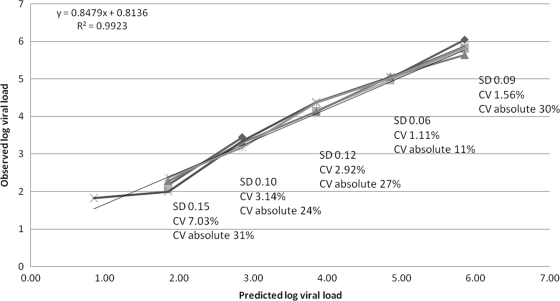
The Cobas TaqMan human immunodeficiency virus type 1 (HIV-1) assay (Roche Molecular Systems, Branchburg, NJ) technology combines the extraction of total nucleic acids on the Cobas AmpliPrep (CAP) with real-time PCR on the Cobas TaqMan analyzer (CTM), using hydrolysis probe technology (5, 6, 9). This technology replaces the CAP-Cobas Amplicor version 1.5 assay (CAP-CA) (gag) based on endpoint PCR, which is limited by its dynamic range, 400 to 750,000 copies/ml for the standard assay and 50 to 100,000 copies/ml for the ultrasensitive assay (1, 5, 6). The CAP-CTM HIV-1 v2.0 assay uses primers targeting both gag and long terminal repeat regions of the HIV genome and has a reportable range of 20 to 1.0E+7 HIV RNA copies/ml (8). This replaces the CAP-CTM HIV-1 assay, the previous version (termed here CAP-CTM HIV-1 v1.0) of the assay, targeting only the gag region (9) with a dynamic range of 40 to 1.0E+7 copies/ml. The performance of the new CAP-CTM HIV-1 v2.0 assay on the CAP-CTM docked platform, which has capacity for 96 assays (although the CTM 48 or CTM 96 could be used), was evaluated in a high-throughput laboratory receiving predominantly HIV-1 subtype C (7) samples in South Africa. The automated docked platform has an initial capacity for 72 samples with continuous feeding, which allows 168 samples (1 ml/sample) to be processed per 8-h shift. In a 24-h laboratory, this system can analyze 399 samples on one automated docked platform. Precision (within-run analysis) and linearity using regression analysis for the CAP-CTM HIV-1 v2.0 were measured on a specially prepared pooled plasma sample which was further divided into six aliquots. The pool mean viral load (VL) was 716,000 RNA copies/ml (log 5.85) determined by CAP-CTM HIV-1 v2.0, which was serially diluted to log 1.0. Following serial dilution, the lowest predicted value was 7.2 RNA copies/ml. Of 36 diluted tubes analyzed, two were invalid due to internal control failures, two further samples had “target not detected” at the predicted 7.2 copies/ml, and two were “target detected but not quantifiable” (<20 copies/ml) at this dilution. The equation of the fitted regression line was y = 0.848x + 0.814 with R2 = 0.992. The standard deviation (SD) using log values and percent coefficient of variation (CV) (using both log and absolute values) are illustrated in Fig. 1.
FIG. 1.
Precision and linearity of the CAP-CTM v.2 over the range log 6.0 to log 2.0. The horizontal axis is the predicted log VL values at each dilution, and the vertical axis is the observed log VL values for each replicate. The six lines shown represent the six replicates. The SD and %CV on the log values are represented for each dilution as well as the %CV for the absolute VL values. The equation of the line is also represented on the plot.
One hundred thirty-seven plasma samples collected in EDTA tubes from consenting patients at a routine antiretroviral therapy clinic in the Charlotte Maxeke (previously Johannesburg Academic) Hospital, Johannesburg, South Africa, were used for the pilot accuracy analysis (ethics clearance no. M061105). Testing of accuracy and agreement was conducted using the Bland-Altman (2) and percent similarity (10) method comparison tools. For accuracy determination across the dynamic range, CAP-CTM HIV-1 v2.0 was compared to the CAP-CTM HIV-1 v1.0 (on the CTM 48 platform) and the Abbott RealTime HIV-1 assay using the automated m2000sp/m2000rt platform (Abbott Molecular Inc., Des Plaines, IL). In contrast to the Roche assays, the Abbott RealTime HIV-1 assay targets the pol/IN region of HIV-1 by using partially double-stranded probes (4) with a dynamic range of 40 to 1.0E+7 copies/ml. A comparison of the CAP-CTM HIV-1 v1.0 and CAP-CTM HIV-1 v2.0 to the CAP-CA version 1.5 was also included, but here the real-time VL results were capped to the limited dynamic range of the standard CAP-CA (400 to 750,000 RNA copies/ml). All analyses were performed on log10-transformed copy/ml values, and all 137 samples were processed and analyzed using the CAP-CTM HIV-1 v2.0 in a single 8-h shift.
Seventy-five samples generated a “target not detected” result on any of the three real-time assays (a reflection of successful antiretroviral treatment at the clinic). The CAP-CTM HIV-1 v1.0 could generate a “target detected” or “target detected but not quantifiable” result in 16% (n = 12) of these samples, with a range of 40 to 146 copies/ml. The CAP-CTM HIV-1 v2.0 could generate a “target detected” or “target detected but not quantifiable” result in 54.6% (n = 41) of the samples, with a range of 20 to 364 copies/ml, and the Abbott RealTime HIV-1 assay could generate such a result in 26.6% (n = 20) of samples with values of <40 copies/ml.
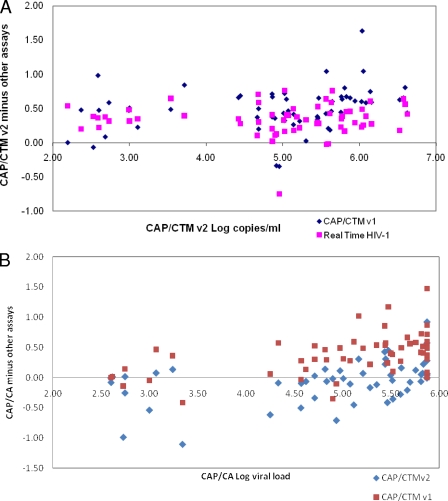
Method comparison for the remaining 62 samples that generated VL values on all three assays is represented in Table 1 and further illustrated in Bland-Altman plots (Fig. 2). There was overall good agreement (3.5% to 4.4% similarity CV) between the existing assays tested in this study and the CAP-CTM HIV-1 v2.0. The greatest bias (least accuracy), however, was found between CAP-CTM HIV-1 v1.0 and CAP-CTM HIV-1 v2.0 with a positive bias of log 0.48 copies/ml. This difference is also clearly visible on the scatter plots in Fig. 2 and does not appear to be restricted to any one range in VL values but is found across all values. The variance of these differences (precision), however, was small, ∼0.35 copies/ml, showing good precision. An effect of the CAP-CTM HIV-1 v2.0 reading higher than the CAP-CTM HIV-1 v1.0 and the Abbott RealTime HIV-1 assay is that 47% and 23%, respectively, of the clinical samples are >log 0.5 copies/ml different in reportable results. This also resulted in 4.8% samples that would be considered to have clinically relevant differences (>log 1.0 copies/ml) between the CAP-CTM HIV-1 v1.0 and CAP-CTM HIV-1 v2.0. The clinical significance of switching to the CAP-CTM HIV-1 v2.0 assay would have to be evaluated in laboratories currently reporting VL results for monitoring using the CAP-CTM HIV-1 v1.0. Even with the lower limit of detection of <20 copies/ml, the majority of results will read higher with CAP-CTM HIV-1 v2.0. Interestingly the CAP-CTM HIV-1 v2.0 had the smallest bias and reads on average lower than the CAP-CA endpoint detection assay, with the overall lowest number of outliers of any of the comparisons performed in this pilot study. A switch to CAP-CTM HIV-1 v2.0 for laboratories currently performing the CAP-CA would therefore not cause laboratories to experience clinically relevant outliers.
TABLE 1.
Bland-Altman and percent similarity statistical parameters for measuring agreement between all assays for n = 62
| Comparison | Bland-Altman parameter
|
% Similarity
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Bias (copies/ml) | SD of difference (copies/ml) | Limits of agreement | Distance over limits (copies/ml) | No. (%) of outliers
|
|||||
| >Log 0.5 copies/ml | >Log 1.0 copies/ml | Mean | SD | CV | |||||
| CAP-CTM HIV-1 v.2 vs CAP-CTM HIV-1 v.1 | 0.48 | 0.33 | 1.14; −0.18 | 0.66 | 29 (46.8) | 3 (4.8) | 95 | 4.18 | 4.4 |
| CAP-CTM HIV-1 v.2 vs RealTime HIV-1 | 0.33 | 0.25 | 0.82; −0.16 | 0.49 | 14 (22.5) | 0 | 97 | 3.66 | 3.79 |
| CAP-CA vs CAP-CTM v.2 | −0.04 | 0.32 | 0.6; −0.69 | 0.65 | 8 (12.9) | 1 (1.6) | 101 | 4.1 | 4.07 |
| CAP-CA vs CAP-CTM v.1 | 0.34 | 0.35 | 1.03; −0.36 | 0.69 | 20 (32) | 3 (4.8) | 97 | 3.34 | 3.45 |
FIG. 2.
Bland-Altman overlay scatter plots. (A) CAP-CTM HIV-1 v2.0 versus CAP-CTM HIV-1 and versus Real-Time HIV-1 (n = 62). The vertical axis is the difference (CAP-CTM HIV-1 v2.0 minus other assays), and the horizontal axis is the CAP-CTM HIV-1 v2.0 log VL copies/ml. (B) CAP-CA versus CAP-CTM HIV-1 v1.0 and CAP-CTM HIV-1 v2.0. The vertical axis is CAP-CA minus other assays, and the horizontal axis is CAP-CA log VL copies/ml. The symbol keys indicate the assays for each plotted point.
Thus, in summary the CAP-CTM HIV-1 v2.0 shows improved sensitivity over the existing CAP-CTM HIV-1 v1.0 assay, with more quantifiable results down to 20 HIV RNA copies/ml. The clinical impact of this improved sensitivity for patients being monitored on antiretroviral treatment programs remains to be determined. There was acceptable precision with log variation of ≤0.15 copies/ml even in the log 2.0 range (3). The positive bias of 0.48 copies/ml between CAP-CTM HIV-1 v1.0 and CAP-CTM HIV-1 v2.0 needs further investigation. The variability of this difference (SD), however, is acceptable and shows good precision (reflected by a smaller distance between the limits of agreement) and overall good performance compared to the Abbott RealTime HIV-1. The application of this new CAP-CTM HIV-1 v.2 on the fully automated CAP-CTM 96 docked platform has potential for HIV VL monitoring in high-throughput settings. Although not investigated in this study, the CAP-CTM HIV-1 v2.0 also has the capability of detecting HIV group O per the manufacturer's instructions but requires further field evaluation for HIV-1 subtypes other than subtype C.
Acknowledgments
Thanks to Reiner Babiel from Roche Diagnostics Lty, Rotkreuz, Switzerland, and Eveline Akkers from Roche Products Pty (Ltd), Isando, South Africa, for technical support.
This publication was made possible by the generous support of the American people through the U.S. Agency for International Development.
The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the U.S. government.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Berger, A., L. Scherzed, M. Sturmer, W. Preiser, H. W. Doerr, and H. F. Rabenau. 2002. Evaluation of the Cobas AmpliPrep/Cobas Amplicor HIV-1 Monitor Ultrasensitive Test: comparison with the Cobas Amplicor HIV-1 Monitor test (manual specimen preparation). J. Clin. Virol. 25(Suppl. 3):S103-S107. [DOI] [PubMed] [Google Scholar]
- 2.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed] [Google Scholar]
- 3.Brambilla, D., S. Granger, and J. Bremer. 2000. Variation in HIV RNA assays at low RNA concentration, abstr. 774. Seventh Conf. Retroviruses Opportunistic Infect., San Francisco, CA.
- 4.Huang, S., J. Salituro, N. Tang, K. C. Luk, J. Hackett, Jr., P. Swanson, G. Cloherty, W. B. Mak, J. Robinson, and K. Abravaya. 2007. Thermodynamically modulated partially double-stranded linear DNA probe design for homogeneous real-time PCR. Nucleic Acids Res. 35:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwok, S., and J. J. Sninsky. 1993. PCR detection of human immunodeficiency virus type 1 proviral DNA sequence, p. 309-315. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, DC.
- 6.Oliver, A. R., S. F. Pereira, and D. A. Clark. 2007. Comparative evaluation of the automated Roche TaqMan real-time quantitative human immunodeficiency virus type 1 RNA PCR assay and the Roche Amplicor version 1.5 conventional PCR assay. J. Clin. Microbiol. 45:3616-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papathanasopoulos, M. A., G. M. Hunt, and C. T. Tiemessen. 2003. Evolution and diversity of HIV-1 in Africa—a review. Virus Genes 26:151-163. [DOI] [PubMed] [Google Scholar]
- 8.Roche Molecular Systems. 2008. Cobas AmpliPrep-Cobas TaqMan HIV-1 test, v.2.0, package insert. Roche Molecular Systems, Branchburg, NJ.
- 9.Schumacher, W., E. Frick, M. Kauselmann, V. Maier-Hoyle, R. van der Vliet, and R. Babiel. 2007. Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 38:304-312. [DOI] [PubMed] [Google Scholar]
- 10.Scott, L. E., J. S. Galpin, and D. K. Glencross. 2003. Multiple method comparison: statistical model using percentage similarity. Cytometry 54B:46-53. [DOI] [PubMed] [Google Scholar]