Abstract
A predominant kgp biovar colonized subgingival sites and buccal and tongue mucosa in 45 of 56 adults in an isolated community. The presence of biovars 381, W83, and W83v, but not HG66, correlated with the Porphyromonas gingivalis load at diseased sites. Biovars W83 and W83v poorly colonized tongue and buccal mucosa.
Porphyromonas gingivalis is a well-documented periodontal pathogen with two surface cysteine proteinases, a lysine (Kgp) and an arginine (RgpA) gingipain, both containing C-terminal hemagglutinin domains that facilitate colonization of the oral cavity by the bacterium (9, 16). Rearrangements within the genome of P. gingivalis have created strain-specific, virulence-related genes (5, 13). For instance, the fimA gene coding for the fimbrial subunit has been extensively used to categorize clinical isolates. Association of the P. gingivalis fimA genotype II with periodontitis has been observed in different cohorts from Japan (1), Germany (2), Brazil (10), and Switzerland (20). While fimA genotype II is the most prevalent gene, the fimA genotype I from ATCC 33277/381 strains has been reported to be the second most prevalent genotype among German periodontitis patients, while the fimA genotype IV gene (from strain W83) is the second most prevalent gene among Japanese periodontitis patients (1). This suggests that differences in frequency of fimA alleles depend on geographic location and/or ethnic origin. However, a recent report highlights the role of gingipains and not fimbriae in preferential binding to matrix proteins, a process that could facilitate colonization of the host (9). Previously we reported that P. gingivalis could be categorized by differential PCR into four kgp biovars: HG66, 381, W83, and W83v, a domain variant of the W83 kgp gene (12). Using paired subgingival samples from the most- and least-diseased sites, we showed that the majority of periodontitis patients in our study were colonized by one predominant biovar with only trace amounts of a second detectable in a few subjects. The frequency of occurrence of biovars HG66, 381, W83, and W83v was 8:11:6:5, respectively (12). In support of this observation is a recent study by Inaba and colleagues (4), who reported that fimA genotype II clinical isolates from nine Japanese periodontitis patients all possessed the HG66 biovar, while isolates from two patients corresponded to biovars 381 and W83 or biovars 381 and W83v.
In this report, we describe the distribution of P. gingivalis biovars on buccal mucosa and dorsum of the tongue, in addition to healthy and diseased subgingival sites, in relation to P. gingivalis load. Site-specific distribution of biovars in three healthy and three diseased gingival sites was also studied in eight subjects colonized with a trace amount of a second biovar.
The study comprised 56 adult Aborigines diagnosed with localized periodontitis with a gender ratio of 33 females (age range, 21 to 67 years; median age, 40 years) to 23 males (age range, 22 to 68 years; median age, 43 years). The Aboriginal Medical Research Council of New South Wales and the Executive of the Durri Aboriginal Medical Service in the North Coast region of New South Wales endorsed the proposal to analyze the molecular epidemiology of oral infection in an isolated, high-risk, high-disease-burden community with preserved family units and no clinical intervention. Informed consent was obtained from each subject. The project was also approved by the Human Ethics Review Committee of the Sydney West Area Health Service.
After removal of supragingival plaque, subgingival plaque was collected from each subject with a sterile curette using a standardized approach whereby the curette was placed in the base of the sulcus or pocket and raised vertically against the tooth surface to remove a sample of ∼10 μl. Curette sampling by two experienced calibrated clinicians gave a more accurate representation of pocket microbiota than paper point sampling (18).
Samples were taken from sites adjacent to areas probed for diagnosis and included sites around three teeth that represented the healthiest gingiva (≤3-mm probing depth with no bleeding or suppuration) and three sites that represented the most-diseased sites (≥4-mm probing depth with bleeding and/or suppuration). The categorization of healthy and diseased site sampling was based on indices of gingival inflammation (gingival index score), clinical attachment levels, gingival recession, bleeding on probing, and probing depth for each tooth (14). Attachment loss in diseased sites ranged from 2 to 6 mm. Radiographic facilities were not available for the study.
Buccal mucosal swabs were collected with a cotton bud rotated from the mid-cheek third molar to the corner of the lip, while tongue swabs were similarly taken from lateral border to lateral border across the anterior dorsum of the tongue. Samples were collected in tubes containing 200 μl phosphate-buffered saline and snap-frozen with dry ice in preparation for extraction of DNA using a QIAamp DNA minikit (Qiagen, Clifton Hill, Victoria, Australia). P. gingivalis load and total bacterial load were quantified, and identification of kgp biovars was determined as previously described (8, 11, 12). Briefly, total bacterial and P. gingivalis loads were determined using the universal primer (300 nM) and probe (175 nM) set or the P. gingivalis-specific primers and probe (100 nM), respectively, using the TaqMan PCR core reagents kit (Applied Biosystems) and an ABI-PRISM 7700 sequence detection system (8, 12). The amount of DNA in each sample was measured against P. gingivalis DNA standards in the range 3.6 fg to 3.6 ng and converted to cell numbers based on one cell containing 2.37 fg of DNA. P. gingivalis load was expressed as per ml for subgingival plaque and for mucosal or tongue samples as previously used (3). For identification of kgp biovars, differential PCR was carried out with HotStarTaq master mix (Qiagen, Australia) using the HG66 biovar-specific primer set HG66F and HG66R (amplicon size, 1.9 kb), the 381 biovar-specific primer set kgpF and 381R (amplicon size, 3.4 kb), and the W83/W83v biovar-specific primer set kgpF and W83R (amplicon size, 3.6 kb for W83 and 2.2 kb for W83v) (12). Total bacterial load (described above) was used as a reference marker for bacterial biomass recovered to verify the sampling technique (15). This standard was preferred over total recovered protein as inflamed sites contain high levels of serum proteins (6, 19) that could have distorted the analysis.
Biovar-specific PCR was carried out using DNA samples isolated from the buccal mucosa, the tongue, and two individually pooled DNA samples isolated from three subgingival sites that represented the three healthiest or the three most-diseased sites, respectively. Due to low loads of P. gingivalis in some of the subjects, it was only possible to detect the kgp biovar(s) present in the subgingival plaque of 45 subjects using our visual gel-based system (12). Thus, kgp biovar 381 was detected in 26 subjects, HG66 in 17 subjects, and biovars W83 and W83v in 7 and 3 subjects, respectively. For 8 of the 45 subjects in which biovars were detected (strong visual band after 40 cycles of PCR), trace amounts of a second biovar (weak visual band after 40 cycles of PCR) were also detected. In the eight subjects possessing two kgp biovars, further analysis showed that the predominant biovars in the pooled samples also predominated at individual sites (Table 1). Similarly, for those subjects harboring P. gingivalis in subgingival plaque and where P. gingivalis biovars could be determined, the tongue and buccal mucosa were colonized by the same predominant biovar present in subgingival plaque. Biovar 381 was detected on the buccal and tongue mucosa of 8 individuals, HG66 on 12 individuals, and biovar W83v on 1 individual. These observations were in keeping with previous reports suggesting that soft tissue surfaces harboring periodontal pathogens could serve as reservoirs for infection (7, 17). However, in none of the seven subjects colonized subgingivally by biovar W83 could this biovar be detected on either the buccal mucosa or the dorsum of the tongue. The use of total bacterial load as a reference marker for bacterial biomass across the four sites healthy, diseased, mucosal, and tongue indicated that the absence of biovars W83 and W83v on mucosal and tongue sites was not due to sampling error, as total bacterial loads were not significantly different from those sites colonized by HG66 or 381 biovars (data not shown).
TABLE 1.
Site-specific distribution of P. gingivalis kgp biovars in subjects colonized with trace amount of a second biovar
| Subject | Biovar found ina:
|
|||||
|---|---|---|---|---|---|---|
| Healthy subgingival sites
|
Diseased subgingival sites
|
|||||
| Site 1 | Site 2 | Site 3 | Site 1 | Site 2 | Site 3 | |
| 1 | 381 | 381, W83 | 381 | 381, W83 | HG66 | |
| 2 | W83v | W83v | W83v, 381 | |||
| 3 | HG66, 381 | HG66, 381 | HG66, 381 | HG66, 381 | ||
| 4 | 381 | 381 | W83v | W83v, 381 | ||
| 5 | HG66 | HG66 | HG66 | 381, HG66 | HG66 | |
| 6 | HG66 | 381 | 381, HG66 | HG66, 381 | HG66, 381 | |
| 7 | HG66 | 381 | 381 | HG66 | HG66, 381 | HG66, 381 |
| 8 | HG66 | HG66 | HG66, W83 | HG66 | HG66 | HG66 |
A biovar in boldface represents the predominant biovar identified in pooled DNA samples isolated from six subgingival sites representing the three healthiest and three most-diseased sites from a given subject.
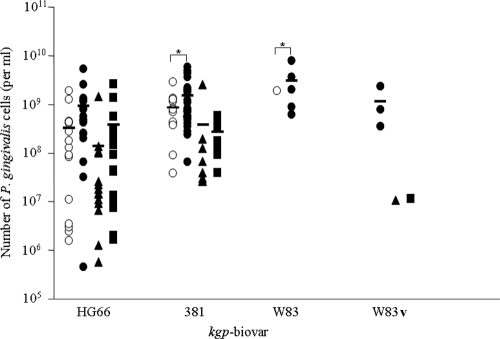
Based on analysis using generalized linear mixed-effects models (GLMEM) (21) for the 45 subjects, there was significantly less detection of biovars 381, W83, and W83v in healthy subgingival sites, buccal mucosa, and tongue than for diseased gingival sites (P < 0.0001). Significant correlation was also observed between P. gingivalis load and the presence of the 381, W83, and W83v biovars (P < 0.0001). For the HG66 biovar, the presence in diseased gingival sites showed no significant correlation to either the presence or absence of this biovar in clinically healthy subgingival sites (P = 0.3640) or the tongue surface (P = 0.0737) (Fig. 1). In summary, the isolated adult Aboriginal community suffering from chronic periodontitis demonstrated a distribution frequency for the kgp biovars HG66 and 381 close to that observed for the multiethnic Sydney metropolitan population (12). The fact that the kgp biovars W83 and W83v were poor colonizers of the dorsum of the tongue and buccal mucosa, while the kgp biovars HG66 and 381 were capable of colonizing a variety of mucosal environments, might explain the high frequency of these biovars in populations suffering from periodontal diseases. It remains to be determined if this behavior is related directly to structural variation in the hemagglutinin domain of Kgp.
FIG. 1.
Relationship between P. gingivalis load and kgp biovar. Subgingival plaque from the least-diseased (open circles) and the most-diseased sites (closed circles), as well as swabs of buccal mucosa (closed triangles) and the dorsum of the tongue (closed squares) from each subject, was assayed for the number of P. gingivalis (expressed as per ml for subgingival plaque and for mucosal or tongue samples) and kgp biovar. The mean value for each group is represented by a solid bar, and statistical comparison based on GLMEM (21) between the groups was analyzed. The asterisk denotes significance (P < 0.0001). It should be noted that due to the limited number of samples in the biovar W83 data set, the GLMEM model recognizes that the level of significance may be unstable.
Acknowledgments
This study was supported by the Australian National Health and Medical Research Council (grant no. 219185).
Essential support was provided by the staff of the Durri Aboriginal Medical Service. We thank Cheryl Chapple and Ky-Anh Nguyen, Institute of Dental Research, Westmead Hospital, for collection of clinical samples and Karen Byth, Westmead Millennium Institute, Westmead Hospital, and Manish Arora, Faculty of Dentistry, University of Sydney, for statistical analysis of the data.
Footnotes
Published ahead of print on 12 August 2009.
REFERENCES
- 1.Amano, A., I. Nakagawa, K. Kataoka, I. Morisaki, and S. Hamada. 1999. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J. Clin. Microbiol. 37:1426-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beikler, T., U. Peters, S. Prajaneh, K. Prior, B. Ehmke, and T. F. Flemmig. 2003. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur. J. Oral. Sci. 111:390-394. [DOI] [PubMed] [Google Scholar]
- 3.Boutaga, K., A. J. van Winkelhoff, C. M. J. E. Vandenbroucke-Grauls, and P. H. M. Savelkoul. 2003. Comparison of real-time PCR and culture for detection of Porphyromonas gingivalis in subgingival plaque samples. J. Clin. Microbiol. 41:4950-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inaba, H., K. Nakano, T. Kato, R. Nomura, S. Kawai, M. Kuboniwa, K. Ishihara, T. Ooshima, and A. Amano. 2008. Heterogenic virulence and related factors among clinical isolates of Porphyromonas gingivalis with type II fimbriae. Oral Microbiol. Immunol. 23:29-35. [DOI] [PubMed] [Google Scholar]
- 5.Kilian, M., E. V. Frandsen, D. Haubek, and K. Poulsen. 2006. The etiology of periodontal disease revisited by population genetic analysis. Periodontol. 2000 42:158-179. [DOI] [PubMed] [Google Scholar]
- 6.Lamster, I. B., and J. K. Ahlo. 2007. Analysis of gingival crevicular fluid as applied to the diagnosis of oral and systemic diseases. Ann. N. Y. Acad. Sci. 1098:216-229. [DOI] [PubMed] [Google Scholar]
- 7.Mager, D. L., L. A. Ximenez-Fyvie, A. D. Haffajee, and S. S. Socransky. 2003. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30:644-654. [DOI] [PubMed] [Google Scholar]
- 8.Martin, F. E., M. A. Nadkarni, N. A. Jacques, and N. Hunter. 2002. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 40:1698-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McAlister, A. D., A. Sroka, R. E. Fitzpatrick, N. S. Quinsey, J. Travis, J. Potempa, and R. N. Pike. 2009. Gingipain enzymes from Porphyromonas gingivalis preferentially bind immobilized extracellular proteins: a mechanism favouring colonization? J. Periodontal Res. 44:348-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Missailidis, C. G., J. E. Umeda, C. Ota-Tsuzuki, D. Anzai, and M. P. Mayer. 2004. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol. Immunol. 19:224-229. [DOI] [PubMed] [Google Scholar]
- 11.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 12.Nadkarni, M. A., K.-A. Nguyen, C. C. Chapple, A. A. DeCarlo, N. A. Jacques, and N. Hunter. 2004. Distribution of Porphyromonas gingivalis biotypes defined by alleles of the kgp (Lys-gingipain) gene. J. Clin. Microbiol. 42:3873-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naito, M., H. Hirakawa, A. Yamashita, N. Ohara, M. Shoji, H. Yukitake, K. Nakayama, H. Toh, F. Yoshimura, S. Kuhara, M. Hattori, T. Hayashi, and K. Nakayama. 2008. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 15:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman, M. G., H. H. Takei, P. R. Klokkevold, and F. A. Carranza. 2009. Carranza's clinical periodontology. Elsevier, Inc., Philadelphia, PA.
- 15.Sebastian, A., and L. Larsson. 2003. Characterization of the microbial community in indoor environments: a chemical-analytical approach. Appl. Environ. Microbiol. 69:3103-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sztukowska, M., A. Sroka, M. Bugno, A. Banbula, Y. Takahashi, R. N. Pike, C. A. Genco, J. Travis, and J. Potempa. 2004. The C-terminal domains of the gingipain K polyprotein are necessary for assembly of the active enzyme and expression of associated activities. Mol. Microbiol. 54:1393-1408. [DOI] [PubMed] [Google Scholar]
- 17.Tanner, A. C., B. J. Paster, S. C. Lu, E. Kanasi, R. Kent, Jr., T. Van Dyke, and S. T. Sonis. 2006. Subgingival and tongue microbiota during early periodontitis. J. Dent. Res. 85:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teles, F. R., A. D. Haffajee, and S. S. Socransky. 2008. The reproducibility of curet sampling of subgingival biofilms. J. Periodontol. 79:705-713. [DOI] [PubMed] [Google Scholar]
- 19.Tsai, C. C., C. C. Kao, and C. C. Chen. 1998. Gingival crevicular fluid lactoferrin levels in adult periodontitis patients. Aust. Dent. J. 43:40-44. [DOI] [PubMed] [Google Scholar]
- 20.van der Ploeg, J. R., E. Giertsen, B. Ludin, C. Morgeli, A. S. Zinkernagel, and R. Gmur. 2004. Quantitative detection of Porphyromonas gingivalis fimA genotypes in dental plaque. FEMS Microbiol. Lett. 232:31-37. [DOI] [PubMed] [Google Scholar]
- 21.Venables, W. N., and B. D. Ripley. 2002. Modern applied statistics with S, 4th ed. Springer-Verlag, New York, NY.