Abstract
H9N2 influenza viruses have become established and maintain long-term endemicity in poultry. The complete genomes of seven avian H9N2 influenza viruses were characterized. These seven influenza virus isolates were obtained from live poultry markets in Shanghai, China, in 2002 and from 2006 to 2008. Genetic analysis revealed that all seven isolates had an RSSR motif at the cleavage site of hemagglutinin (HA), indicating low pathogenicity in chickens. Phylogenetic analyses indicated that the seven avian H9N2 viruses belonged to the lineage represented by Duck/Hong Kong/Y280/97 (H9N2), a virus belonging to the Chicken/Beijing/1/94-like (H9N2) lineage, and that they are all quadruple reassortants consisting of genes from different lineages. The six internal genes of the isolates possessed H5N1-like sequences, indicating that they were reassortants of H9 and H5 viruses. All of the viruses had nonstructural (as well as HA and neuraminidase) genes derived from the Duck/Hong Kong/Y280/97-like virus lineage but also had other genes of mixed avian virus origin, including genes similar to those of H5N1 viruses (Gs/GD-like). The infected chickens showed no signs of disease. These results show the genetic and biological diversity of H9N2 viruses in Shanghai and support their potential role as pandemic influenza agents.
Avian influenza A viruses of subtype H9N2 were first detected in the United States in 1966 (12). In North America, there are no reports of H9N2 virus-associated disease in chickens to date, although these viruses can be found in wild ducks and have caused a number of outbreaks in turkeys (10, 13, 24, 25). In Asia, H9N2 viruses were detected only in apparently healthy ducks in limited surveillance studies of live poultry markets and farms in Hong Kong from 1975 to 1985. However, in the early 1990s, H9N2 viruses became more prevalent in domestic poultry (26). Reports from Korea, South Asia, Middle Eastern countries, South Africa, and Europe since the late 1990s indicate widespread distribution of H9N2 viruses (1, 4, 14, 19, 20, 23). Previous studies revealed that H9N2 influenza viruses from poultry could occasionally be transmitted from poultry to mammalian species, including humans and pigs (3, 17, 21, 22, 29). The Chicken/Beijing-like (Ck/Bei-like) and G1-like viruses were initially found in specimens from both a human and pigs in the late 1990s and were also observed in 2003 and recently in Hong Kong (5). Genetic analyses demonstrated that the human H9N2 influenza virus isolate from 2003 was a novel reassortant and most likely originated directly from local live poultry markets (3). These recent interspecies transmission events suggest that current H9N2 influenza virus variants are still potentially infectious for humans.
During surveillance of live poultry markets in Shanghai from September 2006 to December 2008, the only influenza viruses isolated were H9. It was reported that at least two lineages of H9N2 viruses, represented by A/Duck/Hong Kong/Y280/97 (Dk/HK/Y280) and A/Quail/Hong Kong/G1/97 (Qa/HK/G1), had become established in domestic poultry in Asia (8, 15, 21). Furthermore, human cases of H9N2 virus infection have been reported in Hong Kong and other locations in south China in the late 1990s and early 2000s (3, 9, 17, 22). Follow-up serological surveillance suggests that the incidence of human infections with H9N2 viruses might be higher than previously anticipated (7, 9, 22). The prevalence of H9N2 viruses throughout the world, along with their ability to infect mammals and humans, increases concern about their pandemic potential.
Because of the ongoing concern about the transmission of H9N2 viruses to mammals and humans, continued surveillance of H9N2 viruses from live poultry markets is needed.
MATERIALS AND METHODS
Sampling and virus isolation.
From September 2006 to December 2008, 136 H9N2 influenza virus strains were isolated from 7,438 chickens sampled in live poultry markets in Shanghai, China. These specimens were paired tracheal and cloacal swabs. Most of the chickens sold in Shanghai live poultry markets originated in the Shanghai suburbs. Seven influenza A viruses were isolated by using 10-day-old specific-pathogen-free chicken eggs and were identified as subtype H9N2 by a hemagglutination inhibition test and sequencing of the neuraminidase (NA) gene.
Genetic and phylogenetic analysis.
Viral gene sequencing was carried out as follows. In brief, viral RNA was directly extracted from infected allantoic fluids using RNeasy minikits (Qiagen, Chatsworth, CA), and reverse transcriptions were carried out under standard conditions using a random primer. PCR was performed using specific primers for eight genes (primer sequences are available on request). PCR products were purified with the QIAquick PCR purification kit (Qiagen, Inc.), cloned into the pMD18-T vector (TaKaRa, Dalian, China), and then sequenced using synthetic oligonucleotides (Invitrogen).
Sequence analysis.
Sequence data were compiled and edited using the Lasergene sequence analysis software package (DNAStar, Inc., Madison, WI). Multiple-sequence alignment was carried out by using CLUSTAL W, and the unrooted phylogenetic trees were generated by the distance-based neighbor-joining method using MEGA, version 3.1. Bootstrap values were calculated based on 1,000 replicates of the alignment.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study are available from GenBank under accession numbers GQ335463 to GQ335518.
RESULTS
Prevalence of H9N2 influenza viruses in chickens.
A total of 7,438 samples were collected from chickens from September 2006 to December 2008. In general, H9N2 viruses were isolated from apparently healthy chickens year-round in live poultry markets in Shanghai, China. There were 136 strains of H9N2 influenza viruses isolated from 7,438 chicken samples (overall isolation rate, 1.8%), but the isolation rate had no relationship with the season, in contrast with a previously published finding that it was usually higher in the winter than in the summer (30). In the same period, one H9 virus from 2002 and two H9 viruses each from 2006, 2007, and 2008 were genetically analyzed, and all of them were identified as H9N2.
Phylogenetic analysis.
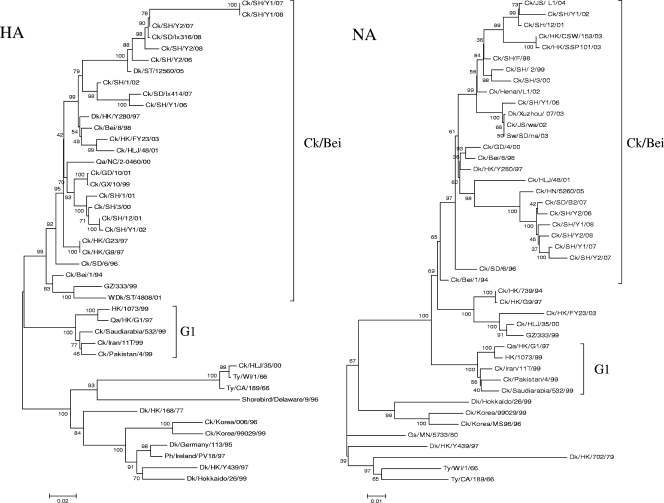
Phylogenetic analysis of the H9 hemagglutinin (HA) gene shows that all of the seven H9N2 viruses are incorporated into the lineage represented by Dk/HK/Y280/97 (Fig. 1). These seven viruses were derived from Ck/Bei/1/94 viruses. The homologies of the nucleotide sequences and deduced amino acid sequences of the HA genes of these seven isolates were 88.6% to 99.6% and 89.1% to 99.1%, respectively. Phylogenetic analysis of the NA gene shows that all of the NA genes from the seven H9N2 viruses tested, like their HA genes, join and are closely related to the Dk/HK/Y280/97 lineage. Also, no Qa/HK/G1/97-like N2 virus was detected (Fig. 1).
FIG. 1.
Phylogenetic tree of the HA (positions 115 to 999) and NA (positions 59 to 1235) genes of the H9N2 avian influenza viruses in Shanghai, China, in 2002 and from 2006 to 2008. The unrooted phylogenetic trees were generated by the distance-based neighbor-joining method using MEGA, version 3.1. The reliability of the trees was assessed by bootstrap analysis with 1,000 replications; only bootstrap values of >90% are shown. Abbreviations: BJ and Bei, Beijing; CA, California; Ck, chicken; Dk, duck; GD, Guangdong; Gf, guinea fowl; Gs, goose; GX, Guangxi; GZ, Guangzhou; HK, Hong Kong; HLJ, Heilongjiang; JS, Jiangshu; NC, Nanchang; NJ, Nanjing; Pg, pigeon; Ph, pheasant; Qa, quail; SD, Shandong; SH, Shanghai; ST, Shantou; Ty, turkey; VNM, Vietnam; WDk, wild duck; WI, Wisconsin; YN, Yunnan.
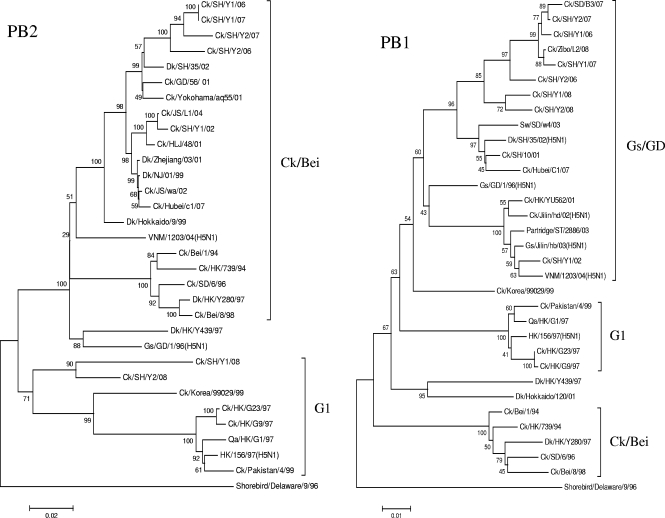
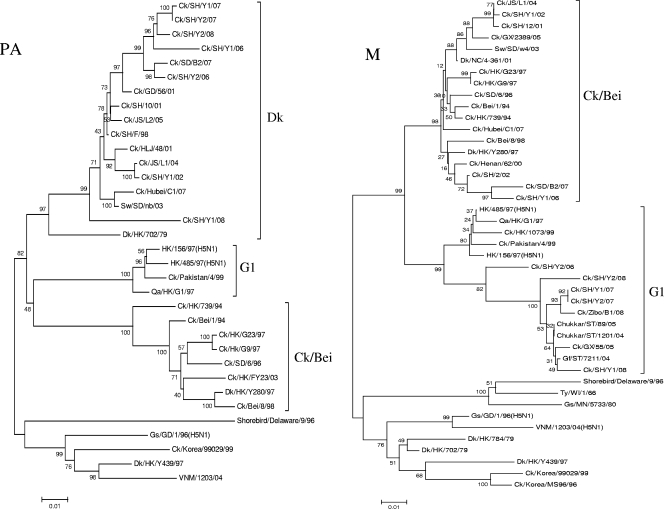
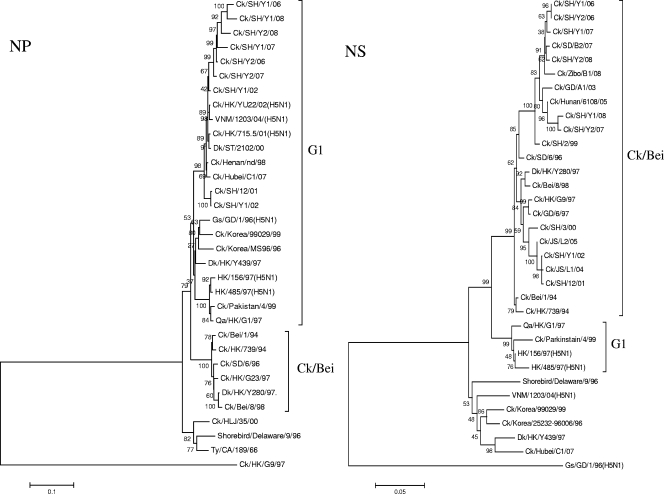
Phylogenetic analyses of the six internal genes revealed that these genes had more diversified sources than the surface genes and that the seven H9N2 viruses had undergone extensive reassortment to generate multiple novel reassortants or genotypes (Table 1; Fig. 2). It was noted that the NP genes of the seven H9N2 viruses shared the same clade with Qa/HK/G1/97. The PB1 genes of these seven H9N2 viruses clustered with Gs/GD/1/96 (H5N1). The PA genes are mainly of aquatic avian origin. The PB2 genes of the five isolates from 2002, 2006, and 2007 are closely related to those of Ck/Bei/1/94-like viruses; however, the PB2 genes of the two isolates from 2008 are G1-like. Phylogenetic analysis shows that the NS genes of the seven H9 influenza viruses isolated in 2002 and 2006 to 2008 were located in the Ck/Bei/1/94 virus lineage. In contrast to the NS gene tree, the M genes of Ck/SH/Y1/02 and Ck/SH/Y1/06 are closely related to the Ck/Bei/1/94 lineage, while those of the other five viruses are closely related to the Qa/HK/G1/97 lineage.
TABLE 1.
Comparison of two key sites of HA gene sequences and genotyping of H9 subtype influenza viruses isolated from chickens in a suburb of Shanghai, China, in 2002 and from 2006 to 2008
| Virusa | Amino acids in the RBS at:
|
Lineagec at the following gene segment:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Selected locationsb | Left edge (aa 232-237) | PB2 | PB1 | PA | HA | NP | NA | M | NS | |
| Ck/SH/Y1/02 | YWTNTLY | NGQQGR | Ck/Bei | Gs/GD | Dk | Ck/Bei | G1 | Ck/Bei | Ck/Bei | Ck/Bei |
| Ck/SH/Y1/06 | YWTNVLY | NGQQGR | Ck/Bei | Gs/GD | Dk | Ck/Bei | G1 | Ck/Bei | Ck/Bei | Ck/Bei |
| Ck/SH/Y2/06 | YWTNVLY | NGLQGR | Ck/Bei | Gs/GD | Dk | Ck/Bei | G1 | Ck/Bei | G1 | Ck/Bei |
| Ck/SH/Y1/07 | YWTNALY | NGLQGR | Ck/Bei | Gs/GD | Dk | Ck/Bei | G1 | Ck/Bei | G1 | Ck/Bei |
| Ck/SH/Y2/07 | YWTNVLY | NGLQGR | Ck/Bei | Gs/GD | Dk | Ck/Bei | G1 | Ck/Bei | G1 | Ck/Bei |
| Ck/SH/Y1/08 | YWTNVLY | NGLQGR | G1 | Gs/GD | Dk | Ck/Bei | G1 | Ck/Bei | G1 | Ck/Bei |
| Ck/SH/Y2/08 | YWTNALY | NGLQGR | G1 | Gs/GD | Dk | Ck/Bei | G1 | Ck/Bei | G1 | Ck/Bei |
SH, Shanghai.
From left to right, aa 109, 161, 163, 191, 198, 202, and 203.
Ck/Bei, Ck/Beijing/1/94-like; Dk, duck; G1, Qa/HK/G1/97-like; Gs/GD, Gs/GD-like.
FIG. 2.
Phylogenetic trees of the PB2 (positions 32 to 2228), PB1 (positions 182 to 1080), PA (positions 23 to 1257), M (positions 270 to 765), NP (positions 2 to 1445), and NS (positions 34 to 693) genes of the H9N2 avian influenza viruses in Shanghai, China, in 2002 and from 2006 to 2008. The method used is described in the legend to Fig. 1. Virus abbreviations are defined in the legend to Fig. 1.
Taken together, the phylogenetic analyses of all eight gene segments of these seven H9 influenza viruses isolated from chickens from a Shanghai suburb revealed that they were quadruple reassortants (Table 1). The differences in their receptor binding sites (RBS) and the left edge of the RBS on the HA gene are also seen in Table 1. The right edges of the RBS (amino acid [aa] 146 to 150) were all GTSKA. Potential glycosylation sites 1 (aa 29 to 31), 2 (aa 141 to 143), 3 (aa 218 to 220), 4 (aa 298 to 300), 5 (aa 305 to 307), 6 (aa 492 to 494), and 7 (aa 551 to 553) were all the same: NST, NVS, NRT, NTT, NVS, NGT, and NGS, respectively. Their proteolytic cleavage sites (aa 335 to 341) were RSSR↓GLF.
All of the surface genes (HA and NA) of these H9N2 influenza viruses were derived from the Ck/Bei-like virus lineage. Overall, the available phylogenetic information from the seven viruses indicated that H9N2 viruses in the Shanghai suburbs were diverse and in a dynamic state.
Alignment studies.
The deduced amino acid sequences of the H9 HA genes were aligned and compared with those of representative H9 viruses. All seven of these isolates, except for two H9N2 viruses (Chicken/Shanghai/Y1/02 and Chicken/Shanghai/Y1/06), had the amino acid residue leucine (L) at position 226 (H3 numbering). But all had glycine (G) at position 228 at the RBS. Amino acid L at position 226 is found in H9N2 viruses of the Dk/HK/Y280/97 and Ck/HK/G9/97 lineages isolated in Hong Kong and their more recent descendants isolated from chickens (2). The L-226-containing or G-228-containing H9N2 viruses displayed human virus-like cell tropisms. The H9 subtype viruses isolated from ducks in the 1970s, as well as the earliest viruses of the Dk/HK/Y280/97 lineage, such as Ck/Bei/1/94 and Ck/HK/739/94, have glutamine (Q) at position 226. The Chicken/Shanghai/Y1/02 and Chicken/Shanghai/Y1/06 viruses also have Q at amino acid residue 226, which was in accordance with the isolation year, because they were isolated earlier. It is also noted that all seven of these H9 viruses isolates from the Shanghai suburbs maintain the Arg-Ser-Ser-Arg (RSSR) motif at the connecting peptide of their HA, representing low pathogenicity in chickens. In fact, their host were all healthy clinically. Compared with those of the Ck/Bei/1/94 virus, aa 63 to 65 were defective in the NA genes of both the Chicken/Shanghai/Y1/02 and the Chicken/Shanghai/Y1/06 virus. However, aa 62 to 64 were defective in the other five isolates. The relationship between the loss of three amino acids at different sites and the biological characteristics of isolates remains to be determined.
DISCUSSION
H9N2 viruses are among the most commonly occurring viruses in domestic poultry populations, with several outbreaks reported in Asia and North America since 1990. Subtype H9N2 viruses are classified as low-pathogenicity viruses both by molecular characterization and by pathotyping. Viruses of this subtype are unique among low-pathogenicity avian influenza viruses in that they infect a wide variety of species, including chickens, quail, turkeys, ducks, geese, pigs, and humans (2, 21, 27). Because H9N2 viruses infect a number of species, they may be significant donors of genetic material to emerging human pathogens (9, 14). There is no evidence to suggest that subtype H9N2 viruses can be transmitted from human to human (28). However, they could emerge as human pathogens through reassortment in intermediate hosts, such as pigs (21), and in avian species, or through direct adaptation in the human host (8). The abilities of subtype H9N2 viruses to spread widely in domestic poultry populations (2, 6), to cross species barriers, and potentially to have an impact on emerging viruses warrant further studies of the evolution of H9N2 viruses in avian species.
In the present study, we genetically characterized seven H9N2 viruses isolated from chickens in our surveillance in 2002 and from 2006 to 2008. This provided convincing evidence that these viruses have been generating quadruple reassortants. The seven isolates fell into three genotypes. These three reassortant H9N2 genotypes were detected in chickens in 2002 and early 2006 (Ck/SH/Y1/02 and Ck/SH/Y1/06), late 2006 and 2007 (Ck/SH/Y2/06 and Ck/SH/Y1/07), and 2008 (Ck/SH/Y1/08 and Ck/SH/Y2/08), respectively. Phylogenetic analyses of the internal genes of these H9N2 viruses showed that some of them are closely related to the internal gene complex found in H5N1 viruses (Gs/GD/1/96). This information suggested that these viruses may share molecular determinants that facilitate their replication in mammalian species. Both Eurasian H5N1 and H9N2 viruses have proven their ability to directly infect humans. However, unlike the H5N1 viruses, H9N2 viruses are currently widespread in domestic poultry and thus remain a potential source of further human infections and, possibly, a new pandemic strain.
The connecting peptides of the HAs of the seven viruses indicated that they are unlikely to have been highly pathogenic for poultry. In fact, the seven viruses were isolated from healthy chickens. The more detailed analysis showed that aa 337 of Chicken/Shanghai/Y1/02 and Chicken/Shanghai/Y1/06 was Ser (AGC). For the other five isolates, aa 337 was Ser (AGT). These two kinds of codons may mutate to AGA, which can change the cleavage site to one characteristic of high pathogenicity in chickens [(R/K)X(K/R)R].
The deduced amino acid sequences of the viruses were aligned. Except for those of Chicken/Shanghai/Y1/02 and Chicken/Shanghai/Y1/06, the HAs of all viruses tested had Leu at position 226 in the RBS (H3 numbering); those of the former two viruses had Glu at position 226. In addition, all the HAs all have glycine (G) at position 228 (18). The findings of the present investigation suggested that the five H9N2 reassortant viruses isolated later contained a receptor-binding profile (19, 21) that favors infection of humans. Moreover, it was reported that aa 627 and aa 701 of the PB2 gene influence the host of the H9 virus. These two sites of the seven isolates are all E and D, showing that these viruses are of avian origin (11, 16).
The vaccine used in Shanghai has been the inactivated H9N2 virus (Ck/Shandong/6/96) since 2003. All the gene segments of the vaccine strain were analyzed phylogenetically with our isolates. The vaccine strain is a nonreassortant Ck/Bei/1/94 virus. Its protective efficacy depends mainly on the antigenic relationship between the vaccine strain and H9N2 virus strains currently circulating in Shanghai poultry. But the present phylogenetic results provided some information for the selection of vaccine strains. Our study showed that the isolation rate had no relationship with season. The use of inactivated vaccine may be a cause.
The fact that H9N2 influenza viruses are not highly pathogenic for poultry makes them more, rather than less, likely to be of pandemic relevance. In fact, viruses that are less pathogenic for poultry have a greater opportunity to become widespread, since they do not raise concern and permit their hosts to survive unhindered. Continuing surveillance of both animal and human influenza viruses seems to be the best option for detecting and interrupting this kind of development.
Acknowledgments
This work was supported by a grant from the National High Technology Research and Development Program of China (program 863) (grant 2007AA02Z407), the Shanghai Scientific Agricultural Tackling Program (grant 2007(3-4)), and the National Key Technology R&D program (grant 2006BAD06A16-4).
We gratefully acknowledge Yu Hai (Shanghai Veterinary Research Institute) for technological help with sequence analyses.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Alexander, D. J. 2003. Report on avian influenza in the Eastern Hemisphere during 1997-2002. Avian Dis. 47:792-797. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. J. 2000. A review of avian influenza in different bird species. Vet. Microbiol. 74:3-13. [DOI] [PubMed] [Google Scholar]
- 3.Butt, K. M., G. J. Smith, H. Chen, L. J. Zhang, Y. H. Leung, K. M. Xu, W. Lim, R. G. Webster, K. Y. Yuen, J. S. Peiris, and Y. Guan. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 43:5760-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameron, K. R., V. Gregory, J. Banks, I. H. Brown, D. J. Alexander, A. J. Hay, and Y. P. Lin. 2000. H9N2 subtype influenza A viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology 278:36-41. [DOI] [PubMed] [Google Scholar]
- 5.Centre for Health Protection. 20 March 2007. Girl recovered from influenza A virus (H9N2). Centre for Health Protection, Department of Health, Hong Kong, SAR, China.
- 6.Chen, B. L., Z. J. Zhang, and W. B. Chen. 1994. Isolation and identification of avian influenza virus. Chin. J. Vet. Med. 10:3-5. [Google Scholar]
- 7.Cheng, X., J. Liu, J. He, and F. Shan. 2002. Virological and serological surveys for H9N2 subtype of influenza A virus in chickens and men in Shenzhen City. Chin. J. Exp. Clin. Virol. 16:319-321. [PubMed] [Google Scholar]
- 8.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 96:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, Y., J. Li, X. Cheng, M. Wang, Y. Zhou, C. Li, F. Cai, H. Miao, Y. Zhang, J. Guo, L. Huang, and D. Bei. 1999. Discovery of men infected by avian influenza A (H9N2) virus. Chin. J. Exp. Clin. Virol. 13:105-108. [PubMed] [Google Scholar]
- 10.Halvorson, D. A., D. D. Frame, A. J. Friendshuh, and D. P. Shaw. 1997. Outbreaks of low pathogenicity avian influenza in USA, p. 36-46. In Proceedings of the Fourth International Symposium on Avian Influenza. U.S. Animal Health Association, Rose Printing Company, Tallahassee, FL.
- 11.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 12.Homme, P. J., and B. C. Easterday. 1970. Avian influenza virus infections. I. Characteristics of influenza A-turkey-Wisconsin-1966 virus. Avian Dis. 14:66-74. [PubMed] [Google Scholar]
- 13.Kawaoka, Y., T. M. Chambers, W. L. Sladen, and R. G. Webster. 1988. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology 163:247-250. [DOI] [PubMed] [Google Scholar]
- 14.Lee, C. W., C. S. Song, Y. J. Lee, I. P. Mo, M. Garcia, D. L. Suarez, and S. J. Kim. 2000. Sequence analysis of the hemagglutinin gene of H9N2 Korean avian influenza viruses and assessment of the pathogenic potential of isolate MS96. Avian Dis. 44:527-535. [PubMed] [Google Scholar]
- 15.Li, K. S., K. M. Xu, J. S. M. Peiris, L. L. M. Poon, K. Z. Yu, K. Y. Yuen, K. F. Shortridge, R. G. Webster, and Y. Guan. 2003. Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans? J. Virol. 77:6988-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Z., H. Chen, P. Jiao, G. Deng, G. Tian, Y. Li, E. Hoffmann, R. G. Webster, Y. Matsuoka, and K. Yu. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 79:12058-12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654-9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matrosovich, M. N., S. Krauss, and R. G. Webster. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156-162. [DOI] [PubMed] [Google Scholar]
- 19.Naeem, K., A. Ullah, R. J. Manvell, and D. J. Alexander. 1999. Avian influenza A subtype H9N2 in poultry in Pakistan. Vet. Rec. 145:560. [DOI] [PubMed] [Google Scholar]
- 20.Nili, H., and K. Asasi. 2002. Natural cases and an experimental study of H9N2 avian influenza in commercial broiler chickens of Iran. Avian Pathol. 31:247-252. [DOI] [PubMed] [Google Scholar]
- 21.Peiris, J. S., Y. Guan, D. Markwell, P. Ghose, R. G. Webster, and K. F. Shortridge. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. Ip, R. W. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916-917. [DOI] [PubMed] [Google Scholar]
- 23.Perk, S., A. Panshin, E. Shihmanter, I. Gissin, S. Pokamunski, M. Pirak, and M. Lipkind. 2006. Ecology and molecular epidemiology of H9N2 avian influenza viruses isolated in Israel during 2000-2004 epizootic. Dev. Biol. (Basel) 124:201-209. [PubMed] [Google Scholar]
- 24.Sharp, G. B., Y. Kawaoka, D. J. Jones, W. J. Bean, S. P. Pryor, V. Hinshaw, and R. G. Webster. 1997. Coinfection of wild ducks by influenza A viruses: distribution patterns and biological significance. J. Virol. 71:6128-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp, G. B., Y. Kawaoka, S. M. Wright, B. Turner, V. Hinshaw, and R. G. Webster. 1993. Wild ducks are the reservoir for only a limited number of influenza A subtypes. Epidemiol. Infect. 110:161-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shortridge, K. F. 1992. Pandemic influenza: a zoonosis? Semin. Respir. Infect. 7:11-25. [PubMed] [Google Scholar]
- 27.Tang, X. Y., G. B. Tian, C. S. Zhao, J. F. Zhou, and K. Z. Yu. 1998. Isolation and characterization of prevalent strains of avian influenza viruses in China. Chin. J. Prev. Vet. Med. 1:1-5. [Google Scholar]
- 28.Uyeki, T. M., Y. H. Chong, J. M. Katz, W. Lim, Y. Y. Ho, S. S. Wang, T. H. Tsang, W. W. Au, and S. C. Chan. 2002. Lack of evidence for human-to-human transmission of avian influenza A (H9N2) viruses in Hong Kong, China 1999. Emerg. Infect. Dis. 8:154-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, C., W. Fan, R. Wei, and H. Zhao. 2004. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003 (H9N2) virus. Microbes Infect. 6:919-925. [DOI] [PubMed] [Google Scholar]
- 30.Xu, K. M., G. J. D. Smith, J. Bahl, L. Duan, H. Tai, D. Vijaykrishna, J. Wang, J. X. Zhang, K. S. Li, X. H. Fan, R. G. Webster, H. Chen, J. S. M. Peiris, and Y. Guan. 2007. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J. Virol. 81:10389-10401. [DOI] [PMC free article] [PubMed] [Google Scholar]