Abstract
In the development of obesity, the source of excess energy may influence appetite and metabolism. To determine the effects of differences in diet composition in obesity, mice were fed either a high-carbohydrate diet (HC; 10% fat energy) or a high-fat energy–restricted diet (HFR; 60% fat energy) over 18 wk in weight-matched groups of mice. To identify obesity-associated genes with persistently altered expression following weight reduction, mice were fed either a standard low-fat diet (LF; 10% fat energy), an unrestricted high-fat diet (HF; 60% fat energy), or a HF diet followed by weight reduction (WR). Mice fed a HF diet had significantly greater gonadal fat mass and higher whole blood glucose concentrations than mice fed an HC diet. Of the mice fed a high-fat diet, total body weight and serum insulin concentrations were greater in HF than in HFR. Microarray analysis revealed that HF vs. HC feeding resulted in global differences in adipocyte gene expression patterns. Although we identified genes whose expression was altered in both moderately and severely obese mice, there were also a large number of genes with altered expression only in severe obesity. Formerly obese, WR mice did not differ significantly from lean controls in total body weight or physiological measures. However, microarray analysis revealed distinctly different patterns of adipocyte gene expression. Furthermore, there were 398 genes with altered expression in HF mice that persisted in WR mice. Genes with persistently altered expression following obesity may play a role in rebound weight gain following weight reduction.
Introduction
The incidence of obesity is increasing at an alarming rate in developed countries (1,2). Aside from total energy intake, differences in diet composition may play an important role in the development of obesity and its complications, such as diabetes and cardiovascular disease. Dietary variety and consumption of restaurant foods have both been associated with greater body fat and studies of rats have revealed different metabolic patterns depending on the source of excess energy (3,4). However, the relationship between dietary composition and physiology remains poorly defined (5) and studies have not systematically evaluated the effects of high-carbohydrate feeding vs. high-fat feeding on metabolism.
Given the high prevalence of obesity, there has been interest in identifying factors associated with successful weight reduction. Most human studies have demonstrated that obese individuals typically return to pretreatment weight within 5 y of successful weight reduction, with a substantial proportion exceeding pretreatment weight (6–8). Expansion of the adipocyte population and secretion of small molecules from adipocytes may be involved in the failure to maintain weight loss; however, many factors responsible for rebound weight gain after weight reduction have yet to be identified.
Adipocyte-secreted factors influence diverse processes such as appetite and energy balance, lipid metabolism, insulin sensitivity, and inflammation (9,10) and visceral adiposity has been shown to be more directly related to cardiovascular risk than subcutaneous fat (11,12). We hypothesized that the visceral fat depot may be differentially responsive to diet composition and may produce factors not yet identified that may contribute to difficulty in maintaining weight loss. Using an oligonucleotide microarray, we performed a comprehensive evaluation of adipocyte gene expression in obesity as a function of diet composition (high fat vs. high carbohydrate). We then characterized gene expression in adipocytes derived from weight-reduced mice, identifying genes that may be involved in rebound weight gain following weight reduction.
Materials and Methods
Animals
Male C57BL/6J mice (Jackson Laboratory) were selected because these mice develop obesity and insulin resistance in response to high-fat feeding (13,14). Mice were obtained at 5–6 wk of age and housed 5 animals per cage. Animals were maintained on a 12-h-light/-dark cycle in a temperature-controlled room. Animal care and experimental manipulations were approved by the Institutional Animal Care and Use Committee at the Johns Hopkins University School of Medicine.
We recorded body weight on arrival and at weekly intervals in the morning. Food intake was recorded twice weekly. All mice were given commercially prepared diets (Research Diets; Supplemental Table 1). After 1 wk of acclimatization, mice were divided into 4 groups: mice fed a low-fat diet (LF;7 10% energy as fat; catalog no. D12450B); mice fed a high-fat diet (HF; 60% energy as fat; no. D12492); mice fed a high-carbohydrate diet (HC) consisting of LF supplemented with a liquid formula, prepared by dissolving the powdered LF in water to a concentration of 103.5 g/L, thus providing 1674 kJ/L; and mice fed a HF/restricted diet (HFR) provided in sufficient quantity to match total body weight to the HC group. For wk 1, mice in the HFR group were given a quantity of food calculated, based in part on pilot studies, to provide the same energy we estimated would be consumed by the mice fed the HC diet. We weighed mice weekly and adjusted the amount of food provided to match total body weight in the HFR and HC groups. Fresh formula was changed daily and provided to mice in the HC group in place of drinking water to increase total energy intake. After 13 wk, HF-fed mice were subdivided into 2 groups. One-half continued to consume the HF ad libitum and the other half was switched to a LF diet and restricted to 2 g per mouse per day to induce weight reduction (WR).
We used a total of 50 mice for this experiment (10 mice per group). Microarray and RT-PCR data are reported on 42 mice. Data on the remaining 8 mice are not available due to inadequately prepared labeled cRNA or degraded RNA.
Insulin tolerance testing and fasting insulin concentrations
Insulin tolerance testing was performed at wk 17. After food deprivation overnight, mice were administered 0.75 units/kg insulin (recombinant human regular insulin; Eli Lilly) intraperitoneally (i.p.). Venous tail blood was obtained for glucose determination by glucometer (Accu-Chek Advantage; Roche Diagnostics) at baseline and at 20, 40, and 60 min after insulin injection. The following week, following overnight food deprivation, mice were anesthetized with Avertin (2, 2, 2 tribromoethanol in tert-amyl alcohol; Sigma-Aldrich) 0.5 mg/g i.p. ~100 μL blood was obtained by mandibular bleed, centrifuged at 4000 × g; 5 min at 4°C and serum was stored at −80°C for subsequent insulin concentration determined by RIA (Linco Research).
Adipocyte isolation and RNA purification
After 18 wk, mice were killed by CO2 asphyxiation and gonadal fat pads were harvested and weighed. Adipocytes were purified from adipose tissue according to established methods (15). Gonadal fat pads were weighed and transferred to vials containing 6 mL Krebs ringer bicarbonate HEPES buffer with 5% bovine fraction V albumin (Millipore), 200 nmol/L adenosine (Sigma), and 10 mg collagenase type 1 powder (Worthington Biochemical). Tissue was minced with sharp scissors and samples were incubated in a shaking 37°C water bath for 1 h. Samples were passed through a 500-μm Nitex mesh (Sefar Filtration) into 50-mL conical tubes, centrifuged at 200 × g for 10 min at room temperature and washed in Krebs ringer bicarbonate HEPES/albumin/adenosine (KRBH) buffer followed by 2 cycles of centrifugation at 200 × g for 1 min at room temperature and washing with KRBH buffer.
Immediately after the final wash, total RNA was extracted from adipocytes using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. For each sample, 200 μL of adipocytes was homogenized in 1 mL of Trizol and remaining cells were frozen at −80°C. The quality of total RNA samples was assessed using an Agilent BioAnalyzer (Agilent Technologies) and was equivalent across all samples analyzed.
Oligonucleotide microarray
A 0.5-μg aliquot of total RNA from each sample was labeled using the Illumina TotalPrep RNA Amplification kit (Ambion) as previously described (16). A total of 0.85 μg of biotin-labeled cRNA was hybridized for 16 h to Illumina’s Sentrix MouseRef-8 Expression BeadChips (Illumina). The arrays were washed, blocked, and then hybridized. Biotinylated cRNA was detected with streptavidin-Cy3 and quantitated using Illumina’s BeadStation 500GX Genetic Analysis Systems scanner. Image processing and data extraction were performed using BeadStudio ver. 15 (Illumina) according to previously described methods (17).
RT-PCR confirmation
RT-PCR was performed with the iScript OneStep RT-PCR kit with SYBR Green (Bio-Rad) using the MyiQ Single Color Real-Time PCR Detection system with an iCycler thermocycler (Bio-Rad). RNA for RT-PCR was obtained from the same samples used for the microarray. Reactions were optimized for each primer set (Supplemental Table 2; 18) and PCR products were separated on a 1% agarose gel to verify product size. For every reaction, a standard curve was generated using serial dilutions of mouse adipocyte RNA. Primer concentration was 200 nmol/L for all reactions. All reactions included a dissociation curve analysis at the end of the amplification to confirm a single product at the expected melting temperature. Relative quantitation of gene expression was performed using the threshold cycle. Differences in mRNA expression were calculated after normalizing to 18S expression.
Data analysis
Values for weights, glucose, and insulin concentrations are expressed as means ± SD. Results of RT-PCR are expressed as relative differences in gene expression. Statistical analyses were performed using a t test for RT-PCR data. Repeated-measures ANOVA or 1-way ANOVA with Student-Newman-Keuls post test was performed for all other comparisons. Statistical analyses were performed using GraphPad InStat version 3.0 for Windows NT (GraphPad Software). Differences were considered significant at P < 0.05.
Microarray data were analyzed using DIANE 6.0, a spreadsheet-based microarray analysis program. Raw microarray data were subjected to Z normalization and tested for significant changes as previously described (19). Genes were determined to be differentially expressed after calculating the Z-ratio and false discovery rate (20). Individual genes with P-value < 0.01, Z-ratio > 2, and false discovery rate < 0.3 were considered significantly changed. Ingenuity pathways analysis (IPA) (Ingenuity Systems) was used to identify biological networks with the greatest number of differentially expressed genes. For each network or pathway, probability scores were calculated using the right-tailed Fisher’s exact test. A full discussion of the statistical methodologies employed can be found in Calvano et al. (21)
Results
Weights and food consumption
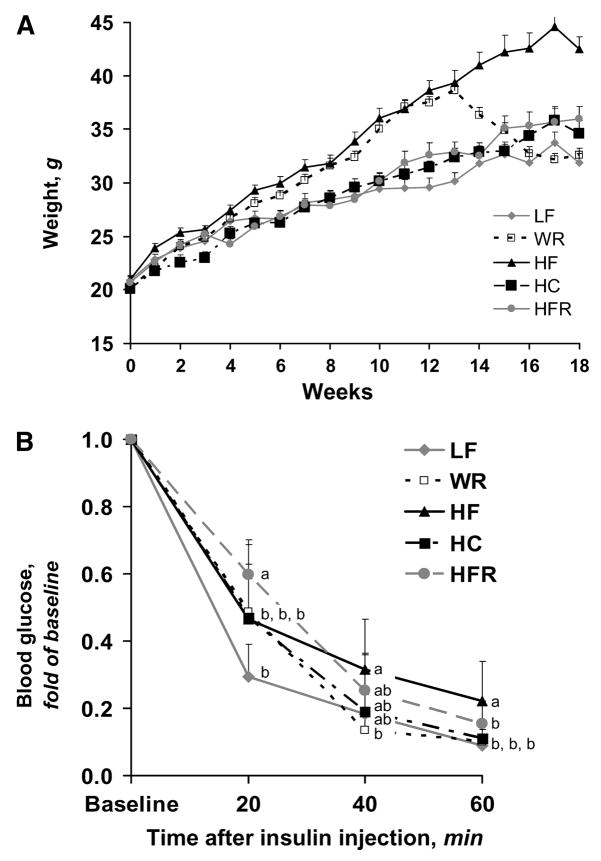
At the conclusion of the study (18 wk), total body weight did not differ between the LF and WR groups despite a 28% difference prior to weight reduction (Fig. 1A; Table 1). Total body weight remained similar between HC and HFR through the duration of the study and did not differ at the time of tissue harvest. Whereas solid food consumption and solid food energy intake were lower in the HC group than in the LF group (Supplemental Fig. 1), consumption of formula in the HC group instead of water resulted in greater total body weight in HC than in LF. Body weight was significantly greater in HF than all other groups (P < 0.001) and body weight was greater in HFR than in LF (P < 0.05). Despite equivalent total body weights, gonadal fat mass was greater in HFR than in HC (Table 1; P < 0.05). Interestingly, although total body weight was greater in HF than in HFR (P < 0.001), both groups had equivalent gonadal fat mass. As expected, gonadal fat mass was greater in HF than in both LF and WR (P < 0.01).
FIGURE 1.
Weekly total body weights (A) and proportional decrease in blood glucose concentrations following administration of 0.75 units/kg insulin i.p. to mice fed LF, HF, HFR, HC, or WR diet after overnight food deprivation (B). Weight at wk 18 and baseline blood glucose concentrations are reported in Table 1. Values are means + SD, n = 10. Means at a time without a common letter differ, P < 0.05.
TABLE 1.
Total body mass, gonadal fat mass, serum insulin, and whole blood glucose concentrations in mice fed a LF, HF, HFR, HC, or WR diet1
| Weight | Gonadal fat | Insulin2 | Glucose2 | |
|---|---|---|---|---|
| g | pmol/L | mmol/L | ||
| LF | 31.9 ± 2.7c | 1.21 ± 0.37b | 63.7 ± 5.2b | 5.57 ± 1.10b |
| HF | 42.5 ± 3.8a | 2.23 ± 0.51a | 145 ± 87.0a | 7.85 ± 1.72a |
| HFR | 36.0 ± 3.6b | 2.02 ± 0.34a | 62.6 ± 8.5b | 7.07 ± 1.45a |
| HC | 34.6 ± 3.7bc | 1.60 ± 0.44b | 57.9 ± 7.7b | 5.24 ± 0.48b |
| WR | 32.6 ± 1.9bc | 1.54 ± 0.30b | 73.3 ± 17.3b | 5.68 ± 1.03b |
Values are means ± SD, n = 6–10. Means in a column without a common letter differ, P < 0.05.
Concentrations were measured after overnight food deprivation.
Glucose metabolism
Insulin and glucose concentrations were measured in all groups of mice after overnight food deprivation. Serum insulin concentrations were higher in HF than in all other groups (P < 0.01; Table 1). Although serum insulin concentrations did not differ between LF, HC, and HFR, whole blood glucose concentrations were higher in HFR compared with LF (P < 0.05) and HC (Table 1; P < 0.01). Whole blood glucose concentrations were also higher in HF than LF, WR, and HC (P < 0.001) but not HFR. Thus, differences in glucose and insulin concentrations appeared to be more closely related to adiposity rather than total body weight.
Following insulin administration, whole blood glucose concentrations were lower at 20 min in LF compared with HFR, HF, HC, and WR (Fig. 1B; P < 0.01), suggesting greater insulin sensitivity in LF than in all other groups. This physiologic difference between LF and WR indicates there was incomplete recovery from prior obesity. Whole blood glucose concentrations 60 min after insulin administration were lower in LF, WR, and HFR (P < 0.05) compared with HF. The latter comparison demonstrates there was an effect of degree of obesity on insulin sensitivity in these mice.
Gene expression
We identified genes differentially expressed in HF vs. LF, HFR vs. HF, HFR vs. LF, HFR vs. HC, WR vs. LF, and WR vs. HF (Table 2; Supplemental Table 3; Supplemental Summary of Analysis). IPA demonstrated altered gene expression in a broad range of cellular functions (Supplemental Table 4). Interestingly, the most highly involved pathway involved PPAR signaling, which was downregulated in WR compared with LF.
TABLE 2.
Numbers of adipocyte genes that were differentially expressed in mice fed a LF, HF, HFR, HC, or WR diet1
| HF vs. LF | HFR vs. HF | HFR vs. LF | HFR vs. HC | WR vs. LF | WR vs. HF | |
|---|---|---|---|---|---|---|
| n | ||||||
| Upregulated | 526 | 298 | 396 | 372 | 390 | 368 |
| Downregulated | 280 | 531 | 352 | 394 | 227 | 538 |
P < 0.01 for all genes with significantly different expression between 2 groups.
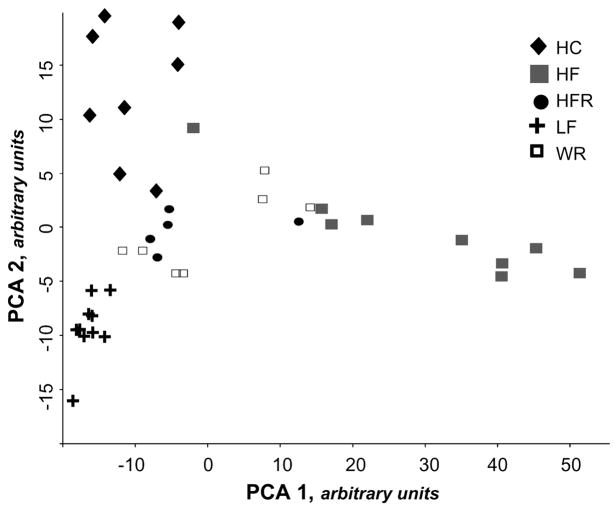
Principal components analysis (PCA) and hierarchical clustering demonstrated global differences in adipocyte gene expression between groups (Fig. 2; Supplemental Fig. 2). Despite equivalent body weight at the time of study, HC and HFR did not overlap in PCA or hierarchical clustering and both groups displayed distinct patterns of gene expression from LF controls. Analysis of HF, LF, and WR revealed separation between the 3 groups, indicating that diet-induced obesity and weight reduction resulted in distinct patterns of adipocyte gene expression.
FIGURE 2.
PCA of adipocyte gene expression was performed in DIANE 6.0 to identify clustering within groups. Each data point represents overall adipocyte gene expression data for a single mouse, n = 5–10.
To evaluate changes in gene expression due to the degree of obesity, we compared adipocyte gene expression in HFR and HF. We identified 587 genes that were not altered in HFR vs. LF but were altered in HFR vs. HF, indicating that expression of these genes did not change with moderate obesity (HFR) but did change with severe obesity (HF). In both PCA and hierarchical clustering, there was clear separation between HF and HFR.
We generated a list of genes that were differentially expressed in both WR vs. LF and HF vs. LF. We found that there were 268 genes upregulated and 130 genes downregulated in HF that were similarly differentially expressed in WR. Thus, for a large number of genes, altered expression in diet-induced obesity persisted even after weight reduction had been achieved and maintained in steady state. We used IPA to identify altered pathways (Supplemental Table 4) and to sort genes by location to compile a list of gene products that localize to the extracellular compartment. This resulted in identifying 59 genes differentially expressed in HF and WR (compared with LF) that code for extracellular proteins (Table 3).
TABLE 3.
Genes that localize to the extracellular compartment that were differentially expressed in adipocytes derived from mice fed a LF or WR diet1
P < 0.01 for all genes listed.
Quantitative RT-PCR
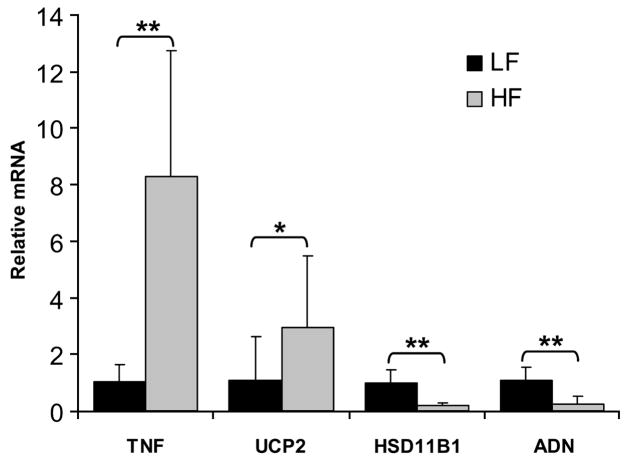
Four genes with differential expression in HF vs. LF were analyzed by RT-PCR to validate results from the microarray experiment (Fig. 3). Targets were selected based on robust fold-change in expression and relevance to adipocyte metabolism as identified in previously published studies. Of the 4 targets studied, 2 increased in HF compared with LF and 2 decreased in HF compared with LF. RT-PCR results were consistent with microarray data for all targets evaluated. Gene expression in WR was intermediate for all 4 genes (data not shown), consistent with the PCA and hierarchical clustering, which indicated that the pattern of adipocyte gene expression in WR was unique compared with that of HF and LF.
FIGURE 3.
Relative mRNA concentrations for tumor necrosis factor (TNF), uncoupling protein 2 (UCP2), hydroxysteroid 11-β dehydrogenase type 1 (HSD11B1), and adipsin (ADN) in mice fed a LF diet compared with mice fed a HF diet. Values were normalized to 18S ribosomal subunit expression. A 40-ng sample of total RNA per reaction was used to quantitate all templates except 18S RNA, where 160 pg per reaction was used. Values are means + SD, n = 5–10. Asterisks indicate that means differ: *P < 0.05, **P < 0.01.
Discussion
In this study, the source of excess energy played a role in the phenotype of obese mice. When matched for total body weight, HF feeding was associated with higher fasting blood glucose concentrations and greater gonadal fat mass than HC feeding. Similarities in insulin concentrations between HFR and HC may be a result of correlation with weight or, alternatively, may be due to mild β cell toxicity induced by HF, as reported in other studies (22,23).
Several studies have identified differences in physiology and metabolism in high-fat vs. high-carbohydrate–induced obesity in rodents (3,24,25). Using a comprehensive microarray, we discovered that differences in diet composition affect the expression of a large number of genes. The clustering of gene expression data demonstrates that diet and pattern of adipocyte gene expression are associated. There are many signals originating from adipocytes that play a role in total body insulin sensitivity and hypothalamic control of appetite and the patterns of adipocyte gene expression we have identified may play a role in these complex biological processes.
Our data confirm that changes in degree of obesity are associated with changes in physiologic measures such as fasting insulin concentrations and insulin sensitivity. Not surprisingly, we found that as obesity increases, the altered expression of a number of genes that occurs in moderate obesity (HFR) persists into severe obesity (HF). However, we also determined that there are a large number of genes whose expression levels in moderately obese mice are not different from those in lean mice but are altered only in severe obesity. PCA and hierarchical clustering also demonstrate distinct segregation of HF and HFR mice by adipocyte gene expression. Thus, increasing obesity is associated with qualitative changes in gene expression, with unique patterns of gene expression in HFR and HF.
By merging WR and HF gene expression lists, we were able to identify factors that could play a role in failure to maintain weight loss after energy restriction. By the end of the study, WR mice had maintained weight in a steady state equivalent to the weight of lean controls for 2 wk. Despite near normalization of measured physiologic variables, WR mice displayed a pattern of adipocyte gene expression markedly different from that of either the obese, HF group or the lean, LF mice. PCA and hierarchical clustering revealed broad differences in adipocyte gene expression among the WR, HF, and LF mice. It remains unclear why a number of obesity-related genes continue to have altered expression following weight reduction. A number of different epigenetic phenomena may be involved, including DNA methylation or DNA/protein acetylation (26).
A number of genes we identified as persistently altered following weight reduction have previously been described as playing a role in appetite and adipose tissue development, thus confirming the physiological importance of our findings. Interestingly, a number of factors in this list are cytokines that have been implicated in cachexia related to illness and inflammation. These findings are consistent with previous studies by Clement et al. (27), which showed differential expression of 100 inflammation-related transcripts in adipose tissue of obese individuals that had been on a very low-energy diet for 1 mo. A number of factors that remain upregulated following weight loss are known to act locally in obesity. For example, colony-stimulating factor 1 has been shown to be upregulated in adipose tissue of human subjects who gain weight with overfeeding (28). Matrix metalloproteinases 2, 12, and 14 are strongly induced in adipose tissue in obesity (29) and matrix metalloproteinase 2 is elevated in obese mice (30).
As many signals related to appetite and satiety originate from adipose tissue, we hypothesized that genes differentially expressed in HF and remaining differentially expressed after diet could play a role in the rebound weight gain that often follows dieting and weight loss. Two proteins in Table 3 have been shown to affect appetite in rodents. Tenascin c (TNC) is an extracellular matrix protein expressed in the central nervous system. TNC-knockout mice were found to have lower neuropeptide Y (NPY) mRNA concentrations in limbic structures of the brain (31). As NPY is known to be orexigenic, one could speculate that increased circulating concentrations of TNC could result in increased NPY concentrations and increased appetite. Additional studies are required to determine whether these factors are consistently elevated postobesity and if they act on the hypothalamus to alter appetite.
To our knowledge, this study is unique in demonstrating that in mice of equivalent body weight, differences in diet composition lead to broad differences in adipocyte gene expression. Altered adipocyte gene expression as a consequence of diet composition may be adaptive to a diet high in saturated fat but may also be responsible for increasing the risk of diabetes and other obesity-related disorders. Although other investigators have shown changes in gene expression in adipocytes and adipose tissue in obesity (32–34), we have demonstrated differences in metabolism and adipocyte gene expression between formerly obese mice and weight-matched controls. Furthermore, this study has demonstrated that there are genes with altered expression in obesity that do not correct following weight reduction. Although a small number of adipocyte-secreted factors are known to be involved in appetite and energy balance, our findings reveal a number of adipocyte-secreted factors with persistently altered expression following weight reduction that may play a role in rebound weight gain following weight loss.
Supplementary Material
Acknowledgments
We thank Daniel Diaczok for assistance with tissue harvesting, William H. Wood III for array hybridization, and Yonqing Zhang for data analysis.
Footnotes
Supported by NIH DK55831 (to D.W.C.) and T32 DK007751 (to R.S.M.). This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
Author disclosures: R. Miller, K. Becker, V. Prabhu, and D. Cooke, no conflicts of interest.
Supplemental Tables 1–4, Supplemental Figures 1 and 2, and a Summary of Analysis are available with the online posting of this paper at jn.nutrition.org.
Abbreviations used: HC, high-carbohydrate/-energy; HF, high-fat; HFR, high-fat–restricted; i.p., intraperitoneally; IPA, ingenuity pathways analysis; KRBH, krebs ringer bicarbonate HEPES/albumin/adenosine; LF, low-fat; WR, weight-reduced; NPY, neuropeptide Y; PCA, principal components analysis; TNC, tenascin c; TNF, tumor necrosis factor.
Literature Cited
- 1.Cowie CC, Rust KF, Byrd-Holt DD, Eberhardt MS, Flegal KM, Engelgau MM, Saydah SH, Williams DE, Geiss LS, et al. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health and Nutrition Examination Survey 1999–2002. Diabetes Care. 2006;29:1263–8. doi: 10.2337/dc06-0062. [DOI] [PubMed] [Google Scholar]
- 2.Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006;295:1549–55. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 3.Dourmashkin JT, Chang GQ, Gayles EC, Hill JO, Fried SK, Julien C, Leibowitz SF. Different forms of obesity as a function of diet composition. Int J Obes (Lond) 2005;29:1368–78. doi: 10.1038/sj.ijo.0803017. [DOI] [PubMed] [Google Scholar]
- 4.Stubbs RJ, Johnstone AM, Mazlan N, Mbaiwa SE, Ferris S. Effect of altering the variety of sensorially distinct foods, of the same macronutrient content, on food intake and body weight in men. Eur J Clin Nutr. 2001;55:19–28. doi: 10.1038/sj.ejcn.1601117. [DOI] [PubMed] [Google Scholar]
- 5.Willett WC. Is dietary fat a major determinant of body fat? Am J Clin Nutr. 1998;67:S556–62. doi: 10.1093/ajcn/67.3.556S. [DOI] [PubMed] [Google Scholar]
- 6.Douketis JD, Macie C, Thabane L, Williamson DF. Systematic review of long-term weight loss studies in obese adults: clinical significance and applicability to clinical practice. Int J Obes (Lond) 2005;29:1153–67. doi: 10.1038/sj.ijo.0802982. [DOI] [PubMed] [Google Scholar]
- 7.Miller WC. How effective are traditional dietary and exercise interventions for weight loss? Med Sci Sports Exerc. 1999;31:1129–34. doi: 10.1097/00005768-199908000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Wadden TA. Treatment of obesity by moderate and severe caloric restriction. Results of clinical research trials. Ann Intern Med. 1993;119:688–93. doi: 10.7326/0003-4819-119-7_part_2-199310011-00012. [DOI] [PubMed] [Google Scholar]
- 9.Rajala MW, Scherer PE. Minireview: The adipocyte–at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology. 2003;144:3765–73. doi: 10.1210/en.2003-0580. [DOI] [PubMed] [Google Scholar]
- 10.Trayhurn P. Endocrine and signalling role of adipose tissue: new perspectives on fat. Acta Physiol Scand. 2005;184:285–93. doi: 10.1111/j.1365-201X.2005.01468.x. [DOI] [PubMed] [Google Scholar]
- 11.Arner P. Not all fat is alike. Lancet. 1998;351:1301–2. doi: 10.1016/S0140-6736(05)79052-8. [DOI] [PubMed] [Google Scholar]
- 12.Pouliot MC, Despres JP, Lemieux S, Moorjani S, Bouchard C, Tremblay A, Nadeau A, Lupien PJ. Waist circumference and abdominal sagittal diameter: best simple anthropometric indexes of abdominal visceral adipose tissue accumulation and related cardiovascular risk in men and women. Am J Cardiol. 1994;73:460–8. doi: 10.1016/0002-9149(94)90676-9. [DOI] [PubMed] [Google Scholar]
- 13.Collins S, Martin TL, Surwit RS, Robidoux J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: physiological and molecular characteristics. Physiol Behav. 2004;81:243–8. doi: 10.1016/j.physbeh.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Surwit RS, Feinglos MN, Rodin J, Sutherland A, Petro AE, Opara EC, Kuhn CM, Rebuffe-Scrive M. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism. 1995;44:645–51. doi: 10.1016/0026-0495(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 15.Smith U, Sjostrom L, Bjornstorp P. Comparison of two methods for determining human adipose cell size. J Lipid Res. 1972;13:822–4. [PubMed] [Google Scholar]
- 16.Cheadle C, Becker KG, Cho-Chung YS, Nesterova M, Watkins T, Wood W, III, Prabhu V, Barnes KC. A rapid method for microarray cross platform comparisons using gene expression signatures. Mol Cell Probes. 2007;21:35–46. doi: 10.1016/j.mcp.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Gunderson KL, Kruglyak S, Graige MS, Garcia F, Kermani BG, Zhao C, Che D, Dickinson T, Wickham E, et al. Decoding randomly ordered DNA arrays. Genome Res. 2004;14:870–7. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 19.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 21.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 22.Zhou YP, Ling ZC, Grill VE. Inhibitory effects of fatty acids on glucose-regulated B-cell function: association with increased islet triglyceride stores and altered effect of fatty acid oxidation on glucose metabolism. Metabolism. 1996;45:981–6. doi: 10.1016/s0026-0495(96)90267-x. [DOI] [PubMed] [Google Scholar]
- 23.Roche E, Maestre I, Martin F, Fuentes E, Casero J, Reig JA, Soria B. Nutrient toxicity in pancreatic beta-cell dysfunction. J Physiol Biochem. 2000;56:119–28. doi: 10.1007/BF03179907. [DOI] [PubMed] [Google Scholar]
- 24.Kahn BB, Pedersen O. Suppression of GLUT4 expression in skeletal muscle of rats that are obese from high fat feeding but not from high carbohydrate feeding or genetic obesity. Endocrinology. 1993;132:13–22. doi: 10.1210/endo.132.1.8419118. [DOI] [PubMed] [Google Scholar]
- 25.Kanarek RB, Marks-Kaufman R. Developmental aspects of sucrose-induced obesity in rats. Physiol Behav. 1979;23:881–5. doi: 10.1016/0031-9384(79)90195-1. [DOI] [PubMed] [Google Scholar]
- 26.Feinberg AP, Cui H, Ohlsson R. DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin Cancer Biol. 2002;12:389–98. doi: 10.1016/s1044-579x(02)00059-7. [DOI] [PubMed] [Google Scholar]
- 27.Clement K, Viguerie N, Poitou C, Carette C, Pelloux V, Curat CA, Sicard A, Rome S, Benis A, et al. Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. FASEB J. 2004;18:1657–69. doi: 10.1096/fj.04-2204com. [DOI] [PubMed] [Google Scholar]
- 28.Levine JA, Jensen MD, Eberhardt NL, O’Brien T. Adipocyte macrophage colony-stimulating factor is a mediator of adipose tissue growth. J Clin Invest. 1998;101:1557–64. doi: 10.1172/JCI2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chavey C, Mari B, Monthouel MN, Bonnafous S, Anglard P, Van Obberghen E, Tartare-Deckert S. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J Biol Chem. 2003;278:11888–96. doi: 10.1074/jbc.M209196200. [DOI] [PubMed] [Google Scholar]
- 30.Ahima RS, Qi Y, Singhal NS, Jackson MB, Scherer PE. Brain adipocytokine action and metabolic regulation. Diabetes. 2006;55 Suppl 2:S145–54. doi: 10.2337/db06-s018. [DOI] [PubMed] [Google Scholar]
- 31.Fukamauchi F, Aihara O, Kusakabe M. Reduced mRNA expression of neuropeptide Y in the limbic system of tenascin gene disrupted mouse brain. Neuropeptides. 1998;32:265–8. doi: 10.1016/s0143-4179(98)90046-4. [DOI] [PubMed] [Google Scholar]
- 32.Baranova A, Collantes R, Gowder SJ, Elariny H, Schlauch K, Younoszai A, King S, Randhawa M, Pusulury S, et al. Obesity-related differential gene expression in the visceral adipose tissue. Obes Surg. 2005;15:758–65. doi: 10.1381/0960892054222876. [DOI] [PubMed] [Google Scholar]
- 33.Moraes RC, Blondet A, Birkenkamp-Demtroeder K, Tirard J, Orntoft TF, Gertler A, Durand P, Naville D, Begeot M. Study of the alteration of gene expression in adipose tissue of diet-induced obese mice by microarray and reverse transcription-polymerase chain reaction analyses. Endocrinology. 2003;144:4773–82. doi: 10.1210/en.2003-0456. [DOI] [PubMed] [Google Scholar]
- 34.Urs S, Smith C, Campbell B, Saxton AM, Taylor J, Zhang B, Snoddy J, Jones VB, Moustaid-Moussa N. Gene expression profiling in human preadipocytes and adipocytes by microarray analysis. J Nutr. 2004;134:762–70. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.