Abstract
By sequence and phylogenetic analyses, the 11 genomic segments of two bovine rotaviruses isolated from clinically infected calves were proven to be derived from the swine-like P[7]G5 genotype. This finding reinforced the hypothesis that interspecies transmission of completely heterologous strains can occur in nature.
Group A rotavirus (GARV), a member of the Reoviridae family, is a major cause of gastroenteritis in young children as well as in many animal species worldwide (3). The rotavirus genome is enclosed in three concentric layers and is composed of 11 segments of double-stranded RNA, which encode six structural proteins (VP1 to -4 and VP6 to -7) and five nonstructural proteins (NSP1 to -5) (3). This unique segmented nature of GARV genomes would enable reassortment between different viruses during coinfection and induce production of progeny viruses with novel or atypical phenotypes as noted above (3).
GARVs have a wide host range, but individual strains appear to be restricted to single hosts (7). In humans, the most common G serotypes are G1, G2, G3, and G9 and two main P genotypes are P[4] and P[8] (12). In pigs, there are at least four major G serotypes (G3, G4, G5, and G11) and two dominant P genotypes (P[6] and P[7]) (13). In cattle, serotypes G6, G8, and G10, in combination with genotypes P[1], P[5], and/or P[11], are the serotypes most commonly found (1). However, there is now increasing evidence that the transmission of GARVs can occur from animal to human as well as from animal to animal either by direct transmission of the virus or by the contribution of one or several genes to reassortants (9, 10). However, there is little evidence for the existence of bovine GARVs bearing all genomic segments of porcine origin.
The aim of this study was to characterize all 11 genomic segments of the two Korean bovine GARV strains. The results showed that both strains had swine-like genomic segments. These results contribute to a better understanding of interspecies transmission of heterologous GARVs.
Two bovine GARV strains (K5 and K8) were isolated from diarrheic fecal samples of two calves from different farms during 2004 and 2005. These strains were propagated and plaque purified three times, using a plaque assay as described elsewhere (11). The viral genomic double-stranded RNA was extracted from the virus-infected TF-104 cells, characterized by reverse transcription-PCR, and sequenced using specific primer pairs (11). Each gene sequence of the two strains was compared with those of other known GARVs (see Table S1 in the supplemental material) by the use of DNA Basic module software (DNAsis MAX, Alameda, CA). Phylogenetic and sequence analyses based on the deduced amino acid alignments were performed as described previously (11).
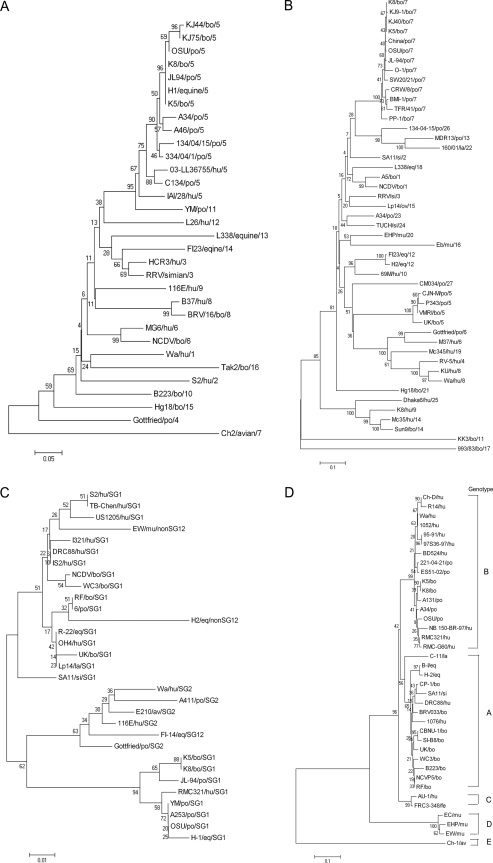
We compared the full-length (354 amino acids) VP7 protein sequence of the two Korean GARV strains with those of the GARV strains representative of all 16 G genotypes (4). Both strains showed the highest (92% to 100%) sequence identity to the porcine G5 GARV strains. Phylogenetically, the VP7 protein of the two strains clustered with porcine G5 GARVs (Fig. 1A). The deduced 292-amino-acid sequence of the VP4 gene of the two strains were compared with those of the representative GARV strains belonging to all 27 known P genotypes (8). The highest (95.2% to 99.0%) identity was found with the P[7] porcine GARVs. Phylogenetic analysis of the VP4 gene of the two strains provided a molecular basis for the findings revealing their close similarity to the P[7] genotype strains, particularly to the porcine P[7] genotype (Fig. 1B).
FIG. 1.
Phylogenetic trees of the VP7 (A), VP4 (B), VP6 (C), and NSP4 (D) proteins of the two Korean bovine rotavirus strains, showing their genetic relationship with the other representative group A rotavirus strains. The genotype for each VP7 (A), VP4 (B), and VP6 (C) protein is indicated as the last number in the strain designation.
Amplification of a 379-bp region of the VP6 gene, which is associated with subgroup (SG)-specific epitopes, allows the classification of GARVs into SGI, SGII, SGI/SGII, and non-SGI/non-SGII groups, according to the diversity within the VP6 gene (6). The deduced 126-amino-acid sequence of the VP6 gene, which includes SG-specific epitopes, showed maximum (96.0% to 98.4%) deduced amino acid identity of two Korean strains to those of the porcine SGI JL94 and YM strains. Phylogenetically, the two strains clustered closest to the porcine JL94 strain, forming a distinct branch within the SGI GARVs, and clustered distantly with the bovine SGI GARVs (Fig. 1C).
By sequence analysis, the NSP4 gene could be classified into five distinct genotypes designated A to E (2). An analysis of the NSP4 protein (292 amino acids) of the two strains showed that they shared higher deduced amino acid identity with the GARV B genotype than with other genotypes: 94.0% to 98.3% deduced amino acid identity with the genotype B porcine GARVs, including the A131, A34, and OSU strains (data not shown). Phylogenetically, the two strains clustered within the porcine NSP4 B genogroup and distantly clustered with the remaining NSP4 genotypes (Fig. 1D).
The partial deduced amino acid sequences of the VP1 (119 amino acids), VP2 (182 amino acids), and VP3 (194 amino acids) genes of the two strains were compared with those of the representative GARVs. For VP1, the highest (99.1 to 100%) deduced amino acid identity of the two strains was observed with the porcine Gottfried strain. Among the strains compared in the phylogenetic tree, the two strains clustered in a separate branch with porcine GARVs, including the YM and Gottfried strains (see Fig. S1A in the supplemental material). For VP2, the highest (92.8%) degree of deduced amino acid identity of the two strains was found with the porcine HP140 strain. The phylogenetic tree showed that the two strains had a fairly close relationship with the porcine HP140 strain and human RMC321, Wa, Beijing, and KU strains (see Fig. S1B in the supplemental material). For VP3, two strains shared the highest (94.4%) deduced amino acid identity with the porcine YM strain. In the phylogenetic tree, these strains clustered most closely with the porcine YM strain (see Fig. S1C in the supplemental material).
The NSP1 (484 amino acids) and NSP2 (332 amino acids) genes of the two strains showed the highest levels of deduced amino acid identity to the porcine OSU strain (99% to 100% identity and 99.7% identity, respectively). The phylogenetic trees of the partial NSP1 (see Fig. S2A in the supplemental material) and NSP2 (see Fig. S2B in the supplemental material) genes showed that the two strains clustered most closely with the porcine OSU strain. The comparison of the NSP3 (350 amino acids) and NSP5 (221 amino acids) genes of the two strains to the other representative strains showed that the sequences of the NSP3 and NSP5 proteins exhibited 98.1% to 99% deduced amino acid identity and 98% deduced amino acid identity, respectively. Phylogenetic analysis confirmed that both genes of the two strains clustered most closely with the porcine OSU strain (see Fig. S2C and D in the supplemental material).
Since G5 strains, which were originally detected and are prevalent in pigs, have been reported to have been found mainly in developing countries (5), they are considered to be emerging pathogens in humans. A previous study (11) reported that bovine GARV strains with the G5 genotype occurred in nature as a novel G genotype in cattle as a result of the natural reassortment between bovine and porcine strains. However, there is little information on the transmission of porcine G5 strains containing the full complement of porcine G5 rotavirus genome segments to heterologous animals and humans in nature. The structural features of each of the 11 genomic segments of the present two bovine strains were remarkably similar to those of the porcine P[7]G5 strains. Moreover, phylogenetic analyses revealed that the 11 genomic segments of these two strains were clustered in all-porcine phylogenetic subclusters. Therefore, the present evidence shows that the transmission of a heterologous strain, in addition to that of a reassortant virus, can also induce gastroenteritis in a heterologous host. To our knowledge, this is the first report of the molecular nature of bovine GARV strains with 11 genomic segments related to porcine strains.
In conclusion, 11 genomic segments of the two bovine GARV strains were all found to be of porcine origin. These findings reinforce the hypothesis that interspecies transmission of a completely heterologous strains can occur in nature. Our findings contribute to the growing body of information about GARV ecology and evolution by providing evidence for cross-species infection.
Nucleotide sequence accession numbers.
The 11 genomic segments of the two bovine GARV strains analyzed have been deposited in GenBank under accession numbers EU541374 (VP1 gene, strain K5), EU541375 (VP1 gene, strain K8), EU541380 (VP2 gene, strain K5), EU541381 (VP2 gene, strain K8), EU541386 (VP3 gene, strain K5), EU541387 (VP3 gene, strain K8), EU541392 (VP4 gene, strain K5), EU541393 (VP4 gene, strain K8), EU541398 (VP6 gene, strain K5), EU541399 (VP6 gene, strain K8), EU541404 (VP7 gene, strain K5), EU541405 (VP7 gene, strain K8), EU542702 (NSP1 gene, strain K5), EU542703 (NSP1 gene, strain K8), EU542708 (NSP2 gene, strain K5), EU542709 (NSP2 gene, strain K8), EU542714 (NSP3 gene, strain K5), EU542715 (NSP3 gene, strain K8), EU542720 (NSP4 gene, strain K5), EU542721 (NSP4 gene, strain K8), EU542726 (NSP5 gene, strain K5), and EU542727 (NSP5 gene, strain K8).
Supplementary Material
Acknowledgments
The study was supported by the National Veterinary Research and Quarantine Services (NVRQS), the Ministry of Agriculture and Forestry, the Korea Science and Engineering Foundation (KOSEF) (grant 2009-0081752), and the Regional Technology Innovation Program of the Ministry of Commerce, Industry and Energy (MOCIE), Republic of Korea. We acknowledge a graduate fellowship from the Korean Ministry of Education and Human Resources Development provided through the Brain Korea 21 project.
Footnotes
Published ahead of print on 26 August 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Alfieri, A. F., A. A. Alfieri, M. A. B. Barreiros, J. P. G. Leite, and L. J. Richtzenhain. 2004. G and P genotypes of group A rotavirus strains circulating in calves in Brazil, 1996-1999. Vet. Microbiol. 99:167-173. [DOI] [PubMed] [Google Scholar]
- 2.Ciarlet, M., F. Liprandi, M. E. Conner, and M. K. Estes. 2000. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotaviruses. Arch. Virol. 145:371-383. [DOI] [PubMed] [Google Scholar]
- 3.Estes, M. K., and A. Z. Kapikian. 2001. Rotaviruses, p. 1917-1974. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA.
- 4.Fukai, K., T. Takahashi, K. Tajima, S. Koike, K. Iwane, and K. Inoue. 2007. Molecular characterization of a novel bovine group A rotavirus. Vet. Microbiol. 123:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gouvea, V., L. de Castro, M. do Carmo Timenetsky, H. Greenberg, and N. Santos. 1994. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J. Clin. Microbiol. 32:1408-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iturriza Gómara, M., C. Wong, S. Blome, U. Desselberger, and J. Gray. 2002. Molecular characterization of VP6 genes of human rotavirus isolates: correlation of genogroups with subgroups and evidence for independent segregation. J. Virol. 76:6596-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA.
- 8.Khamrin, P., N. Maneekarn, S. Peerakome, W. Chan-it, F. Yagyu, S. Okitsu, and H. Ushijima. 2007. Novel porcine rotavirus of genotype P[27] shares new phylogenetic lineage with G2 porcine rotavirus strain. Virology 361:243-252. [DOI] [PubMed] [Google Scholar]
- 9.Martella, V., K. Bányai, M. Ciarlet, M. Iturriza-Gómara, E. Lorusso, S. de Grazia, S. Arista, N. Decaro, G. Elia, A. Cavalli, M. Corrente, A. Lavazza, R. Baselga, and C. Buonavoglia. 2006. Relationships among porcine and human P[6] rotaviruses: evidence that the different human P[6] lineages have originated from multiple interspecies transmission events. Virology 344:509-519. [DOI] [PubMed] [Google Scholar]
- 10.Matthijnssens, J., M. Rahman, V. Martella, Y. Xuelei, S. de Vos, K. de Leener, M. Ciarlet, C. Buonavoglia, and M. Van Ranst. 2006. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J. Virol. 80:3801-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park, S.-H., L. J. Saif, C. Jeong, G.-K. Lim, S.-I. Park, H.-H. Kim, S.-J. Park, Y.-J. Kim, J.-H. Jeong, M.-I. Kang, and K.-O. Cho. 2006. Molecular characterization of novel G5 bovine rotavirus strains. J. Clin. Microbiol. 44:4101-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 15:29-56. [DOI] [PubMed] [Google Scholar]
- 13.Winiarczyk, S., P. S. Paul, S. Mummidi, R. Panek, and Z. Gradzki. 2002. Survey of porcine rotavirus G and P genotype in Poland and in the United States using RT-PCR. J. Vet. Med. B Infect. Dis. Vet. Public Health 49:373-378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.