Abstract
Quantitation of hepatitis C virus (HCV) RNA in plasma and serum samples is a costly procedure in both time and reagents. Additionally, cell-associated viral RNA may not be detected. This study evaluated the accuracy of HCV RNA quantitation in small-volume whole-blood (WB) samples, which would be appropriate for point-of-care diagnostic devices. HCV RNA was extracted from 222 clinical plasma and WB samples of 82 patients with chronic hepatitis C by a specific locked nucleic acid-mediated capture method and quantified by real-time reverse transcription-PCR. The results were compared to the reference plasma viral load determined with the COBAS AmpliPrep/TaqMan (CAP/CTM) HCV test. This assay had an analytical sensitivity of 9 IU per 10-μl sample (95% limit of detection [95% LOD]), a linearity range of 500 to 5 × 106 IU/ml, and was accurate in testing 10 HCV subtypes (<0.22 log10 unit) in plasma. The assay was matrix equivalent for plasma and WB samples (coefficient of determination [R2] of 0.943) and had a specificity of 100% (n = 20) in WB samples. The HCV RNA concentration in clinical WB samples exceeded the estimated hematocrit-corrected plasma viral loads by 0.22 log10 unit, but absolute quantitation results in plasma and WB samples were identical (95% confidence interval, −0.06 to 0.04 log10 unit). The sensitivity in WB samples was 100% (n = 141) for plasma concentrations above the 95% LOD. Quantitation results in 10-μl WB samples correlated linearly with the CAP/CTM HCV plasma test results (R2 = 0.919; n = 140) and did not differ between capillary and venous samples (R2 = 0.960; n = 40). This study shows that HCV RNA quantitation in 10-μl WB samples is appropriate for monitoring viral loads of >900 IU/ml, although the use of WB does not increase the diagnostic sensitivity.
Approximately 130 to 170 million people worldwide are infected with the hepatitis C virus (HCV). The highest prevalence of HCV infection is in developing and transitional economy countries (19) that have limited access to costly HCV quantitation assays. Because the analysis of capillary whole blood (WB) does not require venous blood draw and sample centrifugation, it may allow a point-of-care testing and improve diagnostic and therapeutic HCV management in developing countries.
Although the liver is the main site of HCV replication, HCV RNA has been detected in circulating extrahepatic sites, such as in peripheral blood mononuclear cells (PBMC) (3, 24, 25, 31, 42), in cryoprecipitates (22), and attached to erythrocytes (21) and to platelets (14, 41). HCV RNA lymphotropism may be due to adsorption of viral particles (23, 43) as well as intracellular viral replication (27, 30). Thus, quantitation of HCV RNA in plasma or serum samples may underestimate the total circulating viral load. Furthermore, the detection of HCV RNA in PBMC may serve as an alternative to liver biopsy in the diagnosis of occult hepatitis C infection, defined as the presence of hepatic HCV RNA in the absence of detectable serum HCV RNA and anti-HCV antibodies (4, 5). HCV RNA persistence in PBMC (39) and in WB (38) at the end of interferon treatment may predict a subsequent viral relapse. However, quantitative analysis of viral kinetics in PBMC provides only minor additional predictive information (29). Because PBMC separation, counting, and washing are elaborate, the quantitation of HCV RNA in WB may serve as an alternative. Advantages of WB analysis have been reported for other PBMC-associated viruses, such as human immunodeficiency virus (HIV) (15) and Epstein-Barr virus (13).
However, inconclusive data have been published on whether the use of WB for HCV RNA quantitation provides higher diagnostic sensitivity than the use of plasma or serum. Although one group reported an increase in diagnostic sensitivity (37) with 100- to 1,000-fold HCV RNA concentrations measured in WB (35), these results were not confirmed in subsequent studies (8, 10).
The aim of this study was to evaluate the feasibility of HCV RNA quantitation in 10-μl WB samples and to compare quantitation results with the COBAS AmpliPrep/TaqMan (CAP/CTM) HCV test in plasma. To improve the extraction yield of HCV RNA, a novel sequence-specific oligonucleotide hybridization assay was developed and compared to nonspecific silica filter-based total nucleic acid extraction. Locked nucleic acid (LNA)-modified oligonucleotides have an exceptionally high affinity to cRNA (18) and could serve as a more potent hybridization probe in a chaotropic lysis buffer than DNA (16). The synthetic nucleotide derivate 2′-O-methyl-RNA (2′-OMeRNA) forms stable complexes with RNA that are more resistant to nucleases (9). Thus, DNA oligonucleotides, LNA-modified DNA oligonucleotides, and LNA-modified 2′-OMeRNA oligonucleotides were designed to evaluate HCV RNA extraction efficiency. To interpret quantitation results and diagnostic sensitivity in small-volume WB samples correctly, the assay was validated for analytical sensitivity, precision, linearity, diagnostic specificity, and matrix equivalence for plasma and WB specimens.
MATERIALS AND METHODS
Samples.
Two hundred twenty-two venous or capillary blood samples of 82 outpatients with chronic HCV infection from the Jena University Hospital were obtained after informed consent. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the local Ethical Committee on 27 June 2007 (no. 2040-07/07).
Finger-prick capillary WB samples were collected in K3-EDTA Microvette 100-μl tubes (Sarstedt, Nümbrecht, Germany). Plasma was separated by centrifugation of WB samples in a K3-EDTA Monovette (Sarstedt) at 1,600 × g over 20 min. Residual plasma and whole blood samples were frozen at −80°C. Genotype 1 HCV RNA reference plasma sample number 3443/04 (Paul-Ehrlich Institute, Federal Agency for Sera and Vaccines, Langen, Germany) and the OptiQuant genotype 1 HCV RNA quantitation panel (Acrometrix, Benicia, CA), both calibrated against the World Health Organization (WHO) international standard HCV-RNA 96/790 (33), were obtained for determination of linearity, analytical sensitivity, and precision. A reference genotype panel containing HCV subtypes 1a, 1b, 2a, 2b, 2c, 2i, 3a, 4, 5a, and 6 was obtained from the German National Reference Center for Hepatitis C (University of Essen, Essen, Germany).
HCV quantitation in EDTA plasma samples.
The plasma specimens were tested with the automated CAP/CTM HCV test (Roche Diagnostics, Mannheim, Germany). Briefly, HCV RNA was extracted from a 850-μl plasma sample in the COBAS AmpliPrep system according to the manufacturer's instructions and transferred to the COBAS TaqMan 48 system for real-time reverse transcription-PCR (RT-PCR). HCV RNA quantitation was performed by AmpiLink software version 3.2.2 and expressed in international units (IU) per ml.
HCV RNA capture assay. (i) Extraction.
For HCV extraction, three homologous 25-mer oligonucleotide antisense probes were developed: DNA (Metabion International AG, Martinsried, Germany), LNA-modified DNA (Sigma-Proligo, Paris, France), and LNA-modified 2′-OMeRNA (RiboTask, Odense, Denmark). Oligonucleotide probe sequences that were complementary to nucleotides 68 to 92 of the HCV H77 reference strain (40) were tagged with biotin at their 3′ end. LNA nucleotides were inserted at every second position (Table 1).
TABLE 1.
Oligonucleotide probes and primers designed for the HCV RNA capture assay
| Oligonucleotide probe or primer | Sequence (5′ to 3′)a | Nucleotide positionb
|
|
|---|---|---|---|
| Start | End | ||
| DNA capture probe | AACGCCATGGCTAGACGCTTTCTGC-biotin | 92 | 68 |
| LNA/DNA capture probe | AaCgCcAtGgCtAgAcGcTtTcTgC-biotin | 92 | 68 |
| LNA/2′-OMeRNA probe | AaCgCcAtGgCtAgAcGcUtUcUgC-biotin | 92 | 68 |
| Sense primer | GTGGTCTGCGGAACCGGTGA | 143 | 162 |
| Antisense primer | CGCAAGCACCCTATCAGGCAGT | 309 | 288 |
| Target TaqMan probe A | FAM-CCGAGTAGTGTTGGGTCGCGAAAGG-BHQ-1 | 254 | 278 |
| Target TaqMan probe B | FAM-CCGAGTAGCGTTGGGTTGCGAAAGG-BHQ-1 | 254 | 278 |
| Control TaqMan probe | Cy5-GGAAAGCGCTGGGTTGTGATGAGCC-BHQ-3 | 254 | 278 |
Capital letters refer to DNA or 2′-O-methyl-RNA (2′-OMeRNA); lowercase letters refer to LNA nucleotides.
Nucleotide positions are named with respect to the H77 reference strain of HCV (40).
For indirect capturing, 12.5 pmol of the respective hybridization probe was resuspended in 5 μl nuclease-free water, added to 65 μl lysis/hybridization buffer (4.3 M bicine-buffered guanidine hydrochloride, 20 mM EDTA, 10% [vol/vol] Triton X-100, 0.2% [wt/vol] saponins from quillaja bark [pH 8.5]), and mixed vigorously with 10 μl of WB or plasma. Competitive internal standard (IS) RNA was added to determine assay performance. For RNA denaturation and hybridization, the solution was incubated at 65°C for 5 minutes and then passively cooled for 2 minutes. Streptavidin-coated superparamagnetic polystyrene beads (200 μg) (Dynabeads M-270 Streptavidin, Invitrogen, Carlsbad, CA) were washed twice in 20 μl Tris-Tween buffer (10 mM Tris-HCl, 0.05% [vol/vol] Tween 20 [pH 7.5]), resuspended in 20 μl Tris-Tween buffer, and added to the solution. For binding of biotin-labeled hybrids to the streptavidin matrix, the suspension was incubated at room temperature for 10 minutes with shaking. Afterwards the beads were collected by a magnetic separator (CD3002; Cortex Biochem, Promega, Madison, WI), washed two times in 150 μl washing buffer (150 μM ammonium chloride, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0]) with 0.1% (wt/vol) sodium dodecyl sulfate, two times in 150 μl washing buffer with 0.05% (vol/vol) Tween 20, once in 50 μl washing buffer plus 0.05% (vol/vol) Tween 20, and resuspended in 15 μl deionized water for quantitative real-time reverse transcription-PCR (qRT-PCR).
(ii) Amplification and detection.
Reverse transcription, amplification, and detection were performed as a one-step nonnested RT-PCR in the Smart Cycler II system (Cepheid, Sunnyvale, CA) with the RNA UltraSense one-step quantitative RT-PCR kit (Invitrogen, Carlsbad, CA) in a reaction mixture volume of 75 μl. Primers and probes (Metabion International, Planegg-Martinsried, Germany) were designed to amplify a 287-nucleotide region within the HCV 5′-untranslated region downstream of the capture site and were used at the following concentrations: sense primer and antisense primer at 600 nM each, two carboxyfluorescein (FAM)- or Black Hole Quencher 1 (BHQ-1)-labeled target TaqMan probes at 100 nM each, and a cyanine 5 (Cy5)- or BHQ-3-labeled control TaqMan probe at 200 nM (Table 1). After reverse transcription at 50°C for 30 min and denaturation at 95°C for 2 min, 45 cycles of denaturation (95°C, 5 s) and annealing or extension (67°C, 45 s) were performed. Reporter fluorescence signals of FAM (target) and Cy5 (control) were analyzed by Cepheid Smart Cycler software 2.0d.
(iii) Internal standard RNA.
An artificial IS RNA sequence was designed from a partial sequence of the HCV isolate HC-C2 (subtype 1b [GenBank accession number D10934], nucleotide positions 46 to 332 with respect to the HCV H77 strain [40]) to estimate the assay's extraction and amplification quality. The TaqMan probe binding site was generated by nucleotide permutation. Primer and capture probe binding sites remained unchanged to allow for similar extraction and amplification conditions for viral RNA and IS. Transformed Escherichia coli DH5α containing the IS gene cloned into the pCR 2.1 TOPO vector (Invitrogen, Carlsbad, CA) was purchased from Eurofins MWG Operon (Ebersberg, Germany). For in vitro transcription, plasmid DNA was linearized with the HindIII restriction enzyme (Invitrogen) and transcribed with the Ambion MegaScript T7 RNA polymerase kit (Applied Biosystems, Darmstadt, Germany). The transcript size (415 nucleotides) was confirmed by ethidium bromide-stained 2% agarose gel electrophoresis.
Extraction efficiency for different capture probes.
HCV RNA was extracted from 34 plasma and WB specimens with each of the three hybridization probes and with the Roche high pure viral nucleic acid kit according to the manufacturer's instructions (July 2005). To adjust for qRT-PCR sample input volumes, 33.33 μl of plasma or WB samples were diluted in 200 μl of nuclease-free phosphate-buffered saline (pH 7.4). Briefly, after lysis of diluted samples in carrier RNA-supplemented binding buffer and proteinase K at 72°C over 10 min, the crude lysate was applied to the glass-fiber fleece filter tube for binding of nucleic acids, rinsed with inhibitor removal buffer, washed two times, and eluted in 50 μl of nuclease-free, sterile, double-distilled water prewarmed to 70°C. Fifteen microliters of eluate (equivalent to total nucleic acids in 10-μl specimen) was examined by qRT-PCR. Cycle threshold (CT) values of quantifiable results were tested for normal distribution, linear correlation, and difference to the reference extraction method.
HCV capture assay validation.
HCV capture assay validation was performed with LNA-modified 2′-OMeRNA hybridization probes.
(i) Analytical sensitivity.
HCV genotype 1 reference samples (PEI 3443/04 and Acrometrix OptiQuant) were diluted with HCV-negative plasma (Roche) to concentration levels of 50, 100, 250, 500, 800, 1,000, and 2,963 IU/ml and tested in 5 to 15 replicates over 18 days. Probit analysis for tested log-transformed concentration levels was performed to determine the 95% and 50% limit of detection (95% LOD and 50% LOD).
(ii) Linear range.
Six panel members of the OptiQuant plasma panel containing 50 to 5,000,000 IU/ml HCV RNA (genotype 1) were analyzed in five replicates over 3 days. Linearity was evaluated by the absolute difference of observed and nominal log-transformed HCV titers, the coefficient of determination (R2), and comparison of the linear and cubic regression.
(iii) Precision.
Intra-assay and interassay variability were determined in five to seven replicates in plasma samples and in capillary WB samples. The coefficients of variation (CV) were calculated by the proposed formula for log10-normal distributed values: %CV = 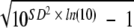 where SD is the standard deviation (12, 20).
where SD is the standard deviation (12, 20).
(iv) Accuracy.
Aliquots of the hepatitis C virus genotype panel (National Reference Center, Essen, Germany) were tested in triplicate and compared to CAP/CTM HCV test results (10-fold dilution, tested in duplicate due to sample volume). The means, standard deviations, and mean differences of log-transformed viral loads were calculated.
(v) Matrix equivalence.
HCV RNA-positive plasma was spiked and divided into 38 pairs of WB and plasma samples from HCV RNA-negative donors (1:10). HCV concentration in matrix spike sample pairs ranging from 908 IU/ml to 2.88 × 106 IU/ml (median, 7.47 × 104 IU/ml) was analyzed by within-run analysis. The mean deviations and linear correlations were calculated for log-transformed viral loads, and Student's t test was used to determine statistical differences.
(vi) Diagnostic specificity in WB samples.
Capillary finger-prick EDTA WB samples from 20 HCV-negative self-declared healthy individuals were obtained after informed consent and analyzed with the HCV capture assay.
Clinical performance of the HCV capture assay in WB and plasma samples.
The HCV RNA concentration of 222 clinical samples was examined by CAP/CTM HCV test in 850-μl plasma samples and by HCV capture assay in 10-μl whole-blood or plasma samples.
(i) Diagnostic sensitivity and specificity in WB samples.
To discriminate between analytical sensitivity and diagnostic sensitivity, samples were categorized with respect to CAP/CTM HCV plasma viral load as follows: <15 IU/ml, 15 to 900 IU/ml, and >900 IU/ml. The overall specificity and subset sensitivities for plasma and WB were calculated. Differences in diagnostic sensitivity in WB and plasma were analyzed by McNemar's test.
(ii) HCV RNA quantitation in plasma samples.
To determine concordance between quantitation results of the HCV capture assay and CAP/CTM at >900 IU/ml (LOD), a linear Passing-Bablok regression (28) and the CUSUM test for linearity were performed.
(iii) HCV RNA quantitation in venous WB samples.
Sample pairs that were tested with the HCV capture assay and returned quantifiable results in both specimens were included to test two hypotheses. Hypothesis A is that WB samples contain HCV RNA amounts that are not accessible by plasma analysis. Hypothesis B is that the total HCV RNA concentration in WB samples exceeds the concentration in plasma samples. For hypothesis A, an estimated WB viral load (cexp) was calculated assuming HCV RNA is present only in the plasma but not in the cellular fraction: cexp = cplasma × (1 − hematocrit) with c representing the viral load in IU/ml and (1 − hematocrit) representing the ratio of plasma in the particular WB sample. Student's t test was performed to test both hypotheses after verification of log-normal distribution with a one-sample Kolmogorov-Smirnov (KS) test. The distributions and mean log deviations between plasma and (corrected) WB samples were calculated.
To determine the correlation of HCV RNA quantitation results in 10-μl WB samples with the reference plasma method (CAP/CTM), Passing-Bablok regression, CUSUM test for linearity, and R2 were calculated for all quantifiable results. Outliers, defined by a log deviation of >1.96-fold SD, had to be confirmed by the HCV RNA capture assay in plasma.
(iv) HCV RNA quantitation in capillary WB samples.
To examine the correlation between venous and capillary WB specimens, 40 sample pairs from 28 patients with quantifiable HCV RNA as measured by HCV capture assay were analyzed. Passing-Bablok regression and R2 were calculated for log-transformed measurement results. All statistical calculations were performed using the SPSS 16.0 software (SPSS Inc., Chicago, IL).
RESULTS
Extraction efficiencies for different capture probes.
Thirty-four plasma and WB samples with plasma viral loads between 144 IU/ml and 7.79 × 106 IU/ml (median, 1.98 × 105 IU/ml) by CAP/CTM were included for analysis of extraction efficiency. Thirty of 34 plasma samples were identified as positive by HCV capture assay with DNA capture probes, 33/34 with LNA-modified DNA capture probes (LNA/DNA), and 34/34 with LNA-modified 2′-OMeRNA capture probes (LNA/2′-OMeRNA). In WB samples, analytical sensitivity was 32/34 with DNA, 34/34 with LNA/DNA, and 34/34 with LNA/2′-OMeRNA, respectively. After RNA was extracted with the Roche high pure viral nucleic acid kit, HCV RNA was detected in all 34 plasma samples and in all 34 WB samples.
The 29 sample pairs that were detected by all four extraction methods were selected for quantitative analysis. A one-sample KS test confirmed normal distribution of measured CT values; therefore, Student's t test for paired samples was performed. CT values were significantly higher after DNA hybridization compared to total nucleic acid extraction in both plasma samples (95% confidence interval [95% CI] of ΔCT, 1.3 to 2.6; P < 0.001) and WB samples (95% CI of ΔCT, 1.4 to 2.5; P < 0.001). CT values after hybridization with LNA/DNA capture probes and silica-based extraction did not differ in plasma samples (95% CI of ΔCT, −0.8 to 0.1; P = 0.092) but were lower in WB samples (95% CI of ΔCT, −0.8 to 0.0; P = 0.044). CT values after hybridization with LNA/2′-OMeRNA were significantly lower in both plasma samples (95% CI of ΔCT, −1.0 to −0.2; P = 0.003) and WB samples (95% CI of ΔCT, −1.0 to −0.1; P = 0.012), indicating a better extraction efficiency. The Pearson correlation coefficients (r) between extraction with the silica-based protocol and HCV capture assay in plasma samples were 0.879 for DNA, 0.951 for LNA/DNA, and 0.961 for LNA/2′-OMeRNA probes; in WB samples, they were 0.908, 0.953, and 0.945, respectively. Because the best qualitative and quantitative results were attained with LNA/2′-OMeRNA capture probes, these probes were used for subsequent assay validation and clinical analyses.
HCV RNA capture assay validation. (i) Analytical sensitivity.
Four out of 95 measurements were invalid due to nonamplification of the IS RNA. The HCV RNA capture assay was positive in 0/5 for a HCV RNA plasma concentration of 50 IU/ml, 3/14 for 100 IU/ml, 8/13 for 250 IU/ml, 11/15 for 500 IU/ml, 14/15 for 800 IU/ml, 14/14 for 1,000 IU/ml, and 15/15 for 2,963 IU/ml, respectively. The 95% LOD was 899 IU/ml (9 IU per 10-μl sample), and the 50% LOD was 218 (2 IU per 10-μl sample) in probit analysis.
(ii) Linear range.
All 36 plasma samples and negative controls tested valid. All five samples with 50 IU/ml HCV RNA and one of five samples with 500 IU/ml HCV RNA were not detected. The mean observed titer deviated from −0.12 to 0.15 log10 IU/ml from the nominal titer; the maximum deviation was 0.29 log10 unit in single measurements. The linear regression between log-transformed observed and nominal viral loads was f(x) = 0.991x + 0.039 (R2 = 0.990) with x as the log10-transformed plasma viral load. The third-order polynomial regression resulted in a quadratic regression curve f(x) = −0.054x2 + 1.503x − 1.074 (R2 = 0.994). The maximum difference between the linear and polynomial fit was 0.12 log10 unit. Thus, assay linearity was estimated to be between 5 × 102 and 5 × 106 IU/ml.
(iii) Precision.
All 39 samples subjected to precision analysis tested valid. The CV in plasma was 17% for 5 × 106 IU/ml (SD, 0.07 log unit), 19% for 5 × 105 IU/ml (SD, 0.08 log unit), 16% for 5 × 104 IU/ml (SD, 0.07 log unit), and 30% for 5 × 103 IU/ml (SD, 0.13 log unit) for genotype 1 in between-run analysis of five replicates each. The CV was 21% as measured by within-run analysis (genotype 1) (mean of 1.84 × 105, SD of 0.09 log unit; n = 6). For capillary WB samples subjected to within-run analysis, the CV was 25% (genotype 1) (mean, 1.08 × 105 IU/ml; SD, 0.11 log unit; n = 6), and the between-run CV was 33% (genotype 2) (mean, 1.04 × 106 IU/ml; SD, 0.14 log unit; n = 7).
(iv) Accuracy for different genotypes.
The means and the standard deviations for log-transformed measurement results for each subtype and mean log deviation to CAP/CTM results are displayed in Table 2. The maximum differences between the HCV capture assay and CAP/CTM quantification were found for subtype 3a (0.21 log unit) and genotype 4 (0.19 log unit).
TABLE 2.
Accuracy of HCV RNA quantitation results for different HCV genotypesa
| HCV subtype of the genotype reference panel | Mean log viral load | SD of log viral load | Mean log difference to the mean CAP/CTM resultb |
|---|---|---|---|
| 1a | 4.98 | 0.10 | −0.01 |
| 1b | 4.74 | 0.03 | 0.12 |
| 2a | 5.69 | 0.04 | 0.08 |
| 2b | 4.94 | 0.13 | 0.08 |
| 2c | 5.50 | 0.03 | 0.04 |
| 2i | 4.67 | 0.20 | 0.18 |
| 3a | 5.55 | 0.15 | 0.21 |
| 4 | 5.20 | 0.15 | 0.19 |
| 5a | 4.71 | 0.16 | 0.05 |
| 6 | 4.65 | 0.05 | 0.07 |
HCV RNA was determined by the HCV RNA capture assay on 10-μl plasma samples and expressed in log10 IU/ml (n = 3 for each HCV subtype).
The difference to the mean COBAS AmpliPrep/COBAS TaqMan (CAP/CTM) result was calculated by subtraction of log-transformed results.
(v) Matrix equivalence.
The mean difference of measurement results between plasma and WB samples with identical HCV RNA concentrations was 0.04 log10 unit, in support of WB; however, this difference was not statistically significant (95% CI, −0.02 to 0.11 log10 unit; P = 0.203). In 6/38 sample pairs, the mean differed more than 0.3 log10 IU/ml. The linear regression had a slope of 1.00 and an intercept of 0.04 with R2 = 0.943.
(vi) Diagnostic specificity in WB samples.
All 20 tested capillary WB samples from healthy donors tested as valid negatives in single measurements.
Clinical performance of the HCV capture assay for plasma samples.
Two hundred twenty-two plasma samples were analyzed by the CAP/CTM HCV test. Forty-six samples tested HCV RNA negative, 165 tested HCV RNA positive (≥15 IU/ml), and eight tested below the analytical sensitivity in plasma (<15 IU/ml) (36). Three samples could not be quantified because of repeatedly invalid results by CAP/CTM.
(i) Diagnostic sensitivity and specificity in WB samples.
Table 3 displays the sensitivity of the HCV RNA capture assay in plasma and WB samples with respect to the CAP/CTM HCV test plasma results. The diagnostic sensitivity was 100% (n = 147) in plasma samples and 100% (n =141) in WB samples for samples with >900 IU/ml. In samples with plasma viral loads between 15 and 900 IU/ml, the diagnostic sensitivity was 76% (13/17) for the HCV RNA capture assay on plasma samples and 58% (10/17) for WB samples without statistical difference by McNemar's test (P = 0.250).
TABLE 3.
Frequencies of qualitative HCV capture assay test results in 10-μl aliquots of 219 clinical plasma and whole-blood specimens
| Sample type and HCV capture assay result for HCV RNA | No. (%) of samples with the following amt of HCV RNA in plasma by the CAP/CTMa HCV test:
|
Total no. of samples | |||
|---|---|---|---|---|---|
| 0 (not detected) | <15 IU/ml | 15 to 900 IU/ml | >900 IU/ml | ||
| WB samples (10 μl) | |||||
| Detectedb | 1 (2) | 0 (0) | 10 (59) | 141 (100) | 152 |
| Not detected | 42 (98) | 8 (100) | 7 (41) | 0 (0) | 57 |
| Not tested | 3 | 0 | 0 | 7 | 10 |
| Plasma samples (10 μl) | |||||
| Detectedb | 1 (2) | 2 (25) | 13 (76) | 147 (100) | 163 |
| Not detected | 45 (98) | 6 (75) | 4 (24) | 0 (0) | 55 |
| Not tested | 0 | 0 | 0 | 1 | 1 |
CAP/CTM, COBAS AmpliPrep/COBAS TaqMan.
Five WB samples and six plasma samples gave inconclusive test results by the HCV capture assay (50% positive test results in duplicate samples), and these samples were included in the HCV RNA detected test results.
HCV RNA capture assay results were concordantly positive in both types of specimens for 151/211 samples; 54 were concordantly negative. Five samples were positive in plasma samples but not in the WB samples, and one sample was positive in the WB sample but negative in the plasma sample. The overall diagnostic sensitivity did not differ significantly between WB and plasma samples (P = 0.219, McNemar's test). The diagnostic specificity was 98% for WB and plasma samples, because one sample of each specimen set tested negative in the CAP/CTM assay but positive in the HCV capture assay. Repetition of measurements did not confirm positive capture assay results; therefore, a cross-contamination error cannot be excluded.
(ii) HCV RNA quantitation in plasma.
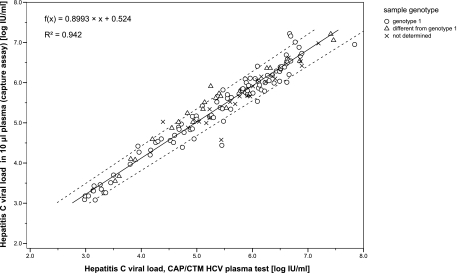
One hundred forty-six plasma samples showed concentrations of >900 IU/ml in the HCV RNA capture assay; 127 of them were quantified by single measurement and 19 in replicates. Figure 1 displays the correlation of quantitation results between the HCV RNA capture assay (10-μl plasma samples) and the CAP/CTM HCV test (850-μl plasma samples) as a scatter plot of log-transformed viral loads. The Passing-Bablok regression equation was f(x) = 0.8993x + 0.524 (R2 = 0.942). The regression slope was 0.8667 to 0.9289 (95% CI); the intercept was 0.3476 to 0.7047 (95% CI). The CUSUM test excluded a significant deviation from linearity (P > 0.10).
FIG. 1.
Correlation of HCV RNA quantitation results in 10-μl plasma samples and the CAP/CTM HCV plasma test. The viral loads are expressed in log10 IU/ml; the genotype is indicated if the information was available. The Passing-Bablok regression, 95% confidence intervals for slope and intercept, and coefficient of determination (R2) for all quantifiable results (n = 146) are indicated.
(iii) HCV RNA quantitation in venous WB samples.
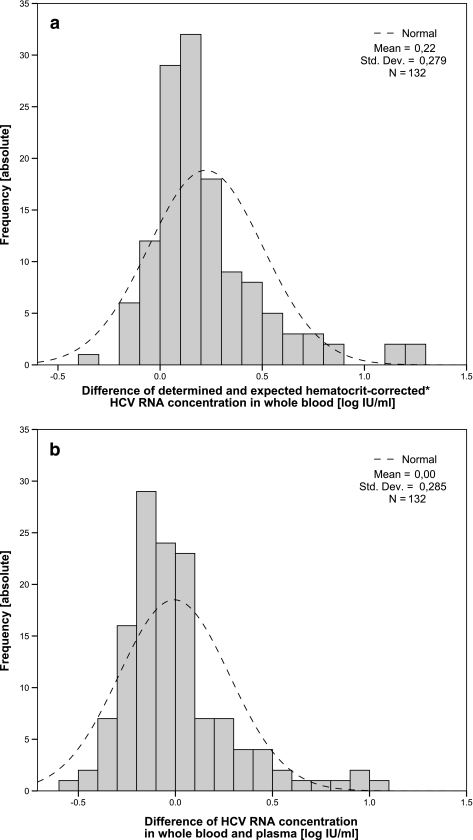
One hundred thirty-two pairs of plasma and WB specimens had quantifiable viral loads (>900 IU/ml) in the HCV RNA capture assay (118 quantified by single measurement; 14 in replicates). Hematocrits of samples ranged from 0.30 to 0.53. The one-sample KS test confirmed log-normal distribution (P > 0.05). The HCV RNA concentration in WB significantly exceeded the estimated viral load cplasma(1 − hematocrit) by 0.22 log10 unit (95% CI, 0.18 to 0.27 log10 unit; P < 0.001), thus indicating the presence of HCV RNA in the non-plasma fraction of WB (a histogram of the differences shown in Fig. 2a). The absolute HCV RNA concentrations in 10-μl WB and 10-μl plasma samples did not differ significantly (P = 0.804), with a mean difference of <0.01 log10 unit (95% CI, −0.06 to 0.04 log10 unit). In 79/132 samples, the HCV concentration was higher in plasma samples than in WB samples. Within an estimated bias of ±0.3 log10 unit, HCV RNA levels were identical or greater than those in plasma samples in 122/132 WB samples (a histogram of the differences shown in Fig. 2b).
FIG. 2.
Histogram of absolute frequency for HCV viral load deviation between measured whole-blood viral load and the estimated hematocrit-corrected whole-blood viral load (a) and measured whole-blood viral load and measured plasma viral load (b). All HCV RNA quantitation results were analyzed with the HCV RNA capture assay in 10-μl specimens and expressed in log10 IU/ml. The dashed line refers to the log-normal distribution curve. The estimated whole-blood viral load refers to the viral load that would be observed if the cellular fraction were free of HCV RNA. It was calculated by multiplication of viral load (in IU/ml) with (1 − hematocrit). Std. Dev., standard deviation.
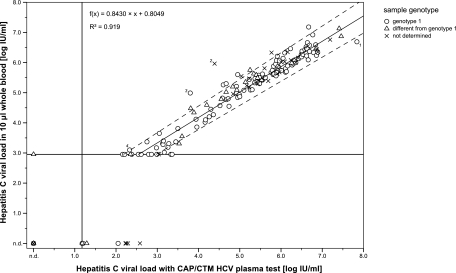
All quantifiable WB HCV RNA capture assay results were tested for correlation with the CAP/CTM HCV plasma test. The Passing-Bablok regression equation was f(x) = 0.8430x + 0.8049 with x as the log10-transformed plasma viral load. There was no significant deviation from linearity (P < 0.10 in the CUSUM linearity test). R2 was 0.919 for results with >900 IU/ml. The scatter plot in Fig. 3 displays the correlation of WB capture assay results and the CAP/CTM HCV test for all analyzed sample pairs (n = 209), the respective genotype, the regression line for quantifiable results (n = 140), and the 95% CI for the slope (0.8008 to 0.8846) and intercept (0.5731 to 1.0198).
FIG. 3.
Comparison of HCV RNA quantitation results in 10-μl whole-blood samples with CAP/CTM HCV plasma test. The viral loads are expressed in log10 IU/ml; the genotype is indicated if the information was available. The Passing-Bablok regression, 95% confidence intervals for slope and intercept, and coefficient of determination (R2) are shown for all quantifiable results (>900 IU/ml; n = 140). The vertical line represents the limit of detection for CAP/CTM, while the horizontal line is the 95% LOD for the capture assay. The numbers 1 to 4 with four points indicate sample pairs that fulfill the outlier criteria.
With the outlier criterion deviation of >1.96-fold SD (0.68 log10 unit), four sample pairs were defined. For one pair of specimens (outlier 1, see Fig. 3), HCV RNA was higher in plasma samples than in WB samples, while for the other three pairs of specimens, it was vice versa. The HCV concentration in plasma sample 1 was 7.84 log IU/ml in plasma with the reference method, 6.69 log IU/ml in WB, and 6.95 log IU/ml in plasma with the HCV RNA capture assay. An inhibition of the CAP/CTM quantitation standard (quantitation standard CT of 36.5) indicated an overestimation of plasma viral load. Outliers 2 and 3 were confirmed by intramethod comparison; the reference plasma viral load and intermethod and intramethod differences between plasma and WB samples were 4.39, 1.58, and 0.94 log10 unit, respectively, for outlier 2 and 3.80, 1.19, and 1.02 log10 unit, respectively, for outlier 3. Outlier 4 was below the HCV capture assay's limit of quantification in plasma samples but above the limit of quantification in WB samples and could therefore not be confirmed by intramethod analysis.
(iv) HCV RNA quantitation in capillary WB samples.
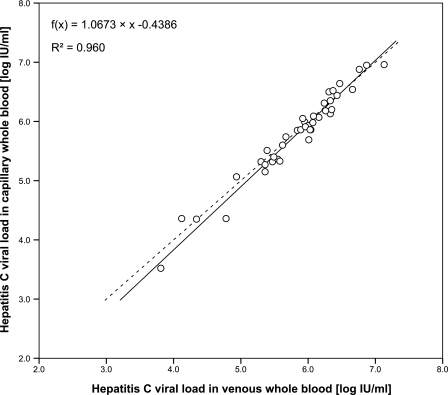
The correlation between quantitation results in capillary and venous WB samples is demonstrated in a scatter plot in Fig. 4. The Passing-Bablok regression equation was f(x) = 1.0673x − 0.4386 (R2 = 0.960) for log-transformed viral loads. Deviation between capillary and venous WB samples ranged from −0.42 to 0.24 log10 IU/ml. Thirty-eight of 40 sample pairs deviated less than 0.3 log10 unit. Thirty-six of 40 HCV RNA quantitation results in capillary WB samples were greater than 105 IU/ml, thus indicating an overrepresentation of samples with rather high viral loads.
FIG. 4.
Correlation of HCV RNA quantitation results in 10-μl capillary whole-blood samples and in 10-μl venous whole-blood samples expressed in log10 IU/ml. The Passing-Bablok regression and the coefficient of determination (R2) are indicated (n = 40).
DISCUSSION
Determination of hepatitis C viral load kinetics is the essential parameter for the therapeutic management of chronic hepatitis C (26). Because quantitation of HCV RNA in plasma or serum samples is a time-consuming and costly procedure performed in central laboratories, a HCV RNA quantitation assay for 10-μl samples appropriate for capillary WB point-of-care diagnostic devices was developed.
The findings of this study demonstrate that HCV RNA quantitation in 10-μl WB samples correlate very well (R2 = 0.919) with the quantitation results of the fully automated CAP/CTM HCV plasma test for plasma concentrations above 900 IU/ml. The observed correlation coefficient (r = 0.958) is located within the reported range of correlation of the CAP/CTM HCV test to other commercially available HCV quantitation plasma assays, such as the Roche COBAS Amplicor Monitor 2.0 (r, 0.92 to 0.97) (34). The application as a qualitative assay for screening purposes is restricted by the analytical sensitivity of the assay, since it has been reported that every 40th analyzed plasma sample contains less than 600 IU/ml HCV RNA (11). Even though sensitivity and precision decrease with sample volume reduction (6), a 95% LOD of 9 IU per 10-μl sample (900 IU/ml) and a variability below 0.13 log10 IU/ml in plasma was demonstrated despite manual handling.
Correct interpretation of differences in diagnostic sensitivity for plasma and WB samples requires validation of the analytical sensitivity and demonstration of matrix equivalence. We demonstrated matrix equivalence for artificially created plasma and WB samples because of the lack of commercially available standardized WB samples and showed a direct proportion (regression slope of 1.00) without significant deviation between both types of specimens (mean deviation of 0.04 log10 unit; P = 0.203).
The HCV RNA concentration in clinical WB samples significantly exceeded the estimated (hematocrit-corrected) concentration by 0.22 log10 unit. This indirectly indicates that significant amounts of HCV RNA cannot be mobilized into the plasma fraction by means of standard WB centrifugation. Whether this amount of HCV RNA is intracellular or can be concentrated by ultracentrifugation, as suggested previously for occult HCV infection (1), has not been validated. An indirect arithmetic correction was applied to determine the amount of nonplasma HCV RNA, because direct analysis of the cellular fraction is highly dependent on the stringency of washing and may remove cell-associated HCV RNA (32).
This study demonstrates that the additional cellular fraction of HCV RNA on average compensates only for the loss of plasma volume, because the mean HCV RNA concentration in 10-μl WB samples did not differ from 10-μl plasma samples by the same method (P = 0.804). In only 4/132 clinical samples did HCV RNA in WB samples exceed the concentration in plasma by an order of magnitude. Furthermore, diagnostic sensitivity was not increased by the use of WB; it was 100% for WB and for plasma in plasma samples above the 95% LOD. In our series, the overall sensitivity (regardless of the analytical sensitivity) was identical in plasma and WB samples (90% in WB samples versus 94% in plasma samples). Cook et al. (8) also described equal sensitivities for WB and serum analysis by a competitive RT-PCR method for different extraction methods.
The selection of patients is crucial for correct interpretation of the results. In our study, exclusively anti-HCV-positive patients were recruited regardless of their current therapeutic status. In a different anti-HCV-negative patient population that was HCV RNA positive by liver biopsy specimens, the sensitivity in WB samples was 14%, whereas for PBMC analysis, it was 57% (4). These data suggest PBMC as a possible origin of HCV RNA in the non-plasma fraction in patients with occult hepatitis C. The relevance of PBMC-associated HCV RNA in patients that are HCV RNA positive in plasma seems rather small. Less than 8% of PBMC from chronically HCV-infected patients were shown to contain HCV RNA: 0.02 to 0.15% by nested RT-PCR (42), 0.01% by in situ hybridization (24), and 0.2% to 8.1% by in situ RT-PCR (25). On average, HCV RNA concentrations in PBMC of therapy-naïve patients range from 10−5 to 30 IU/cell with a median of <0.01 IU/cell as illustrated by Pugnale and coworkers (29) in their Fig. 2. Assuming a PBMC concentration of 1.5 × 106 to 5 × 106 cells per ml of WB, the approximate median amount of additional PBMC-derived HCV RNA is 30,000 IU per ml WB (range, 15 IU/ml to 1.5 × 108 IU/ml). Because approximate median PBMC-derived HCV RNA and plasma-derived HCV RNA are of the same order of magnitude in therapy-naïve patients (104 to 107 IU/ml), a significant increase in WB viral load due to PBMC-associated HCV RNA seems unlikely. Furthermore, during antiviral therapy, the median viral decline in plasma and in PBMC does not differ (29), thus indicating a direct proportion of plasma and WB viral load in HCV RNA-positive patients. Our findings suggest that HCV RNA that is presumably associated to other cellular fractions, such as erythrocytes, does not significantly increase the WB viral load. Although erythrocyte-associated HCV RNA has been detected by an in situ PCR technique (21), it was not confirmed by Sadallah, Hess, and colleagues (32), who demonstrated erythrocyte association for human immunodeficiency virus type 1 (15).
We observed a proportional bias between HCV RNA capture assay results in WB and CAP/CTM results in plasma as indicated by a slope of 0.843 (significantly different from 1) in the Passing-Bablok regression, thus underestimating WB viral loads >1.3 × 105 IU/ml as analyzed by Bland-Altman blotting (2) (data not shown). The same observation was made for plasma specimens (slope of 0.8993; cutoff of 1.59 × 105 IU/ml), although the assay linearity was shown to be at least 5 × 106 IU/ml plasma. Therefore, this bias is neither matrix dependent nor due to nonlinearity of the capture assay. An overestimation of high plasma viral loads by the CAP/CTM HCV test is possible, as it has also been reported by the French National Reference Center for Viral Hepatitis (7) in their analysis of serial dilutions.
The specificity of our HCV RNA quantitation protocol was not compromised by the use of WB samples. The risk of artifactual amplification of human cellular nucleic acids was decreased by the use of real-time PCR and a sequence-specific extraction. Because thermal stability of RNA/DNA hybrids is decreased under chaotropic conditions (16), LNA-modified 2′-OMeRNA hybridization probes were applied for HCV RNA extraction due to superiority to a commercially available silica-based spin filter protocol (Roche high pure nucleic acid kit). Capture probes, primers, and TaqMan probes were designed to hybridize in the highly conserved 5′-untranslated region of HCV. Although mismatches between the capture probe and reported HCV sequence theoretically occur in 20.5% of assays, according to 337 reported HCV sequences in the Los Alamos hepatitis C sequence database (17), all tested genotypes were equally detected in plasma with a maximum deviation of 0.21 log10 unit. In addition, HCV RNA quantities in capillary and venous WB samples were identical (R2 = 0.960; n = 40). Thus, capillary HCV RNA viral load monitoring with this novel method may be a promising alternative to venous plasma analysis for monitoring early HCV viral load kinetics with plasma viral loads above 900 IU/ml.
Acknowledgments
Tony Bruns was a full-time employee of Clondiag GmbH, Jena, Germany from 2007 to 2008. Katrin Steinmetzer and Eugen Ermantraut are currently full-time employees of Clondiag GmbH Jena and shareholders of Inverness Medical Innovation.
Footnotes
Published ahead of print on 19 August 2009.
REFERENCES
- 1.Bartolomé, J., J. M. López-Alcorocho, I. Castillo, E. Rodríguez-Iñigo, J. A. Quiroga, R. Palacios, and V. Carreño. 2007. Ultracentrifugation of serum samples allows detection of hepatitis C virus RNA in patients with occult hepatitis C. J. Virol. 81:7710-7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed] [Google Scholar]
- 3.Bouffard, P., P. H. Hayashi, R. Acevedo, N. Levy, and J. B. Zeldis. 1992. Hepatitis C virus is detected in a monocyte/macrophage subpopulation of peripheral blood mononuclear cells of infected patients. J. Infect. Dis. 166:1276-1280. [DOI] [PubMed] [Google Scholar]
- 4.Carreño, V., I. Castillo, J. Bartolomé, E. Rodríguez-Iñigo, N. Ortiz-Movilla, S. de Lucas, and M. Pardo. 2004. Comparison of hepatitis C virus RNA detection in plasma, whole blood and peripheral blood mononuclear cells of patients with occult hepatitis C virus infection. J. Clin. Virol. 31:312-313. [DOI] [PubMed] [Google Scholar]
- 5.Castillo, I., M. Pardo, J. Bartolomé, N. Ortiz-Movilla, E. Rodríguez-Iñigo, S. de Lucas, C. Salas, J. A. Jiménez-Heffernan, A. Pérez-Mota, J. Graus, J. M. López-Alcorocho, and V. Carreño. 2004. Occult hepatitis C virus infection in patients in whom the etiology of persistently abnormal results of liver-function tests is unknown. J. Infect. Dis. 189:7-14. [DOI] [PubMed] [Google Scholar]
- 6.Chen, G., L. Kobayashi, and I. Nazarenko. 2007. Effect of sample aliquot size on the limit of detection and reproducibility of clinical assays. Clin. Chem. 53:1962-1965. [DOI] [PubMed] [Google Scholar]
- 7.Chevaliez, S., M. Bouvier-Alias, R. Brillet, and J. Pawlotsky. 2007. Overestimation and underestimation of hepatitis C virus RNA levels in a widely used real-time polymerase chain reaction-based method. Hepatology 46:22-31. [DOI] [PubMed] [Google Scholar]
- 8.Cook, L., A. M. Ross, G. B. Knight, and V. Agnello. 2000. Use of whole blood specimens for routine clinical quantitation of hepatitis C virus RNA does not increase assay sensitivity. J. Clin. Microbiol. 38:4326-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotten, M., B. Oberhauser, H. Brunar, A. Holzner, G. Issakides, C. R. Noe, G. Schaffner, E. Wagner, and M. L. Birnstiel. 1991. 2′-O-methyl, 2′-O-ethyl oligoribonucleotides and phosphorothioate oligodeoxyribonucleotides as inhibitors of the in vitro U7 snRNP-dependent mRNA processing event. Nucleic Acids Res. 19:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, H. D. J., J. David, P. R. Grant, J. A. Garson, G. M. Chandy, and P. Abraham. 2008. Whole blood as an alternative to plasma for detection of hepatitis C virus RNA. J. Clin. Microbiol. 46:3791-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fytili, P., C. Tiemann, C. Wang, S. Schulz, S. Schaffer, M. P. Manns, and H. Wedemeyer. 2007. Frequency of very low HCV viremia detected a highly sensitive HCV-RNA assay. J. Clin. Virol. 39:308-311. [DOI] [PubMed] [Google Scholar]
- 12.Gleaves, C. A., J. Welle, M. Campbell, T. Elbeik, V. Ng, P. E. Taylor, K. Kuramoto, S. Aceituno, E. Lewalski, B. Joppa, L. Sawyer, C. Schaper, D. McNairn, and T. Quinn. 2002. Multicenter evaluation of the Bayer VERSANT HIV-1 RNA 30 assay: analytical and clinical performance. J. Clin. Virol. 25:205-216. [DOI] [PubMed] [Google Scholar]
- 13.Hakim, H., C. Gibson, J. Pan, K. Srivastava, Z. Gu, M. J. Bankowski, and R. T. Hayden. 2007. Comparison of various blood compartments and reporting units for the detection and quantification of Epstein-Barr virus in peripheral blood. J. Clin. Microbiol. 45:2151-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamaia, S., C. Li, and J. P. Allain. 2001. The dynamics of hepatitis C virus binding to platelets and 2 mononuclear cell lines. Blood 98:2293-2300. [DOI] [PubMed] [Google Scholar]
- 15.Hess, C., T. Klimkait, L. Schlapbach, V. D. Zenero, S. Sadallah, E. Horakova, G. Balestra, V. Werder, C. Schaefer, M. Battegay, and J. Schifferli. 2002. Association of a pool of HIV-1 with erythrocytes in vivo: a cohort study. Lancet 359:2230-2234. [DOI] [PubMed] [Google Scholar]
- 16.Jacobsen, N., P. S. Nielsen, D. C. Jeffares, J. Eriksen, H. Ohlsson, P. Arctander, and S. Kauppinen. 2004. Direct isolation of poly(A)+ RNA from 4 M guanidine thiocyanate-lysed cell extracts using locked nucleic acid-oligo(T) capture. Nucleic Acids Res. 32:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21:379-384. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, R., S. K. Singh, A. A. Koshkin, V. K. Rajwanshi, M. Meldgaard, and J. Wengel. 1998. The first analogues of LNA (locked nucleic acids): phosphorothioate-LNA and 2′-thio-LNA. Bioorg. Med. Chem. Lett. 8:2219-2222. [DOI] [PubMed] [Google Scholar]
- 19.Lavanchy, D. 2009. The global burden of hepatitis C. Liver Int. 29(Suppl.):174-181. [DOI] [PubMed] [Google Scholar]
- 20.Limpert, E., W. A. Stahel, and M. Abbt. 2001. Log-normal distributions across the sciences: keys and clues. BioScience 51:341-352. [Google Scholar]
- 21.Lotz, G., F. Szalay, G. Firneisz, M. Abonyi, G. Lengyel, L. Telegdy, E. Ibrányi, B. Nemes, F. Kury, Z. Schaff, and S. Simon. 2002. Localization of hepatitis C virus RNA on human erythrocytes RT in situ PCR technique. Scand. J. Gastroenterol. 37:578-584. [DOI] [PubMed] [Google Scholar]
- 22.Manzin, A., L. Solforosi, M. Candela, G. Cherubini, G. Piccinini, M. Brugia, A. Gabrielli, and M. Clementi. 1996. Hepatitis C virus infection and mixed cryoglobulinaemia: assessment of HCV RNA copy numbers in supernatant, cryoprecipitate and non-liver cells. J. Viral. Hepat. 3:285-292. [DOI] [PubMed] [Google Scholar]
- 23.Meier, V., S. Mihm, P. B. Wietzke, and G. Ramadori. 2001. HCV-RNA positivity in peripheral blood mononuclear cells of patients with chronic HCV infection: does it really mean viral replication? World J. Gastroenterol. 7:228-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moldvay, J., P. Deny, S. Pol, C. Brechot, and E. Lamas. 1994. Detection of hepatitis C virus RNA in peripheral blood mononuclear cells of infected patients in situ hybridization. Blood 83:269-273. [PubMed] [Google Scholar]
- 25.Muratori, L., D. Gibellini, M. Lenzi, M. Cataleta, P. Muratori, M. C. Morelli, and F. B. Bianchi. 1996. Quantification of hepatitis C virus-infected peripheral blood mononuclear cells by in situ reverse transcriptase-polymerase chain reaction. Blood 88:2768-2774. [PubMed] [Google Scholar]
- 26.National Institutes of Health. 2002. NIH Consensus Statement on Management of Hepatitis C: 2002. NIH Consens. State. Sci. Statements 19:1-46. [PubMed] [Google Scholar]
- 27.Nishiguchi, S., K. Fukuda, S. Shiomi, T. Takeda, T. Kuroki, M. Ogami, H. Morimoto, S. Otani, M. Sakurai, and A. Matsuhisa. 2003. Peripheral blood mononuclear cells are possible extrahepatic replication sites for hepatitis C virus. Hepatogastroenterology 50:1301-1304. [PubMed] [Google Scholar]
- 28.Passing, H., and W. Bablok. 1983. A new biometrical procedure for testing the equality of measurements from two different analytical methods. Application of linear regression procedures for method comparison studies in clinical chemistry. J. Clin. Chem. Clin. Biochem. 21:709-720. [DOI] [PubMed] [Google Scholar]
- 29.Pugnale, P., E. Herrmann, A. U. Neumann, J. Pawlotsky, S. W. Schalm, C. Ferrari, Y. Homburger, S. Zeuzem, F. Negro, and DITTO-HCV Study Group. 2008. Hepatitis C viral kinetics in plasma and peripheral blood mononuclear cells during pegylated interferon-alpha2a/ribavirin therapy. J. Hepatol. 48:932-938. [DOI] [PubMed] [Google Scholar]
- 30.Radkowski, M., J. F. Gallegos-Orozco, J. Jablonska, T. V. Colby, B. Walewska-Zielecka, J. Kubicka, J. Wilkinson, D. Adair, J. Rakela, and T. Laskus. 2005. Persistence of hepatitis C virus in patients successfully treated for chronic hepatitis C. Hepatology 41:106-114. [DOI] [PubMed] [Google Scholar]
- 31.Roque-Afonso, A. M., J. Jiang, F. Penin, C. Tareau, D. Samuel, M. A. Petit, H. Bismuth, E. Dussaix, and C. Féray. 1999. Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets. J. Virol. 73:9213-9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadallah, S., M. Heim, C. Hess, T. Klimkait, M. Battegay, and J. A. Schifferli. 2005. Contrary to HIV, hepatitis C virus is not associated with erythrocytes in vivo. J. Hepatol. 42:150-152. [DOI] [PubMed] [Google Scholar]
- 33.Saldanha, J., N. Lelie, A. Heath, and WHO Collaborative Study Group. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 34.Sarrazin, C., B. C. Gärtner, D. Sizmann, R. Babiel, U. Mihm, W. P. Hofmann, M. von Wagner, and S. Zeuzem. 2006. Comparison of conventional PCR with real-time PCR and branched DNA-based assays for hepatitis C virus RNA quantification and clinical significance for genotypes 1 to 5. J. Clin. Microbiol. 44:729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, W. N., P. Wu, J. Q. Han, M. J. Perino, D. R. LaBrecque, and J. T. Stapleton. 1997. Distribution of hepatitis C virus (HCV) RNA in whole blood and blood cell fractions: plasma HCV RNA analysis underestimates circulating virus load. J. Infect. Dis. 176:20-26. [DOI] [PubMed] [Google Scholar]
- 36.Sizmann, D., C. Boeck, J. Boelter, D. Fischer, M. Miethke, S. Nicolaus, M. Zadak, and R. Babiel. 2007. Fully automated quantification of hepatitis C virus (HCV) RNA in human plasma and human serum by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 38:326-333. [DOI] [PubMed] [Google Scholar]
- 37.Stapleton, J. T., D. Klinzman, W. N. Schmidt, M. A. Pfaller, P. Wu, D. R. LaBrecque, J. Han, M. J. Phillips, R. Woolson, and B. Alden. 1999. Prospective comparison of whole-blood- and plasma-based hepatitis C virus RNA detection systems: improved detection using whole blood as the source of viral RNA. J. Clin. Microbiol. 37:484-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watkins-Riedel, T., P. Ferenci, P. Steindl-Munda, M. Gschwantler, C. Mueller, and M. Woegerbauer. 2004. Early prediction of hepatitis C virus (HCV) infection relapse in nonresponders to primary interferon therapy means of HCV RNA whole-blood analysis. Clin. Infect. Dis. 39:1754-1760. [DOI] [PubMed] [Google Scholar]
- 39.Xu, D., Y. Xie, and Z. Li. 2005. Clearance of HCV RNA in peripheral blood mononuclear cell as a predictor of response to antiviral therapy in patients with chronic hepatitis C. Hepatobiliary Pancreat. Dis. Int. 4:550-553. [PubMed] [Google Scholar]
- 40.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zahn, A., N. Jennings, W. H. Ouwehand, and J. Allain. 2006. Hepatitis C virus interacts with human platelet glycoprotein VI. J. Gen. Virol. 87:2243-2251. [DOI] [PubMed] [Google Scholar]
- 42.Zehender, G., L. Meroni, C. D. Maddalena, S. Varchetta, G. Monti, and M. Galli. 1997. Detection of hepatitis C virus RNA in CD19 peripheral blood mononuclear cells of chronically infected patients. J. Infect. Dis. 176:1209-1214. [DOI] [PubMed] [Google Scholar]
- 43.Zignego, A. L., C. Giannini, M. Monti, and L. Gragnani. 2007. Hepatitis C virus lymphotropism: lessons from a decade of studies. Dig. Liver. Dis. 39(Suppl.):S38-S45. [DOI] [PubMed] [Google Scholar]