One reasonable explanation was that on the one hand, the recombinant HAdV-11 QS acquired the elements essential for virus replication and proliferation from HAdV-14 by means of recombination; on the other hand, it may have retained the key neutralizing antigen epitope (hexon) to escape the immune attack against HAdV-14. HAdV-14 has been reported to cause respiratory illnesses more frequently than HAdV-11a (6, 24). On the basis of that finding, the corresponding rate of seropositivity for HAdV-14 was suggested to be higher than that for HAdV-11a in the whole population. As a result, HAdV-11 QS not only could possess the virulence of HAdV-14 but also could avoid the neutralizing antibody against HAdV-14, which exists much more widely in the population than that against HAdV-11a.
TABLE 2.
Comparisons of amino acid sequences of HAdV-11 QS and strains of subgenus B:2a
| Functional protein | % Similarity
|
|||||
|---|---|---|---|---|---|---|
| HAdV-11a (RKI/BC34) | HAdV-11p (Slobitski) | HAdV-34 (Compton) | HAdV-35 (Holden) | HAdV-14 (de Wit) | SAdV-21 (UN) | |
| Penton | — | 98.3 | 95.3 | 98.5 | 99.4 | 91.3 |
| Hexon | — | 98.4 | 90.8 | 94.2 | 91.1 | 89.9 |
| HVR | 100.0 | 96.8 | 81.4 | 88.0 | 82.2 | 81.9 |
| Fiber | 100.0 | 92.3 | 62.3 | 62.0 | 99.0 | 55.4 |
| pIIIa | — | 99.3 | 99.3 | 99.4 | 99.8 | 91.8 |
| pIX | — | 97.1 | 97.1 | 97.8 | 99.2 | 89.9 |
| pVI | — | 99.1 | 99.1 | — | 98.3 | 87.2 |
| pVIII | — | 99.1 | 99.1 | 99.1 | 99.5 | 95.1 |
| pVII | — | 98.4 | 98.4 | 98.9 | 100.0 | 92.7 |
| pV | — | 98.5 | 98.5 | 98.5 | 100.0 | 85.5 |
| pTP | — | 98.7 | 96.0 | 98.7 | 96.6 | 92.5 |
| pIVa2 | — | 98.4 | 97.9 | 97.9 | 100.0 | 93.0 |
| 100K | — | 98.7 | 99.0 | 98.8 | 99.8 | 85.5 |
| L1-52/55K | — | 98.9 | 99.7 | 99.2 | 100.0 | 95.6 |
| DBP | — | 99.2 | 99.4 | 62.9 | 99.8 | 88.9 |
| DNA polymerase | — | 98.9 | 93.1 | 99.1 | 93.7 | 87.5 |
The name of each prototype strain is included within parentheses after its respective serotype; UN, unknown; HAdV-35 has no available pVI; HAdV-26 has no available pIVa2. The highest similarity value in each row is in boldface. —, no available information. The GenBank accession numbers of the corresponding coding nucleotides are AF532578 for HAdV-11p, AY737797 for HAdV-34, AY128640 for HAdV-35, AY803294 for HAdV-14, and AC_000010 for SAdV-21. The HVR of AdV-11a belonged to strain RKI-2797/04 (GenBank accession number AY16371); the fiber protein of AdV-11a belonged to strain BC34 (GenBank accession number AAA42491).
The statement presented above could also be partly supported by the following clinical evidence. HAdV-11 is a recognized pathogen in urinary tract infections, while HAdV-14 is associated with pharyngoconjunctival fever and acute respiratory disease (34). The recombinant (HAdV-11 QS) showed the ability to cause respiratory tract diseases and not kidney and urinary tract infections, although HAdV-11a also shared a high degree of homology with the prototype HAdV-11 strain (Table 2). In addition, it is worth noting that HAdV-14, which also belongs to subgenus B:2, caused several outbreaks with severe clinical consequences (21, 24). At the same time, HAdV-11a, which has rarely caused severe respiratory system diseases, has been found all around the world in recent years, such as in Turkey, China, and Latin America (7, 14, 36). This coincidence also indicates the probability of recombination from the point of view of an epidemic.
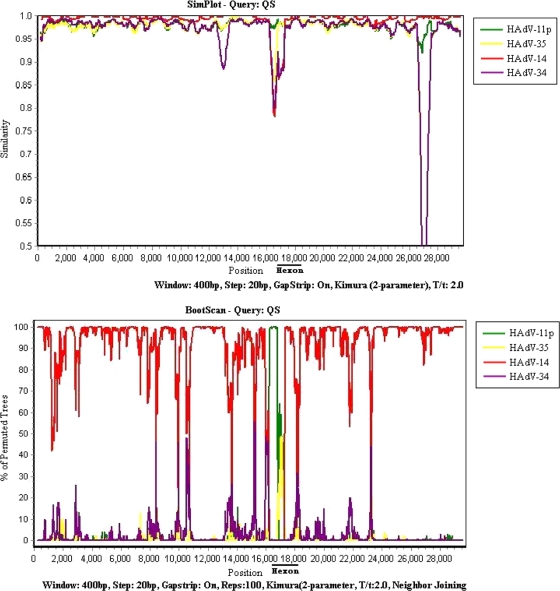
In order to further clarify the possible recombination events, the SimPlot program was used for whole-genome sequence analyses of HAdV-11 QS and the other prototypical subgenus B strains (HAdV-11p, HAdV-14, HAdV-34, and HAdV-35). Similar to the analyses presented above, recombination between HAdV-11 strain QS and HAdV-14 was suggested to exist, except in the hexon gene region (Fig. 2), which further confirmed the recombination between HAdV-11 QS and HAdV-14.
FIG. 2.
Similarities in the whole genome of HAdV strain QS compared to the sequences of the prototypical B:2 species strains of HAdV (HAdV-11p, HAdV-14, HAdV-34, and HAdV-35). The upper graph shows the results of similarity analysis, while the lower graph shows the results of bootscan analysis. The window is 400 bp, and the step is 20 bp. The Kimura model with the Jukes-Cantor correction was used. The vertical axis indicates the percentage of nucleotide similarity and the permuted trees between HAdV QS and the four strains representing the four serotypes of HAdV-B2. The horizontal axis indicates the nucleotide positions in the whole genome.
In addition, a close genetic relationship has been reported to exist between the HAdVs and SAdVs (17, 25). When we selected SAdV-21 for comparison, we found that HAdV-11 QS also exhibited a high degree of amino acid sequence homology (average, >90%) with SAdV-21, except in the fiber region (Table 2), which enabled differentiation between the AdV species and interaction with the receptor on the target cells. This probably indicates a close phylogenetic relationship between HAdV-11 QS and SAdV-21.
In conclusion, this is the first report on recombination between HAdV-11 and HAdV-14, but it is difficult to clarify the exact time and manner of recombination. Whether the emergence of recombination between HAdV-11a and HAdV-14 might increase virulence, thereby posing a new global challenge with regard to acute respiratory diseases in the near future, warrants further investigation. Sentinel virological surveillance for HAdV has thus far not been established in China; both epidemiological and virological surveillance of this uninvestigated respiratory disease pathogen should be strengthened in order to cope with the emerging biosafety incidents caused by the various recombinants. Based on the genomic analyses, HAdV-11 strain QS has a HAdV-14 chassis with a partial HAdV-11 hexon, so it may be a novel adenovirus. This raises the possibility that it should be renamed HAdV-B55 (M. P. Walsh, J. Seto, M. S. Jones, J. Chodosh, W. Xu, and D. Seto, unpublished data).
Acknowledgments
We thank the Shaanxi CDC microbiology laboratory for specimen collection.
This work was supported by grant 2007AA02Z463 from the Ministry of Science and Technology of the People's Republic of China and grants 2009ZX1004-201 and 2009ZX1004-202 from the National Infectious Disease Surveillance Program of the Ministry of Science and Technology of the People's Republic of China.
We have no conflicts of interest to report.
Footnotes
Published ahead of print on 12 August 2009.
REFERENCES
- 1.Adrian, T., G. Schafer, M. K. Cooney, J. P. Fox, and R. Wigand. 1988. Persistent enteral infections with adenovirus types 1 and 2 in infants: no evidence of reinfection. Epidemiol. Infect. 101:503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benko, M., P. Elo, K. Ursu, W. Ahne, S. E. LaPatra, D. Thomson, and B. Harrach. 2002. First molecular evidence for the existence of distinct fish and snake adenoviruses. J. Virol. 76:10056-10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benkö, M., B. Harrach, G. W. Both, W. C. Russell, B. M. Adair, É. Ádam, J. C. de Jong, M. Hess, M. Johnson, A. Kajon, A. H. Kidd, H. D. Lehmkuhl, Q.-G. Li, V. Mautner, P. Pring-Akerblom, and G. Wadell. 2005. Family Adenoviridae, p. 213-228. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 4.Berk, A. J. 2007. Adenoviridae: the viruses and their replication, p. 2355-2394. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 5.Caillet-Boudin, M. L. 1989. Complementary peptide sequences in partner proteins of the adenovirus capsid. J. Mol. Biol. 208:195-198. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H. L., S. S. Chiou, H. P. Hsiao, G. M. Ke, Y. C. Lin, K. H. Lin, and Y. J. Jong. 2004. Respiratory adenoviral infections in children: a study of hospitalized cases in southern Taiwan in 2001-2002. J. Trop. Pediatr. 50:279-284. [DOI] [PubMed] [Google Scholar]
- 7.Chmielewicz, B., J. Benzler, G. Pauli, G. Krause, F. Bergmann, and B. Schweiger. 2005. Respiratory disease caused by a species B2 adenovirus in a military camp in Turkey. J. Med. Virol. 77:232-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garnett, C. T., D. Erdman, W. Xu, and L. R. Gooding. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 76:10608-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, N. M., N. G. Wrigley, W. C. Russell, S. R. Martin, and A. D. McLachlan. 1983. Evidence for a repeating cross-beta sheet structure in the adenovirus fibre. EMBO J. 2:1357-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gustin, K. E., and M. J. Imperiale. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasson, T. B., P. D. Soloway, D. A. Ornelles, W. Doerfler, and T. Shenk. 1989. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 63:3612-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, M. S., II, B. Harrach, R. D. Ganac, M. M. Gozum, W. P. Dela Cruz, B. Riedel, C. Pan, E. L. Delwart, and D. P. Schnurr. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 81:5978-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kajon, A. E., A. S. Mistchenko, C. Videla, M. Hortal, G. Wadell, and L. F. Avendano. 1996. Molecular epidemiology of adenovirus acute lower respiratory infections of children in the south cone of South America (1991-1994). J. Med. Virol. 48:151-156. [DOI] [PubMed] [Google Scholar]
- 15.Kauffman, R. S., and H. S. Ginsberg. 1976. Characterization of a temperature-sensitive, hexon transport mutant of type 5 adenovirus. J. Virol. 19:643-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller, M., T. Tagawa, M. Preuss, and A. D. Miller. 2002. Biophysical characterization of the DNA binding and condensing properties of adenoviral core peptide mu. Biochemistry 41:652-659. [DOI] [PubMed] [Google Scholar]
- 17.Kidd, A. H., D. Garwicz, and M. Oberg. 1995. Human and simian adenoviruses: phylogenetic inferences from analysis of VA RNA genes. Virology 207:32-45. [DOI] [PubMed] [Google Scholar]
- 18.Komoriya, A., L. J. Green, M. Mervic, S. S. Yamada, K. M. Yamada, and M. J. Humphries. 1991. The minimal essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J. Biol. Chem. 266:15075-15079. [PubMed] [Google Scholar]
- 19.Lauer, K. P., I. Llorente, E. Blair, J. Seto, V. Krasnov, A. Purkayastha, S. E. Ditty, T. L. Hadfield, C. Buck, C. Tibbetts, and D. Seto. 2004. Natural variation among human adenoviruses: genome sequence and annotation of human adenovirus serotype 1. J. Gen. Virol. 85:2615-2625. [DOI] [PubMed] [Google Scholar]
- 20.Li, Q. G., J. Hambraeus, and G. Wadell. 1991. Genetic relationship between thirteen genome types of adenovirus 11, 34, and 35 with different tropisms. Intervirology 32:338-350. [DOI] [PubMed] [Google Scholar]
- 21.Louie, J. K., A. E. Kajon, M. Holodniy, L. Guardia-LaBar, B. Lee, A. M. Petru, J. K. Hacker, and D. P. Schnurr. 2008. Severe pneumonia due to adenovirus serotype 14: a new respiratory threat? Clin. Infect. Dis. 46:421-425. [DOI] [PubMed] [Google Scholar]
- 22.Lukashev, A. N., O. E. Ivanova, T. P. Eremeeva, and R. D. Iggo. 2008. Evidence of frequent recombination among human adenoviruses. J. Gen. Virol. 89:380-388. [DOI] [PubMed] [Google Scholar]
- 23.Lutz, P., and C. Kedinger. 1996. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J. Virol. 70:1396-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metzgar, D., M. Osuna, A. E. Kajon, A. W. Hawksworth, M. Irvine, and K. L. Russell. 2007. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J. Infect. Dis. 196:1465-1473. [DOI] [PubMed] [Google Scholar]
- 25.Purkayastha, A., S. E. Ditty, J. Su, J. McGraw, T. L. Hadfield, C. Tibbetts, and D. Seto. 2005. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: implications for gene therapy and vaccine vector development. J. Virol. 79:2559-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosa-Calatrava, M., L. Grave, F. Puvion-Dutilleul, B. Chatton, and C. Kedinger. 2001. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 75:7131-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell, W. C. 2000. Update on adenovirus and its vectors. J. Gen. Virol. 81:2573-2604. [DOI] [PubMed] [Google Scholar]
- 28.Rux, J. J., and R. M. Burnett. 2004. Adenovirus structure. Hum. Gene Ther. 15:1167-1176. [DOI] [PubMed] [Google Scholar]
- 29.Rux, J. J., and R. M. Burnett. 2000. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol. Ther. 1:18-30. [DOI] [PubMed] [Google Scholar]
- 30.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone, D., A. Furthmann, V. Sandig, and A. Lieber. 2003. The complete nucleotide sequence, genome organization, and origin of human adenovirus type 11. Virology 309:152-165. [DOI] [PubMed] [Google Scholar]
- 32.Temperley, S. M., and R. T. Hay. 1992. Recognition of the adenovirus type 2 origin of DNA replication by the virally encoded DNA polymerase and preterminal proteins. EMBO J. 11:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 34.Wold, W. S., and M. S. Howrwitz. 2007. Adenoviridae, p. 2395-2436. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 35.Zhang, W., J. A. Low, J. B. Christensen, and M. J. Imperiale. 2001. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 75:10446-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, Z., Y. Zhang, S. Xu, P. Yu, X. Tian, L. Wang, Z. Liu, L. Tang, N. Mao, Y. Ji, C. Li, Z. Yang, S. Wang, J. Wang, D. Li, and W. Xu. 2009. Outbreak of acute respiratory disease in China caused by B2 species of adenovirus type 11. J. Clin. Microbiol. 47:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zubieta, C., G. Schoehn, J. Chroboczek, and S. Cusack. 2005. The structure of the human adenovirus 2 penton. Mol. Cell 17:121-135. [DOI] [PubMed] [Google Scholar]