Abstract
Most of the members of the genus Bifidobacterium, including the related organism Alloscardovia omnicolens, are inhabitants of the gastrointestinal tract and oral cavity of humans and animals and have been considered nonpathogenic for humans. However, the actual site of isolation and the clinical significance of A. omnicolens and of Bifidobacterium species are unclear. This may be due in part to the difficulties in distinguishing these organisms from other genera such as Actinomyces. To determine the potential disease-causing role of these organisms, we analyzed the clinical significance of 15 A. omnicolens and Bifidobacterium isolates identified by 16S rRNA gene sequencing from a clinical laboratory. All of the organisms in this study were isolated from sterile sites or in significant numbers by standard clinical microbiological culture methods. Our 15 clinical strains fit into only four species: A. omnicolens (five isolates), Bifidobacterium scardovii (four isolates), B. longum (two isolates), and B. breve (four isolates). All five A. omnicolens isolates, one of the B. breve isolates, and three of the four B. scardovii isolates were cultured from urine at 105 CFU/ml. One B. scardovii isolate was from a patient with a genitourinary tract wound infection, two B. longum isolates were from abdominal wounds, and three B. breve isolates were from blood cultures. This study enlarges the spectrum of diseases and clinical sources associated with A. omnicolens and Bifidobacterium species and addresses identification problems.
Many bifidobacteria are reported as commensals in humans, particularly in the gastrointestinal tract and the oral cavity (1, 12). Bifidobacteria are also reported as present in other ecological niches, such as the intestinal tracts of insects and other animals, in sewage, and in food (1, 12). It has been proposed that bifidobacteria are important for the health of the human gastrointestinal tract (2, 5). Some of the species, such as Bifidobacterium breve, B. longum, B. infantis, and B. lactis, are used as probiotics for putative health benefits in the human gastrointestinal tract (2, 5). The actual impact of the ingestion of probiotics is still unclear. It is also not clear whether species which are used as probiotics are different from normal flora species that have been associated with disease.
The clinical significance of bifidobacteria is also unknown. Since these organisms appear to be important for the health of the human gastrointestinal tract, they have long been thought to be nonpathogenic. Three species, B. dentium, B. inopinatum, and B. denticolens, have been found in dental caries, and B. dentium may actually contribute to pathogenicity in these cases (4, 10). B. inopinatum and B. denticolens have since been renamed Scardovia inopinatum and Parascardovia denticolens, respectively (8). B. scardovii has been isolated from human blood, urine, and a hip specimen, but its clinical relevance is unknown (6). Alloscardovia omnicolens is related to bifidobacteria and has been isolated from urine, blood, the oral cavity, a urethral specimen, a tonsil specimen, and a lung and aortic abscess; however, actual clinical significance in these cases is not clear (7). However, Bifidobacterium species and A. omnicolens are difficult to identify and may be missed in specimens by many laboratories. In this study, we correlated the associated diseases and source of site of clinical isolate with the genetic identification for strains of A. omnicolens and Bifidobacterium species and discuss the pathogenic potential of these organisms.
MATERIALS AND METHODS
Bacterial strains.
All of the organisms in this study were isolated from 2000 to 2007 at either the Veterans Affairs Medical Center in Houston, TX, or the Veterans Affairs Puget Sound Health Care System in Seattle, WA. Early in the study, the urine strains were isolated when one of us was investigating the fastidious organisms occurring in urine which were not detected by culture but might affect the leukocyte esterase/nitrate screening tests and might be associated with disease (3). These organisms were presumptively identified as Actinomyces sp., Gardnerella vaginalis, Mobiluncus sp., or unidentified gram-positive rods since Bifidobacterium spp. are usually thought to be anaerobes that do not grow in CO2. The nonurine isolates were identified because they occurred at sterile sites and/or were deemed of possible clinical significance. In each case, patient information associated with each strain sufficient to assess clinical significance was obtained, if available. See Table 1 for a list of the strains used in this study and the clinical sites from which they were isolated.
TABLE 1.
Bifidobacterium and A. omnicolens isolates used in this study, including the sites of isolation and the original identification of each before 16S rRNA gene sequencing
| Isolate | Original identification and method | Clinical source | GenBank accession no. |
|---|---|---|---|
| A. omnicolens Ref104 | Unidentified GPRa (Gram stain and culture characteristics) | Urine | EU743947 |
| A. omnicolens F1074 | GPR most closely resembling Mobiluncus sp. (RapID ANA II code, 471771; API Coryne code, 0450721) | Urine | EU743948 |
| A. omnicolens F2755 | Variable GPR (Gram stain and culture characteristics; RapID ANA II code, 471771; API Coryne code, 0450721) | Urine | EU743949 |
| A. omnicolens F2849 | Lactobacillus sp. (Gram stain and culture characteristics; RapID ANA II code, 471771; API Coryne code, 0450721) | Urine | EU743950 |
| A. omnicolens F3468 | Unidentified GPR (Gram stain and culture characteristics) | Urine | EU743951 |
| B. scardovii F3319 | Actinomyces sp. (API Coryne strip code, not available) | Urine | EU743952 |
| B. scardovii VAMC 6345 | Unknown GPR resembling Actinomyces sp. (Gram stain and culture characteristics) | Urine | EU743953 |
| B. scardovii S2870 | A. israelii (Gram stain and culture characteristics) | Cervix/uterus exudate | EU743954 |
| B. scardovii F961 | Anaerobic GPR, not a known pathogen (Gram stain and culture characteristics; RapID ANA II code, 677673; API Coryne code, 6460361) | Urine | EU743955 |
| B. longum F1519 | Actinomyces sp., not A. israelii (RapID ANA II code, not available) | Peritoneal wound | EU743956 |
| B. longum F492 | Actinomyces sp., possibly A. israelii (RapID ANA II code, 425671; API Coryne code, 2450561) | Appendix wound | EU743957 |
| B. breve S7365 | Bifidobacterium sp. (RapID ANA II code, not available) | Blood | EU743958 |
| B. breve VAMC11797 | Actinomyces sp. (Gram stain and culture characteristics) | Urine | EU743959 |
| B. breve 220119 S38 | Actinomyces sp. (Gram stain and culture characteristics) | Blood | EU743960 |
| B. breve TBAC519720 | Unidentified microaerophilic GPR (Gram stain and culture characteristics) | Blood | EU743961 |
GPR, gram-positive rod.
Culture conditions.
Urine specimens were inoculated onto Columbia agar with 5% sheep blood (BA), Columbia colistin nalidixic acid agar (CNA), modified chocolate agar (CA), and MacConkey agar (Mac) with a 1-μl loop (all media were from Remel, Lenexa, KS). The CA and CNA plates were incubated at 35°C with additional CO2 (7 to 8%) and time (48 h). The BA and Mac plates were incubated at 35°C in air for 18 to 24 h. The CNA and CA plates of negative cultures were reincubated at 35°C with added CO2 (7 to 8%) and read at 7 days. After a direct Gram stain was performed, wound specimens were also inoculated onto BA, Mac, CA, and CNA plates as described above. If no organisms were observed on the direct Gram stain, no growth was reported after 48 h for wound specimens. If organisms were observed on the direct Gram stain, plates were held for up to 7 days under these conditions to recover slow-growing or fastidious organisms. In addition, wound specimens were inoculated onto the following anaerobic media: brain heart infusion agar (double-pour plate with blood), phenylethyl alcohol blood agar with vitamin K, and Bacteroides fragilis isolation agar (all anaerobic media were from Remel). The anaerobic plates were incubated at 35°C in jars in an atmosphere of 18 to 20% CO2/balance N2 generated with the AnaeroPack system (Mitsubishi Gas Company America, New York, NY). Anaerobic plates were checked at 24 h, 48 h, and 5 days for growth; plates were also held, if necessary, for up to 7 days if organisms were observed on the direct Gram stain. Blood culture specimens were submitted to the laboratory in BacT/Alert FN (anaerobic, with charcoal) and BacT/Alert SA (aerobic) media (bioMérieux, Durham, NC) and incubated in a BacT/Alert 3D system (bioMérieux). Positive specimens flagged by the system were Gram stained and inoculated onto BA, Mac, CA, CNA, and brain heart infusion agar and incubated and read the same as wound specimen plates.
Bacterial identification.
Light microscopy and biochemical tests, such as the catalase test, were initially used to identify isolates. In some cases, either a RapID ANA II strip (Remel, Lenexa, KS) or an API Coryne strip (bioMérieux, Marcy l'Etoile, France) were set up in an attempt to identify isolates. Suspected G. vaginalis isolates were stabbed into a Gardnerella agar plate (PML Microbiologicals, Wilsonville, OR) to observe beta-hemolysis.
DNA isolation and 16S rRNA gene sequencing.
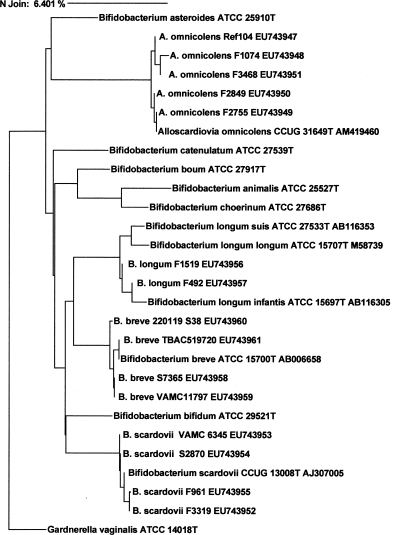
Nucleic acid was extracted from bacterial cultures with the UltraClean Microbial DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA). The initial PCR was performed as previously described (9). Cycle sequencing was performed at a core sequencing facility, and the resulting electropherograms were analyzed by using the MicroSeq database. Comparison strains were sought in the NCBI, MicroSeq, and BIBI databases. The neighbor-joining (NJ) method was used to generate a dendrogram to determine strain relatedness (Fig. 1).
FIG. 1.
Dendrogram relationships by 16S rRNA gene similarity of the A. omnicolens and Bifidobacterium species isolates in this study. Complete names with strain designations in the dendrogram are for sequences in the MicroSeq database. Complete names with strain designations and GenBank accession numbers are for sequences in the NCBI database. Sequences of the strains in this study are indicated by abbreviated names with strain designations and GenBank accession numbers. G. vaginalis is used as the outgroup.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the isolates used in this study are EU743947 to EU743961 (Table 1).
RESULTS
A total of 15 isolates of four species (A. omnicolens, B. scardovii, B. longum, and B. breve) were isolated (Table 1). Only one of these isolates, B. breve strain S7365, was identified correctly to the genus level by phenotypic methods (Table 1). In this study, 16S rRNA gene sequencing was necessary to identify all of the other isolates to both the genus and species levels. We found G. vaginalis and some species of the Actinomyces, Lactobacillus, and Bifidobacterium/Alloscardovia genera are morphologically similar, are catalase negative, grow better or equally well under anaerobic and CO2 atmospheric conditions, and have similar small colonies on blood agar and CNA (see Table 3). We examined whether the Bifidobacterium/Alloscardovia group could be differentiated from G. vaginalis by using the human blood hemolysis test. None of the Bifidobacterium/Alloscardovia isolates tested were beta-hemolytic on Gardnerella agar. Lactobacillus species can be differentiated from Bifidobacterium/Alloscardovia species, G. vaginalis, and Actinomyces species because they are usually resistant to vancomycin and have a more distinct Gram stain morphology (see Table 3). We concluded that it is difficult to phenotypically separate Actinomyces species from Bifidobacterium/Alloscardovia species, and in these cases, 16S rRNA gene sequencing is necessary for full identification.
TABLE 3.
Differential characteristics of some catalase-negative, nonmotile, similarly shaped, gram-positive rodsa
| Organism | Vancomycin | Hemolysis | Hippurate hydrolysis | Nitrate reduction | Esculin hydrolysis | Gram stain morphology |
|---|---|---|---|---|---|---|
| Bifidobacterium sp. | S | − | + | − | + | Short, irregularly shaped rods, may have rudimentary branching |
| A. omnicolens | S | − | − | − | + | Short, irregularly shaped rods, may have rudimentary branching |
| Actinomyces sp. | S | − | V | V | V | Short, irregularly shaped rods, may have rudimentary branching |
| Lactobacillus sp. | R/V | Alpha | − | V | V | Most are long regular-chaining rods; some are C shaped |
| G. vaginalis | S | −b | + | − | NA | Small, gram-variable rods |
There were five A. omnicolens isolates, and all were recovered from urine culture specimens (Table 1). We estimate the frequency of A. omnicolens isolated in urine to be less than 0.2% of the total isolates (3 out of 2,005 total urine isolates in a 1-year period). We reported earlier that about 3% of the urine isolates were in the Actinomyces or Gardnerella group, and when we examined these isolates more thoroughly, we found about 1 in 20 of these actually to be Bifidobacterium sp. or A. omnicolens (3).
Two of the isolates, A. omnicolens F2755 and F2849, were recovered from the same patient approximately 2 months apart (Table 2). This patient had underlying bladder cancer and concurrent urinary tract infection (UTI) (Table 2). A. omnicolens was recovered at >105 CFU/ml from both cultures from this patient and was the only organism isolated in the presence of an increased white blood cell (WBC) count by urinalysis (UA). A. omnicolens is nitrate reduction negative, so it is expected that nitrite would not be detected in urine samples from which this organism is cultured (7). The RapID ANA II panel code for both isolates was 471771 (99.8% identical to Actinomyces israelii) (Table 1). A code of 0450721 (unacceptable profile, closest to G. vaginalis) was obtained with the API Coryne strip for both isolates (Table 1). Treatment information was not available for this patient. A. omnicolens strain F3468 was isolated as the only organism at >105 CFU/ml from a third patient with T-cell lymphoma, although the UA of this patient was normal (Table 2). The patient's physician felt that the patient had a “possible UTI” on presentation, although the patient was not treated for a UTI. In the fourth case, A. omnicolens was recovered as the only organism at >105 CFU/ml but was not associated with a UTI, so the patient was not treated (strain F1074, Table 2). This isolate had the same RapID ANA II and API Coryne strip profiles as isolates F2755 and F2849 (Table 1). No clinical data were available for the other A. omnicolens isolate, strain Ref104, which was described as an unidentified gram-positive rod when it was initially recovered.
TABLE 2.
Bifidobacterium and A. omnicolens isolates, their culture and other laboratory characteristics, and their clinical correlations
| Isolate | Laboratory result(s) | Clinical correlation |
|---|---|---|
| A. omnicolens Ref104 | Not available | Not available |
| A. omnicolens F1074 | >105 CFU/ml; UA normal | Gastroesophageal reflux disease; not felt to be UTI |
| A. omnicolens F2755 | >105 CFU/ml; UA hazy, leukocyte esterase 2+, nitrite negative, no bacteria, 11-30 WBCs/HPFa | Bladder cancer, UTI |
| A. omnicolens F2849 | Same patient as F2755, 2 mo apart; >105 CFU/ml; UA hazy, leukocyte esterase 3+, nitrite negative, no bacteria, 72 WBCs/HPF | Bladder cancer, UTI |
| A. omnicolens F3468 | >105 CFU/ml; UA normal | T-cell lymphoma, possible UTI |
| B. scardovii F3319 | >105 CFU/ml; UA leukocyte esterase trace, nitrite negative, bacterium positive, 5-10 WBCs/HPF | Dementia, possible UTI |
| B. scardovii VAMC 6345 | Not available | Not available |
| B. scardovii S2870 | Not available | Not available |
| B. scardovii F961 | >105 CFU/ml; UA leukocyte esterase trace, nitrite negative, bacterium positive, 0-5 WBCs/HPF | Multiple sclerosis, pancreatic insufficiency; not felt to be UTI |
| B. longum F1519 | 104 CFU, mixed with C. clostridioforme, C. glycolyticum, and P. oris | Tissue from bowel resection, severe abdominal and back pain |
| B. longum F492 | 4+, mixed with C. clostridioforme, B. thetaiotaomicron, and B. fragilis group | Right lower quadrant fasciitis and myositis |
| B. breve S7365 | Not available | History of decubitis ulcers, frequent admissions for recurrent UTIs |
| B. breve VAMC11797 | >105 CFU/ml | Only isolate in urine; probable UTI |
| B. breve 220119 S38 | Blood culture positive, wound culture positive (left lower quadrant abdominal) with Actinomyces at same time | Peritonitis |
| B. breve TBAC519720 | Blood culture positive; isolated with B. vulgatus and Fusobacterium spp. | Stage B prostate cancer, ileal resection; thought to be transient bacteremia |
HPF, high-power field.
There were four B. scardovii isolates, and three were cultured from urine; the fourth was recovered from a cervix/uterus exudate specimen (Table 1). There are no clinical data available for two of the isolates (strains VAMC 6345 and S2870). B. scardovii strain F961 was isolated at >105 CFU/ml from a patient with multiple sclerosis and pancreatic insufficiency (Table 2). The UA of this patient was slightly abnormal (trace leukocyte esterase, bacteria observed in the microscopy sample), but there was no documented UTI. This patient was not treated for a UTI. A profile of 677673 (no identification) was obtained with the RapID ANA II strip, and the API Coryne strip yielded a profile of 6460361 (96.6% Arcanobacterium haemolyticum) (Table 1). B. scardovii strain F3319 was isolated from a patient with an abnormal UA as the only organism recovered at >105 CFU/ml, although treatment information was not available for this patient.
Both B. longum isolates were from patients with mixed bacterial abdominal wound infections (Table 1). In one case (B. longum strain F492), the patient presented with right lower abdominal fasciitis and myositis (Table 2). A gangrenous appendix was removed by open appendectomy 1 day after presentation. Three days after the surgery, the patient's WBC count remained high (18,900/μl) with continued abdominal infection which required surgery. Tissue from the abdominal wall grew B. longum, C. clostridioforme, B. fragilis group, and Bacteroides thetaiotaomicron. The B. longum isolate was present in large numbers (4+). The patient was treated with levofloxacin, metronidazole, and vancomycin and recovered. Isolate F492 had a RapID ANA II code of 425671 (no identification) and an API Coryne profile of 2450561 (99.8% identical to Cellulomonas sp.) (Table 1). The other B. longum isolate (strain F1519) was isolated from a patient with a 2-day history of excruciating abdominal and back pain. His WBC count was 22,600/μl on admission. An exploratory laparotomy was performed that revealed 2 feet of necrotic bowel. B. longum (104 CFU/ml), Clostridium glycolyticum, C. clostridioforme, and Prevotella oris were isolated from peritoneal fluid drawn from the patient. This patient was treated with piperacillin-tazobactam and eventually died from complications of his illness.
Three of the four B. breve isolates were recovered from blood cultures; the fourth (strain VAMC 11797) was isolated as the only organism at >105 CFU/ml from urine from a patient with a probable UTI (Table 2). Of the B. breve strains isolated from blood cultures, one was recovered along with Bacteroides vulgatus and a Fusobacterium species from a patient that had an ileal resection and was thought to represent transient bacteremia (strain TBAC519720). B. breve strain S7365 was isolated from a patient with a history of decubitis ulcers who had been frequently admitted with UTIs. The last B. breve isolate, strain 220119 S38, was recovered from several blood culture bottles from a patient with peritonitis. Treatment information is not available for any of the patients from whom B. breve was recovered.
DISCUSSION
Bifidobacterium species and A. omnicolens have been infrequently isolated from human clinical specimens. These organisms are normal flora of the human gastrointestinal tract and oral cavity, and some strains have been used as probiotics to protect against gut pathogens (1, 2, 5, 12). Beighton et al. analyzed bifidobacteria from human saliva and, interestingly, found B. longum in 10.8% of their subjects and low numbers of A. omnicolens and B. scardovii bacteria (1.6% each) (1). B. breve was not found in human saliva in that study (1). In contrast, from the site of isolation, the source of our strains seems to be gastrointestinal/genitourinary. Antibiotic susceptibility testing was not performed on the isolates in this study, although several strains of bifidobacteria were reported as uniformly susceptible to several antimicrobial agents used to treat gram-positive organisms, such as vancomycin and the beta-lactams (11).
We recovered B. longum and B. breve, which are used as probiotics in humans, but not B. infantum or B. lactis, which are also used as probiotics (2). The B. longum strains were isolated from abdominal wounds with other anaerobes and with C. clostridioforme.
The B. breve strains were isolated from both blood and urine. The B. breve isolate recovered from urine represented a probable UTI. Of the three B. breve isolates recovered from blood cultures, at least one was isolated as the only organism and was also probably isolated from an abdominal wound, representing a true infection including B. breve with other organisms.
While not all of the A. omnicolens and Bifidobacterium isolates in this study were implicated as causative agents of disease, these are organisms that should not be ignored when isolated from clinical specimens. These organisms are not easily identified; they are commonly misidentified as other catalase-negative, gram-positive rods such as Actinomyces sp. Usually, bifidobacteria can be differentiated from Lactobacillus species and G. vaginalis by using simple phenotypic tests such as vancomycin resistance and beta-hemolysis on Gardnerella agar, respectively (Table 3). With more isolates tested, the API Coryne and RapID ANA II systems hold promise, as biochemical profiles were generated, some of which were unique. However, at this time, 16S rRNA gene sequencing is necessary to differentiate bifidobacteria from Actinomyces species and to fully identify bifidobacteria.
Bifidobacteria may be underreported or not recovered, as clinical laboratories may consider them normal flora or not recover these organisms since they grow slowly and are difficult to identify. However, we show that when isolated in high numbers from clinical specimens, these organisms might be considered as potential contributing pathogens. The relationship of clinical isolates to probiotics may also be important.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Beighton, D., S. C. Gilbert, D. Clark, M. Mantzourani, M. al-Haboubi, F. Ali, E. Ransome, N. Hodson, M. Fenlon, L. Zoitopoulos, and J. Gallagher. 2008. Isolation and identification of Bifidobacteriaceae from human saliva. Appl. Environ. Microbiol. 74:6457-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman, T. M., G. L. Plosker, and D. P. Figgitt. 2007. Spotlight on VSL#3 probiotic mixture in chronic inflammatory bowel diseases. Biodrugs 21:61-63. [DOI] [PubMed] [Google Scholar]
- 3.Clarridge, J. E., III. 2003. Occurrence of fastidious organisms identified by 16S rDNA sequencing recovered from urine relative to uropathogens, abstr. C-199, p. 138. In Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 4.Crociani, F., B. Biavati, A. Alessandrini, C. Chiarini, and V. Scardovi. 1996. Bifidobacterium inopinatum sp. nov. and Bifidobacterium denticolens sp. nov., two new species isolated from human dental caries. Int. J. Syst. Bacteriol. 46:564-571. [DOI] [PubMed] [Google Scholar]
- 5.Guandalini, S. 2008. Probiotics for children with diarrhea. J. Clin. Gastroenterol. 42:S53-S57. [DOI] [PubMed] [Google Scholar]
- 6.Hoyles, L., E. Inganas, E. Falsen, M. Drancourt, N. Weiss, A. L. McCartney, and M. D. Collins. 2002. Bifidobacterium scardovii sp. nov., from human sources. Int. J. Syst. Evol. Microbiol. 52:995-999. [DOI] [PubMed] [Google Scholar]
- 7.Huys, G., M. Vancanneyt, K. D'Haene, E. Falsen, G. Wauters, and P. Vandamme. 2007. Alloscardovia omnicolens gen. nov., sp. nov., from human clinical samples. Int. J. Syst. Evol. Microbiol. 57:1442-1446. [DOI] [PubMed] [Google Scholar]
- 8.Jian, W., and X. Dong. 2002. Transfer of Bifidobacterium inopinatum and Bifidobacterium denticolens to Scardovia inopinata gen. nov., comb. nov., and Parascardovia denticolens gen. nov., comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 52:809-812. [DOI] [PubMed] [Google Scholar]
- 9.Mahlen, S. D., and J. E. Clarridge, III. 2009. Oral abscess caused by Campylobacter rectus: case report and literature review. J. Clin. Microbiol. 47:848-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modesto, M., B. Biavati, and P. Mattarelli. 2006. Occurrence of the family Bifidobacteriaceae in human dental caries and plaque. Caries Res. 40:271-276. [DOI] [PubMed] [Google Scholar]
- 11.Moubareck, C., F. Gavini, L. Vaugien, M. J. Butel, and F. Doucet-Populaire. 2005. Antimicrobial susceptibility of bifidobacteria. J. Antimicrob. Chemother. 55:38-44. [DOI] [PubMed] [Google Scholar]
- 12.Turroni, F., A. Ribbera, E. Foroni, D. van Sinderen, and M. Ventura. 2008. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie van Leeuwenhoek 94:35-50. [DOI] [PubMed] [Google Scholar]
- 13.York, M. K., K. L. Ruoff, J. E. Clarridge III, and K. Bernard. 2004. Identification of gram-positive bacteria, p. 3.18.1.1-3.18.1.15. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, 2nd ed. ASM Press, Washington, DC.