Abstract
An important aspect of microarray studies involves the prediction of patient survival based on their gene expression levels. To cope with the high dimensionality of the microarray gene expression data, it is customary to first reduce the dimension of the gene expression data via dimension reduction methods, and then use the Cox proportional hazards model to predict patient survival. In this paper, we propose a variant of Partial Least Squares, denoted as Rank-based Modified Partial Least Squares (RMPLS), that is insensitive to outlying values of both the response and the gene expressions. We assess the performance of RMPLS and several dimension reduction methods using a simulation model for gene expression data with a censored response. In particular, Principal Component Analysis (PCA), modified Partial Least Squares (MPLS), RMPLS, Sliced Inverse Regression (SIR), Correlation Principal Component Regression (CPCR), Supervised Principal Component Regression (SPCR) and Univariate Selection (UNIV) are compared in terms of mean squared error of the estimated survival function and the estimated coefficients of the covariates, and in terms of the bias of the estimated survival function. It turns out that RMPLS outperforms all other methods in terms of the mean squared error and the bias of the survival function in the presence of outliers in the response. In addition, RMPLS is comparable to MPLS in the absence of outliers. In this setting, both RMPLS and MPLS outperform all other methods considered in this study in terms of mean squared error and bias of the estimated survival function.
Introduction
Microarray studies allow researchers to quickly and efficiently perform simultaneous analyses of thousands of genes in a single experiment to gain insight into gene function. Much of the interest on microarray data analysis derives from the potential of identifying the genes that relate to biological processes, the classification of tumor types and tumor stages based on gene expression patterns, and the study of gene interactions. However, because microarray data often include survival information on patients, it is important to analyze patient survival times (response) in terms of their corresponding gene expression levels (predictors). This paper is concerned with dimension reduction methodologies when modeling survival times in the presence of censoring, taking into account the microarray data information.
The major challenge in using microarray data in survival analysis is its large dimensionality, typically in the range of ten to thirty thousand genes, while the number of cases is usually orders of magnitude smaller. Existing statistical methods such as the commonly used linear regression model and survival analysis require less predictors than cases. Furthermore, gene expression levels are often highly correlated, which makes the analysis even more difficult. Several authors have proposed penalized partial likelihood approaches for the Cox Proportional Hazards (PH) model (Cox (1972)) to cope with the high dimensionality of the gene expression data. Li and Luan (2003) used kernel transformations of the Cox partial likelihood in the framework of a penalization method. Gui and Li (2005a) proposed using a threshold gradient descent minimization of the Cox partial likelihood to estimate the regression parameters. Gui and Li (2005b) also proposed a penalized method for the Cox regression based on the Least Angle Regression (LARS) algorithm of Efron (2004). However, Engler and Li (2007) pointed out that there are several drawbacks to these methods. For example, the approach of Li and Luan (2003) does not provide a recipe for the selection of the genes to be included in the prediction of the survival function. In the approach proposed by Gui and Li (2005a), the number of selected genes is sensitive to changes in the threshold parameter. When the penalty function is not strictly convex, as in the case of LARS, and given that the predictors are highly correlated, Gui and Li’s approach (2005b) often identifies only one of the predictors and ignores the others.
Another approach to deal with the high dimensionality of gene expression levels is to employ a two-stage procedure. In stage 1, we reduce the dimension of the microarray data matrix from N x p to N x K whereK < N using dimension reduction methods, and then in stage 2, we apply the regression model in the reduced subspace. Several papers in the literature provide comparison studies among the different dimension reduction methods employing the two-stage procedure. Bura and Pfeiffer (2003) concluded that Sliced Average Variance Estimation (SAVE) is better than Sliced Inverse Regression (SIR) in terms of classification accuracy of tumor classes. Boulesteix (2004) combined Partial Least Squares (PLS) with Linear Discriminant Analysis (LDA). The approach outperforms several classification methods such as Nearest Neighbor (NN), Prediction Analysis of Microarray (PAM) and Support Vector Machines (SVM). Nguyen and Rocke (2004) and Nguyen (2005) concluded that PLS and modified versions of PLS (to incorporate censoring) outperfom Principal Component Analysis (PCA) in terms of percentage of correct classification, and mean squared error of the estimated survival function, where the survival is evaluated using the average of the covariates in the Cox model. According to Dai et al (2006), PLS and Sliced Inversed Regression (SIR) outperform PCA in terms of classification error rates. In Bair et al. (2006), Supervised Principal Component Regression (SPCR) outperforms PCA and PLS in terms of classification error of tumor subtypes. Bolvestad et al (2007) stated that PCA performed slightly better than SPCR in terms of the log-rank test, prognostic index and the deviance in the Cox model. In Zhao and Sun (2007), Correlation Principal Component Regression (CPCR) is as competitive as modified versions of PLS in terms of root mean squared error of prediction of martingale residuals in the Cox model, and in terms of classification accuracy.
However, the performance of the different dimension reduction methods seems to be data-specific. In other words, method A may outperform method B for one dataset, but the opposite may be observed for another dataset. When the number of genes far exceeds the number of cases, no clear-cut winner among the dimension reduction methods can be deduced either in the context of classification or prediction. Furthermore, there is a lack of a large simulation study that compares the different dimension reduction methods, in the presence of outliers, in terms of the mean squared error of the β′s, which are the coefficients of the genes in the Cox PH model, and the mean squared error and bias of the estimated survival function evaluated using the covariates corresponding to the individuals in the Cox regression model.
In this paper, we assess the performance of several dimension reduction methods through a simulation study using the Cox Proportional Hazards regression model at the second stage in the presence of outliers. The competing methods are: Principal Component Analysis (PCA), Modified Partial Least Squares (MPLS) of Nguyen and Rocke (2004), Rank-based Modified Partial Least Squares (RMPLS) (described in section 1), Sliced Inverse Regression (SIR), Univariate Selection (UNIV), Supervised Principal Component Regression (SPCR), and Correlation Principal Component Regression (CPCR). We consider the following measures to compare the methods:
MSE(β): mean squared error of the weights placed on the covariates,
ave(d2): mean squared error of the estimated survival function evaluated using the average of the covariates.
ave(d2.ind): mean squared error of the estimated survival function evaluated using the covariates corresponding to the individuals.
ave(bias): average bias of the estimated survival function evaluated using the average of the covariates.
ave(bias.ind): average bias of the estimated survival function evaluated using the covariates corresponding to the individuals.
Both measures of bias, ave(bias) and ave(bias.ind), are calculated at the deciles of the true survival function.
It turns out that in the presence of outliers in the response, RMPLS outperforms all other methods, including MPLS, in terms of ave(d2) and ave(d2.ind). Also, in terms of ave(bias) and ave(bias.ind), RMPLS outperforms all other methods for small to medium deciles. Furthermore, RMPLS is comparable to MPLS in the absence of outliers in the response for all five measures. In this setting, both RMPLS and MPLS outperform other methods in terms of ave(d2) and ave(d2.ind), and in terms of ave(bias) and ave(bias.ind) for small to medium deciles. In terms of MSE(β), PCA, MPLS, RMPLS and SPCR perform relatively the same in both the presence and absence of outliers, and these methods outperform CPCR and UNIV.
The paper is organized as follows. We describe the Cox proportional hazards model and the dimension reduction methods in section 1. We present a variant of Partial Least Squares in this section, which we refer to as Rank-based Modified Partial Least Squares. The method is insensitive to outlying values in both the predictors and response, and also incorporates the censoring information. In section 2, we describe the simulation procedure for the gene expression values, and the survival and censoring times. In section 3, we provide simulation results for two scenarios: 1) when the number of components, K, is fixed across the methods, and 2) when K is selected using cross-validation for each method. Also, the assessment of the performance of the methods on two real datasets are given in section 3. We provide some conclusions and discussion in section 4.
1. Dimension Reduction Methods
Dimension reduction seeks to reduce the size of the microarray dataset, often in the order of thousands, while trying to retain most of the relevant information contained in the original dataset, according to some criteria. This is typically done by creating a set of orthogonal linear combinations of the gene expression levels and then selecting a subset of these based on some criteria associated with the ability of the elements in this subset to predict the response. A notable example is Partial Least Squares (PLS) which will be described in this section along with several other dimension reduction methods. First, we introduce some notation and describe the Cox PH model.
Notation
Define X to be the N x p matrix of centered gene expression values (i.e., the p columns of X are centered by subtracting the column means from the column values), where N is the number of individuals (patients), and p is the number of genes with N ≪ p. Let y be the N x 1 vector of true survival times, c be N x 1 vector of right-censoring times, and let y and c be independent. What we actually observe is Ti = min(yi, ci), and censoring indicators δi = I(yi ≤ ci) for i = 1, . . . , N (δi = 1 if the true survival time is observed, and δi = 0 if censoring occurs).
Cox Proportional Hazards (PH) Model
One popular regression model that takes into account the censored response is the Cox Proportional Hazards (PH) model. The Cox model is given as:
| (1) |
where h0(t) denotes an unspecified baseline hazard function, and zi is the vector of covariates corresponding to the ith individual, and β′s are the regression coefficients. Here, the parameters β can be estimated by maximizing the partial likelihood, which is given in the expression below (Klein and Moeschberger (2003)):
| (2) |
where D is the number of deaths, t1 < t2 < ⋯ < tD are the ordered death times, z(i) are the covariates corresponding to the individual with survival time ti, and the risk set R(ti) is the set of individuals who are still under study at the time just prior to ti. The partial likelihood in Eq. (2) does not involve the baseline hazard h0(.), and thus, h0(.) can be left unspecified in the estimation of β. Eq. (1) implies the proportionality of the hazard rates assumption, which states that given two individuals with different covariate values, the ratio of the hazard functions for these two individuals does not depend on time. Since the hazard function characterizes the survival function, the PH model can be rewritten in terms of survival function as:
| (3) |
where S0(t) denotes the baseline survival function, which can be estimated by the Kaplan-Meier product limit estimator (Kaplan (1958)) or the Nelson-Aalen estimator (Aalen (1978)). We use the Nelson-Aalen estimator to estimate the baseline survival function in this paper.
When the number of predictors p is larger than the number of individuals N, the parameter estimates obtained from the Cox partial likelihood Eq. (2) are nonunique, unstable and have large variances. To cope with the high dimensionality of the gene expression data, we first use dimension reduction methods to reduce the dimension of the original data from p to K whereK < N, and then apply the Cox regression model in the reduced subspace. In other words, the dimension of the microarray data matrix X is first reduced from N x p to N x K whereK < N using dimension reduction techniques in the first stage. We denote the N x K reduced data matrix by X̃. In the second stage, the reduced data matrix X̃ is used in the multivariate Cox PH regression model.
We now describe the dimension reduction methods.
1.1. Principal Component Analysis (PCA)
PCA is a dimension reduction technique that sequentially constructs orthogonal components by maximizing the variance of the linear combinations of the original predictors. Mathematically, the sequence of the weight vectors is obtained as,
| (4) |
subject to the orthogonal constraints w′kX′Xwj = 0 for all 1 ≤ j < k, where k = 1, . . . , m, and m = min(N, p). The kth Principal Component (PC) is defined as x̃k = Xwk. The constraints w′kX′Xwj = 0 ensure that the PCs are orthogonal.
One approach to derive the Principal Components (PCs) is through the eigenvalue decomposition of the sample covariance matrix, which equals because X is centered. Since S is symmetric, it can be diagonalized by the orthogonal matrix of its eigenvectors,
| (5) |
where the NxN matrix Δ = diag(λ1 ≥ ⋯ ≥ λN) and represent the eigenvalues of S in descending order, and the columns of the pxN orthogonal matrix V = (v1, . . . , vN) are the corresponding eigenvectors that provide the weights (loadings) for the linear combinations. The PCs are x̃k = Xvk, where vk correspond to the columns of V. Since the λ’s are in descending order, the PCs are also ordered in terms of the amount of variation in the original data they account for. In many cases, the first few PCs explain most of the variation in the original data, and thus, we can ignore the rest of the PCs without losing much of the information. Also, because the weight vectors w are constructed so that they are unit vectors, w′w = 1, the proportion of the variation explained by the kth PC is λk/p, and the cumulative proportion for the first K PCs is .
1.2. Modified Partial Least Squares (MPLS)
The Partial Least Squares (PLS) method was first developed by Wold (1966) in econometrics, and later became popular in chemometrics and sensory evaluation (see Geladi (1992)). The objective criterion in PLS is to maximize the covariance between the linear combination of the original predictor variables X and the response variable y. Thus, the weights wk are constructed sequentially as,
| (6) |
subject to the constraints w′kX′Xwj = 0 for all 1 ≤ j < k, where k = 1, . . . , m and m = min(N, p), as in PCA. Unlike PCA which ignores the response y completely in constructing the components, PLS incorporates both the response and the predictors. Another objective function of PLS is:
| (7) |
subject to the constraints w′kX′Xwj = 0 for all 1 ≤ j < k. In the literature, several authors such as Boulesteix and Strimmer (2006) and Rosipal and Kramer (2006) adopt objective function (7) while others such as Nguyen and Rocke (2004) and Nguyen (2005) adopt objective function (6) for PLS. It turns out that the solutions to the two objective criteria (6) and (7) are the same up to a proportionality constant (see De Jong and Phatak (1996) for details).
Several authors have discussed the use of PLS to analyze microarray data (Datta (2001); Nguyen and Rocke (2002, 2004); Nguyen (2005)). Frank and Friedman (1993) pointed out that the statistical properties of PLS are largely unknown despite its numerous applications. For example, there is a lack of theoretical understanding regarding the characteristics of PLS that delineate the conditions under which the method performs well. Naik and Tsai (2000) noted that PLS performs well in the presence of collinearity in single-index models, especially in the case when the covariates are highly correlated. They also showed that the estimates obtained from PLS are consistent up to a scaling constant. A good review of the different algorithms for PLS is given in Boulesteix and Strimmer (2006). We adopt the orthogonal scores algorithm of Marten and Naes (1989) for the simulations in this paper. The algorithm is given below:
The p columns of X and vector y are standardized (mean 0 and variance 1).
Let w̃ = X′y; define the weight vector .
Let t̃ = Xw; define the scores vector .
Find q1 = y′t, and q2 = X′t.
Deflate X and y: X = X – and .
The algorithm is repeated to obtain k weight vectors sequentially.
However, response (survival) outcomes are usually right-censored, and hence, the construction of PLS components, as given above, does not consider censoring information, which induces bias in the estimates. Improvements to this approach were proposed by combining the construction of PLS components and Cox regression model, and hence, incorporating censoring into the construction of PLS components. Park et al (2002) reformulated the Cox model as a standard Poisson regression and derived the PLS components from the formulation of PLS for the generalized linear models. However, Gui and Li (2004) pointed out that Park’s algorithm may fail to converge when the number of covariates is large. They proposed the Partial Cox Regression (PCR), which involves the construction of predictive components by repeated least square fitting of residuals and Cox regression fitting. These components can then be used in the Cox model. We describe one elegant solution proposed by Nguyen and Rocke (2004) that includes the censoring information, denoted by the Modified Partial Least Squares (MPLS).
Nguyen and Rocke (2004) showed that the PLS weights in Eq. (6) can be expressed as,
| (8) |
where vi are the ith eigenvector of X′X. Closed form expressions for the constants θik are given by Nguyen and Rocke (2004), and they depend on the response y only through the dot product , where ui are the eigenvectors of XX′. The dot product ai is the estimated slope coefficient of the simple linear regression of y on ui when X is centered. Thus, Nguyen and Rocke proposed to replace this dot product ai by the slope coefficient obtained from the univariate Cox PH regression of y on ui. In the orthogonal scores algorithm used in this work, the q1’s are precisely the a’s.
1.3. Rank-based Modified Partial Least Squares (RMPLS)
The optimization criterion of PLS maximizes the covariance of a linear combination of the predictors X and the response y. However, the usual covariance or correlation measure is heavily influenced by outliers, and thus, the PLS method is sensitive to outliers. We propose to replace the usual Pearson correlation by the Spearman rank correlation because the Spearman correlation is insensitive to outlying values of both X and y. In the orthogonal scores algorithm given in section 1.2 with standardized X and y, we make the following changes. In step 2 of the algorithm, since Cor(X, y) = X′y, we replace Cor(X, y) with CorR(X, y) where CorR(X, y) denotes the correlation of the ranks between the columns of the matrix X and the vector y. In step 4, q2 can be expressed as . Since Cor(X) = X′X, we make the change . In step 5, we update RX and Ry instead of X and y. To incorporate the censoring information, we use MPLS with these changes, and denote the new approach Rank-based Modified Partial Least Squares (RMPLS).
Theoretical Derivation: We present the weights wk in RMPLS as solutions to an optimization problem. Here, we ignore the censoring for simplicity (censoring is incorporated using the procedure of Nguyen and Rocke (2004)). The criterion of the usual PLS is to to find the weight vector, w, such that w maximizes the covariance of Xw and y. An equivalent statement in terms of the ranks is to find the weight vector, w, such that w maximizes the covariance of RXw and Ry, where Rz denotes the ranks of the vector z. RMPLS explores a different optimization problem. The columns of the data matrix X and the response y are first converted to their ranks and then centered, denoted by RX and Ry respectively. We search for the weight vector w such that w maximizes the covariance of RXw and Ry. The first weight vector, w1, is obtained from the following maximization criterion.
| (9) |
where CovR is the covariance of the ranks, RX is the matrix of the ranks of X (i.e., columns of RX correspond to the ranks of the columns of X), and Ry is the vector of the ranks of y. Here, RX and Ry are centered.
We state the following theorem (without proof) from Mardia (2003), which helps in finding a closed form solution for w1.
Theorem 1: Let a, x be vectors and let B be a symmetric matrix with B > 0. The maximum of a′x subject to x′Bx = 1, is
| (10) |
Further,
| (11) |
where the maximum is attained at .
Using Theorem 1 with B = I, x = w, and , we obtain
| (12) |
The first component is t1 = Xw1. The second weight vector, w2, is obtained from the following maximization criterion,
| (13) |
subject to the constraint w′X′t1 = 0.
Let SX = X′X, and . We can deduce that
| (14) |
where I is a p x p identity matrix, and is a constant. We should note that
In general, the kth weight vector is obtained from the following maximization criterion,
| (15) |
subject to SXwj = 0, for j = 1, . . . , k – 1.
It turns out that wk, k ≥ 2, takes the form
| (16) |
where
| (17) |
where , and ζ1, ζ2, . . . , ζk–1 can be obtained by solving the following system of linear equations for ζ’s
| (18) |
1.4. Sliced Inverse Regression (SIR)
When the number of covariates p is much larger than the number of cases N, the forward regression function E(y|X) is difficult to estimate. The idea of Sliced Inverse Regression, proposed by Li (1991), is to focus instead on the inverse regression function E(X|y), which consists of p one-dimensional regressions, and is easier to estimate. In practice, SIR is implemented by replacing y by its discrete version, denoted by ỹ, which is constructed by slicing the range of y onto H intervals. The slicing can be done by the quantiles of y, so that the number of cases in each slice is not too small. SIR then obtains the projection vectors vk through the eigenvalue decomposition of ∑X|ỹ = Cov(E(X|ỹ)) with respect to ∑x = cov(X),
| (19) |
subject to the constraints vk∑xvk = 1. Here, λk is the kth eigenvalue of ∑X|ỹ in descending order, and the vk is the corresponding eigenvector. Here, for each discretized value of ỹ, E(X|ỹ) denotes the average of the cases of X that correspond to that discretized value of ỹ.
The SIR components x̃k = Xvk, for k = 1, . . . , K (K ≤ min(H – 1, N, p)), are linear combinations of the p original predictors weighted by the projection vectors vk, where the vk’s are derived so that the first few represent directions with maximum variability between the SIR components and the response variable (Dai et al 2006). We should note that SIR does not require any of the usual assumptions on the distribution of y|X, so any model can be applied in the analysis. Similar to PLS, SIR incorporates the response (survival times) in conjunction with gene expression data. Details on SIR can be found in Li (1991), Li et al (1999), Li and Li (2004), Dai et al (2006).
Since SIR is designed for uncensored response, it cannot be applied directly to censored survival data. Li et al (1999) proposed a double slicing procedure to bypass this censoring problem by first partitioning the response y into a censored and an uncensored part, then performing the slicing within those two parts, and finally combining the two parts for the eigenvalue decomposition. Li and Li (2004) pointed out that the implementation of SIR requires the covariance matrix ∑x to be non-singular, which is not the case when p is much larger than N. To resolve this issue, they propose to first reduce the dimension of p to K, whereK < N ≪ p, via a dimension reduction method such as PCA or PLS, and then apply SIR to these K components. We adopt this approach in our simulation study.
1.5. Univariate Selection (UNIV)
Bovelstad et al (2007) first fits a univariate regression model for each gene g, and then tests the null hypothesis βg = 0 vs. the alternative βg ≠ = 0 using the score test. Since the response is censored, we use the Cox model for regression. We arrange the genes according to increasing p-values after testing each gene one-by-one. We then pick out the top-ranked K genes, according to p-values, to include in the multivariate Cox regression model. In this paper, we consider two scenarios for the selection of K: 1)we fix K to be the same for the different dimension reduction methods, and 2) we allow adaptive tuning for K by cross-validation (details given in section 3). Unlike PCA, UNIV ignores the correlation among the genes, which may cause many of the selected genes to have insignificant p-values in the multivariate Cox model as pointed out by Van Wieringen et al (2008).
1.6. Supervised Principal Component Regression (SPCR)
One possible drawback of PCA is that the method completely ignores patient survival. Bair and Tibshirani (2004, 2006) proposed the supervised principal component regression (SPCR), which employs univariate selection (UNIV) to pick out a subset of original gene expression data that is correlated with patient survival, and then apply PCA to the reduced gene expression data. One criterion to pick out the subset of genes is to obtain the λSPCR percent of the top ranked genes according to the p-values from UNIV. In this paper, we choose λ = 20% of the top ranked genes.
1.7. Correlation Principal Component Regression (CPCR)
Sun (1995) proposed a variant of SPCR, called Correlation Principal Component Regression (CPCR). The first step of CPCR is to do Principal Component Analysis (PCA) on the gene expression data matrix X, but retaining all the principal components. In other words, K1 = min(p, N) principal components (PC) are first obtained. In the context of regression, the second step to CPCR involves regressing the response variable y on the first K < K1 PC’s, such that these K PC’s have the highest correlations with y (Sun (1995)). In this paper, we select K using two strategies: 1) we fix K to be the same for the different dimension reduction methods, and 2) we use cross-validation to select K based on the minimization of the squared error of the estimated survival function. Similar to PLS, CPCR takes into account the response variable, while PCA does not.
Since the response is censored, Zhao and Sun (2007) proposed to replace the correlation between the censored response and the PC’s by the p-value obtained from the univariate Cox regression model of the response and each of the PC’s. Thus, in the second step of CPCR, we use UNIV in a univariate Cox model to pick out the top-ranked K PC’s.
2. Simulation Procedure
As mentioned earlier, to cope with the high dimensionality of the microarray gene expression data, we first reduce the dimension of the gene expression from p to K ≪ N ≪ p via dimension reduction methods, and then apply the regression model in the reduced subspace. We follow the simulation setup from Nguyen (2005), which is described in detail in the next subsection. We investigate the performance of several dimension reduction methods in the Cox regression model: PCA, MPLS, RMPLS, SIR, UNIV, SPCR, and CPCR. The results of the simulations are provided in section 3. We now describe the simulation setup.
2.1. Simulation Setup
The simulation procedure described by Nguyen (2005) comprises two main parts: 1) generating gene expression values, and 2) generating the survival and censoring times. We describe these two parts in detail.
2.1.1. Generating gene expression values
Let xij be the ijth entry of the gene expression data matrix X, where i = 1, . . . , N denote the indices for the cases, and j = 1, . . . , p denote the indices for the gene. We generate , for k = 1, . . . , d, where are the component values, and εij ∼ N(με, ) are the noise. The ijth entry of the gene expression data matrix is xij = exp( ). Thus, the gene expressions are generated as a linear combination of the d independent underlying components and an error component. It is clear that xij ∼ LN(ai, ), with parameters , and . As pointed out by Nguyen (2005), the gene expression data matrix is generated so that the first K principal components explain a specified proportion of variability in the data matrix, and the variation explained (TVPE) by the first K principal components is controlled in the simulation by γ = σε/στ. In this simulation setup, we fix d = 6, με = 0, μτ = 5/d, στ = 1, and vary σε so as to capture the desired TVPE, namely 40%, 50%, 60% and 70%. For each TVPE and each p ∈ 100, 300, 500, 800, 1000, 1200, 1400, 1600, we generate 5000 datasets. Since we want to consider p ≫ N, we fix the sample size N = 50. Since r is a set of fixed constants, it is convenient to select rki ∼ Unif(–0.2, 0.2), and we use the same set of r for all the simulations.
Since the true regression parameters, βj with j = 1, . . . , p, are fixed, it is convenient to generate them from an N(0, ) distribution. In these simulations, we fix σπ = 0.2 for all p’s.
2.1.2. Generating survival and censoring times
Once we generate the gene expression data matrix X, we generate the survival time of the ith individual, yi, independently from the censoring time, ci, with (i = 1, . . . , N). In these simulations, we consider an exponential baseline distribution for both the survival and censoring times, with density f0(t) = λe–λt. In other words, y0i ∼ Exp(λy), and c0i ∼ Exp(λc), where y0i and c0i denote the baseline survival and censoring time, respectively, for the ith individual. The survival and censoring time for the ith individual are and , respectively. Here, Xi are the covariates corresponding to the ith individual.
The observed data for the ith individual is Ti = min(yi, ci), and the corresponding censoring indicator is δi = I(yi < ci), with δi = 1 for death event and δi = 0 for censored response. The true censoring rate is under the exponential baseline survival. In the simulation setup, we fix λy = 2, and vary λz to obtain the desired amount of censoring of 1/3 and 1/2. Since both the true and censoring times for the ith individual depend on , fixing σπ = 0.2 will lead to large absolute values of for large p, and thus, the survival times will have outliers for large values of p.
3. Simulation Results
We consider two scenarios for the selection of K for the different methods: 1) K is fixed across the different methods, and 2) K is selected based on the minimization of the cross-validation squared error of the estimated survival function for each method. Since p ≫ N in real microarray data, we choose a sample size of N = 50, and consider the number of genes, p = 100, 300, 500, 800, 1000, 1200, 1400, and 1600. We generate 5000 data sets, and for each dataset, we apply dimension reduction methods in stage 1, and use the data in the reduced subspace to apply the Cox PH model in stage 2. We consider several dimension reduction methods: PCA, MPLS, RMPLS, SIR, UNIV, SPCR, and CPCR.
For scenario 1, we fix K = 3 for all the methods. Since the data matrix is generated so that the first K PCs explain a specified proportion of predictor variability, we set the proportion of variability explained to be 40%, 50%, 60% and 70%. We should note that for SIR, we first reduce the dimension of the data matrix from p to K = 3 via PCA or MPLS, then apply SIR to the reduced subspace and obtain KSIR = 2 SIR components. For Univariate Selection (UNIV), we fit a univariate Cox model for each gene, then obtain K = 3 most important genes according to the rank of the p-values of the coefficient in the univariate Cox model. For Supervised Principal Component Regression (SPCR), we first select λSPCR = 20% of the genes by UNIV, then apply PCA to the λSPCR genes to obtain the K = 3 SPCR components. For Correlation Principal Component Regression (CPCR), we first apply PCA to the original data matrix to obtain λCPCR = min(N, p) PCs, then apply UNIV to the resulted PCs to obtain the K = 3 CPCR components.
For scenario 2, we allow adaptive tuning for each method by use of cross-validation (CV). We exclude SIR from the analysis because the method does not improve PCA or MPLS. Also, for SPCR, we fix λSPCR = 20%, and apply cross-validation to select K.
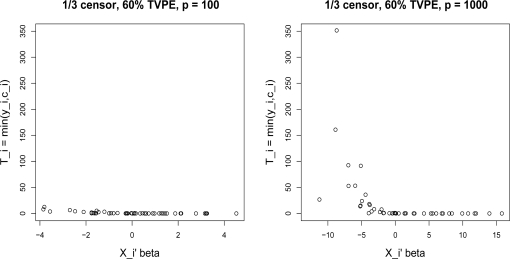
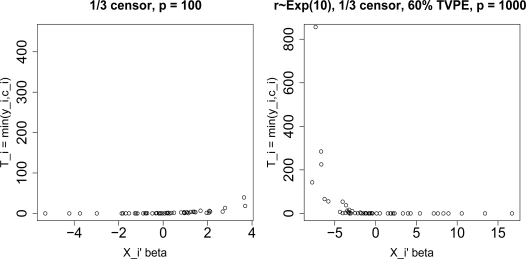
As mentioned in section 2, the survival and censoring times are generated so that for large p, i.e. p ≥ 300, some outliers are generated. Figure 1 shows, for one simulation in the case p = 100, the observed survival times Ti = min(yi, ci) do not have outliers. However, for p = 1000, the Ti have outliers. In these simulations, we want to investigate the effect of outliers in the response on the different dimension reduction methods.
Figure 1:
1/3 censoring with p = 100 and p = 1000 for one simulation run. The observed survival times Ti = min(yi, ci) are plotted against , where i = 1, . . . , N.
3.1. Scenario 1: K is Fixed
We assess the performance of the different methods using the following measures: 1) MSE(β), 2) ave(d2), 3) ave(d2.ind), 4) ave(bias), and 5) ave(bias.ind). The first, third, fourth and fifth measures have not been investigated in the literature, and the second measure has been investigated by Nguyen (2005). We now define these measures.
The first measure, MSE(β), is defined in terms of the weights placed on the genes,
| (20) |
where i = 1, . . . , s indicates the ith simulation, and j = 1, . . . , p indicates the jth gene. For the ith simulation, the p x 1 vector β̂ is obtained by β̂ = W β̂Cox where W is the vector of weights obtained from the dimension reduction step (such as PCA, PLS,. . .), and β̂Cox are the parameter estimates obtained from the Cox model.
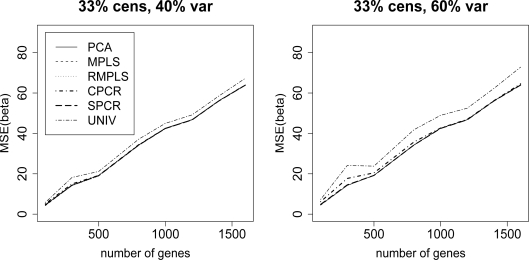
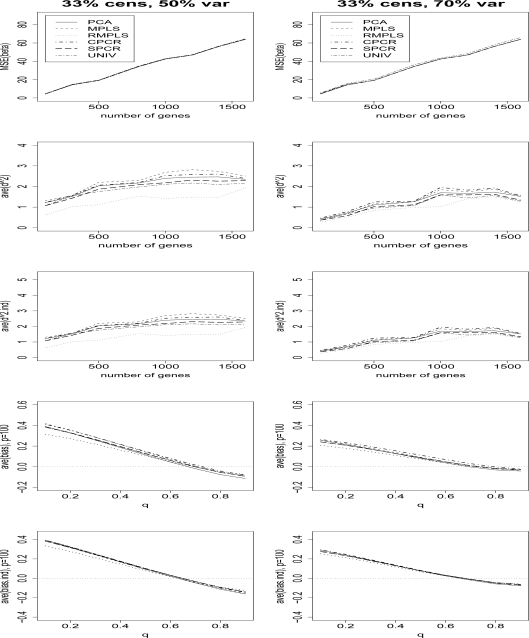
Figure 2 compares the MSE(β) for PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV for censoring rate of 1/3 and TVPE of 40% and 60%. In the case when p is small (p = 100) in the absence of outliers in the response, PCA, MPLS, RMPLS and SPCR perform relatively the same, and they outperform CPCR and UNIV. In the case when p is large (p ≥ 300) in the presence of outliers, we observe the same result as in the case of no outliers.
Figure 2:
Cox model: 1/3 censored. MSE(β) for datasets with 40% and 60% TVPE accounted by the first 3 PCs comparing PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV.
The next two measures, ave(d2) and ave(d2.ind), are in terms of the mean squared error of the estimated survival function. The ave(d2) is defined as:
| (21) |
where for the ith simulation, t corresponds to the observed death time, and
| (22) |
and
| (23) |
Here, both the true and estimated survival is obtained from the average of the covariates X̄ in the ith simulation, denoted by X̄ (i), and Ŝ0 is the Nelson-Aalen estimator of the baseline survival function.
The next measure, ave(d2.ind), measures the mean squared error of survival where the survival function is evaluated using the covariates corresponding to the individuals, rather than the average of the covariates,
| (24) |
where for the ith simulation,
| (25) |
and
| (26) |
where Xn(i) are the covariates corresponding to the nth individual in the ith simulation.
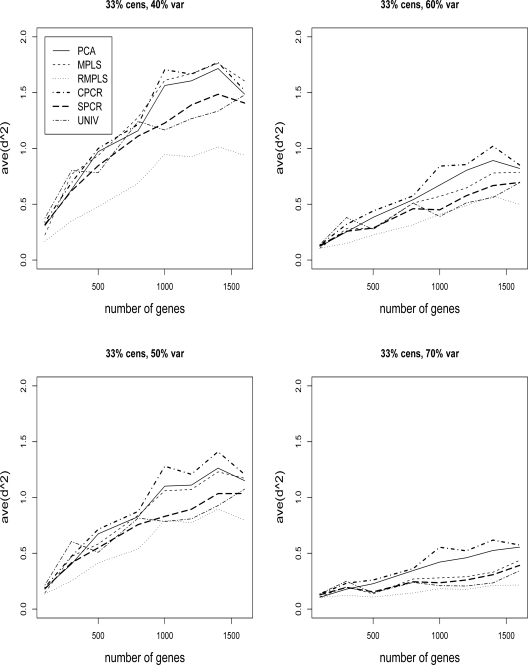
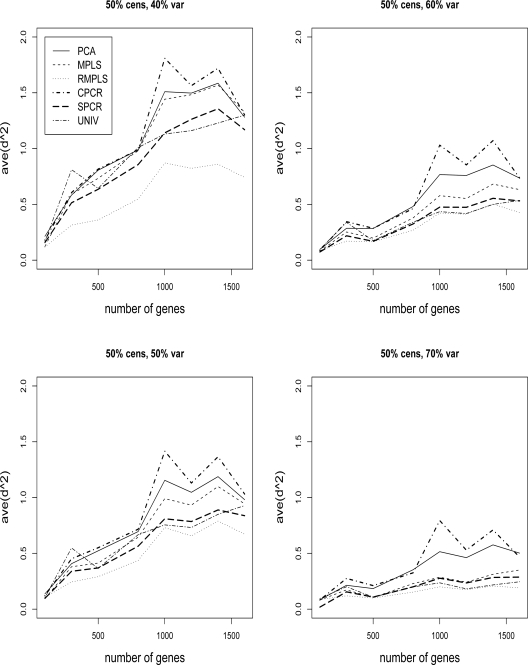
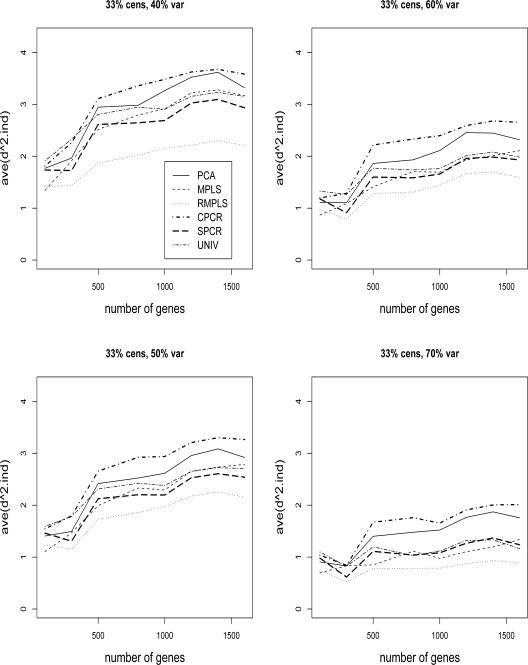
Figures 3 and 4 compare the ave(d2) of survival for PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV for censoring rate of 1/3 and 1/2, respectively, and TVPE of 40%, 50%, 60% and 70%. In the case when p is small (p = 100) in the absence of outliers in the response, RMPLS performs slightly better than MPLS, and both methods outperform PCA for low to moderate TVPE (40% and 50%). SPCR yields close ave(d2) to PCA, and all four methods RMPLS, MPLS, PCA and SPCR outperform both CPCR and UNIV. At high censoring rate of 1/2, the performance of all methods deteriorate because of the small effective sample size. However, the pattern remains the same as in the case of 1/3 censoring. This result is consistent with the findings of Nguyen (2005). In the case when p is large (p ≥ 300) in the presence of outliers, RMPLS substantially outperforms all other methods. MPLS is affected by outliers, since the method performs worse than PCA some of the times. SPCR performs better than PCA. UNIV performs surprisingly well, better than PCA in some instances. CPCR performs relatively worst among all the methods.
Figure 3:
Cox model: 1/3 censored. ave(d2) of survival for datasets with 40%, 50%, 60% and 70% TVPE accounted by the first 3 PCs comparing PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV. The x–axis denotes the number of genes, p, and the y-axis denotes ave(d2).
Figure 4:
Cox model: 1/2 censored. ave(d2) of survival for datasets with 40%, 50%, 60% and 70% TVPE accounted by the first 3 PCs comparing PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV.
Figure 5 compares the ave(d2.ind) of survival for PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV for censoring rate of 1/3, and TVPE of 40%, 50%, 60% and 70%. In the case when p is small (p = 100) in the absence of outliers in the response, RMPLS performs slightly worse than MPLS. Both methods outperform all other methods for all TVPE. Again, similar to the results for the measure ave(d2), SPCR yields close ave(d2.ind) to PCA, and both methods perform better than CPCR. UNIV performs worst among all the considered methods. In the case when p is large (p ≥ 300) in the presence of outliers, RMPLS substantially outperforms all other methods. Again, MPLS is affected by outliers, since the method performs worse than SPCR most of the times. Both SPCR and MPLS outperform PCA. UNIV performs well, better than PCA in some instances. CPCR generally performs worst among all the methods. The results for censoring rate of 1/2 are similar to those for censoring rate of 1/3 (not shown), although the performance of the methods deteriorate due to a high censoring rate.
Figure 5:
Cox model: 1/3 censored. ave(d2.ind) of survival for datasets with 40%, 50%, 60% and 70% TVPE accounted by the first 3 PCs comparing PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV. The x–axis denotes the number of genes, p, and the y-axis denotes ave(d2.ind).
The next two measures, ave(bias) and ave(bias.ind) are in terms of bias of the estimated survival function. Both measures of bias are calculated at the deciles of the true survival function. The ave(bias) is evaluated using the average of the covariates, and the ave(bias.ind) is evaluated using the covariates corresponding to the individuals. We now describe these measures:
| (27) |
where q = 0.1, 0.2, . . . , 0.9. For the ith simulation, correspond to the deciles of the true survival function. In other words, S̄i(tq) = q. The estimated survival is .
The ave(bias.ind) is defined as:
| (28) |
where for the ith simulation and nth individual, so that Sin(tq) = q, and Ŝin(tq) = .
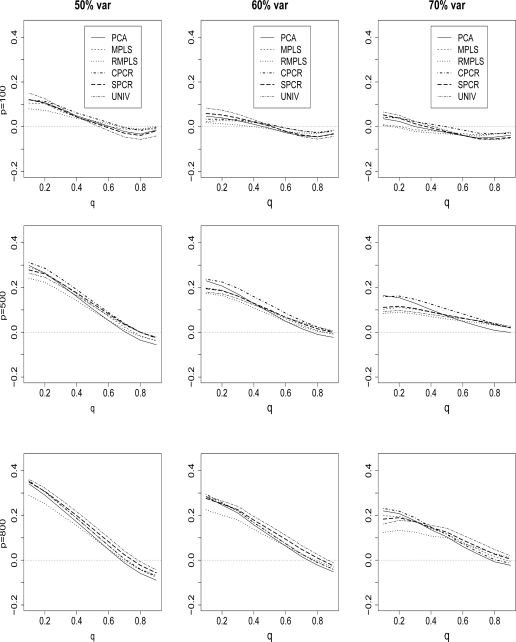
Figure 6 compares the ave(bias) of the estimated survival function for PCA, MPLS, RMPLS, SPCR, CPCR and UNIV for censoring rate of 1/3, p = 100, 500 and 800, and TVPE of 50%, 60% and 70%. The results for the cases p = 300, 1000, 1200, 1400 and 1600 are similar to the results for p = 500, and 800, so we omit these plots. Also, the results for the censoring rate of 1/2 are not shown since they are similar to the results for censoring rate of 1/3. However, at high censoring rate of 1/2, the performance of all methods deteriorate because of the small effective sample size. RMPLS generally outperforms all other methods, including MPLS, for small to medium deciles (q = 0.1, . . . , .5) in both cases when p is small (p = 100) in the absence of outliers in the response or when p is large (p ≥ 300) in the presence of outliers. For large deciles (q = .6, . . . , .9), there is no clear-cut winner among the methods.
Figure 6:
Cox model: 1/3 censored. ave(bias) of survival is plotted against q, the deciles of the true survival function, for datasets with 50%, 60% and 70% TVPE accounted by the first 3 PCs comparing PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV. The x–axis denotes q, the deciles of the true survival function, and the y-axis denotes ave(bias). The rows of the plots are for datasets with dimension p = 100, 500, and 800.
In the case when p is small (p = 100) in the absence of outliers in the response, both RMPLS and MPLS outperform PCA for all deciles (q = .1, . . . , .9). SPCR and CPCR yield close estimates to PCA for the case of 1/3 censoring, and UNIV performs relatively worst. In the case when p is large (p ≥ 300) in the presence of outliers in the response, MPLS is affected by outliers, since the method performs worse than PCA, SPCR, and UNIV some of the times.
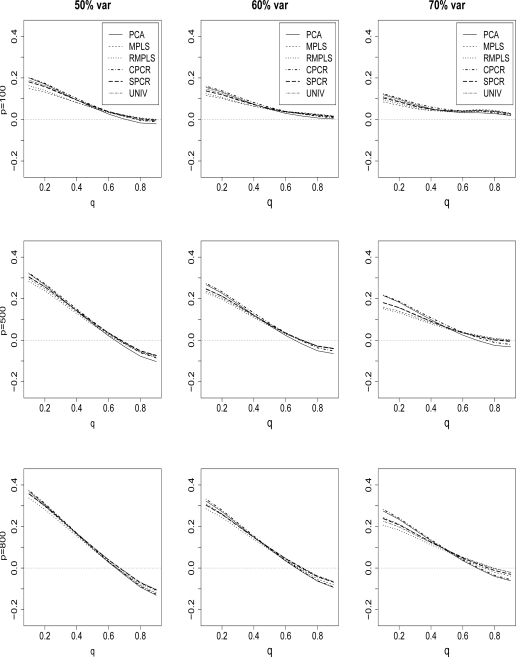
Figure 7 compares the ave(bias.ind) of the estimated survival function for PCA, MPLS, RMPLS, SPCR, CPCR and UNIV for censoring rate of 1/3, p = 100, 500 and 800, and TVPE of 50%, 60% and 70%. Again, the results for the cases p = 300, 1000, 1200, 1400 and p = 1600 are similar to the results for p = 500 and 800, so we omit these plots. Also, the results for censoring rate of 1/2 are not shown. In the case when p is small (p = 100) in the absence of outliers in the response, RMPLS is comparable to MPLS. Both methods outperform all other methods, including PCA, for all TVPE for small to medium deciles (q = 0.1, . . . , .5). Also, SPCR and UNIV perform slightly better than PCA and CPCR. In the case when p is large (p ≥ 300) in the presence of outliers in the response, RMPLS outperforms all other methods, including MPLS, for q = 0.1, . . . , .5. For large deciles q = 0.6, . . . , .9, RMPLS, MPLS, SPCR and UNIV perform relatively the same. Furthermore, RMPLS, SPCR, and UNIV perform slightly better than PCA and CPCR for all deciles.
Figure 7:
Cox model: 1/3 censored. ave(bias.ind) of survival is plotted against q for datasets with 50%, 60% and 70% TVPE accounted by the first 3 PCs comparing PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV. The x–axis denotes q, the deciles of the true survival function, and the y-axis denotes ave(bias.ind). The rows of the plots are for datasets with dimension p = 100, 500, and 800.
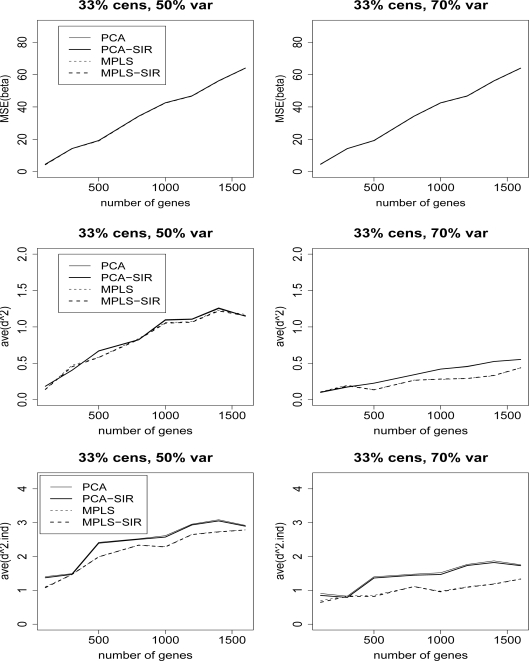
Figure 8 compares the MSE(β), ave(d2), and ave(d2.ind) for methods coupled with SIR (PCA and MPLS) and their un-SIR counterparts for censoring rate of 1/3 and TVPE of 50% and 70% using the baseline exponential survival in the Cox model. SIR does not improve upon the performance of the dimension reduction methods. The results are similar for TVPE of 40% and 60%, censoring rate of 1/2, and the two bias measures (ave(bias) and ave(bias.ind)), so we omit these plots.
Figure 8:
Cox model: 1/3 censored. MSE(β), ave(d2) and ave(d2.ind) for datasets with 50% and 70% TVPE accounted by the first 3 PCs comparing PCA, MPLS, PCA-SIR, and MPLS-SIR. The x–axis denotes the number of genes, p. The top row is the plot of the MSE(β), middle row is ave(d2), and the bottom row is ave(d2.ind).
3.2. Scenario 2: K is Selected by Cross-Validation (CV)
In practice, the number of components is chosen by cross-validation, which leads to different K for different methods. We provide simulation results based on cross-validation as a criterion to select K. We employ a 2–fold CV using the minimization of the squared error of the estimated survival function, denoted by CV (surv.error), for the simulated data to compare the different methods under the Cox model. The CV (surv.error) is defined as:
| (29) |
where i = 1, . . . , s is the index for the simulation run, s = 5000 simulations, m = 1, . . . , M is the index for the fold, M = 2, Dm is the set of death times in the mth fold, –m denotes the estimated survival function for the mth fold, and denotes the estimated survival function when the mth fold is removed. In this setting, for each simulation run, we use a 50 : 50 split of the data into a training set and a test set. Thus, the index m also denotes the test set, and –m denotes the training set. Also, the estimated survival functions are evaluated using the covariates correponding to the individuals, i.e.,
| (30) |
and
| (31) |
where N. = 25 denotes the number of individuals either in test or training set, Ŝm,n is the estimated survival function for the nth individual in the test set, and Ŝ–m,n is the estimated survival function for the nth individual in the training set. Here, .
For each method, a CV (surv.error) is obtained for each value of λ, which is the tuning parameter for that method. Here, λ < min(Nm, N–m). In these simulations, we let λ = 1, 2, . . . , 20. The optimal λ corresponds to K which minimizes CV (surv.error). Table 1 shows the K chosen by 2–fold CV for the different methods using the minimization of the CV (surv.error) under the Cox model with 1/3 censoring and 5000 simulations.
Table 1:
Cox model: 1/3 censored. K chosen by 2–fold CV for the different methods.
| p | 100 | 300 | 500 | 800 | 1000 | 1200 | 1400 | 1600 |
|---|---|---|---|---|---|---|---|---|
| PCA | 3 | 3 | 6 | 4 | 5 | 4 | 7 | 5 |
| MPLS | 1 | 2 | 1 | 2 | 2 | 1 | 2 | 2 |
| RMPLS | 4 | 3 | 2 | 4 | 4 | 2 | 2 | 2 |
| CPCR | 1 | 3 | 1 | 2 | 2 | 3 | 1 | 3 |
| SPCR | 1 | 4 | 4 | 5 | 5 | 2 | 1 | 2 |
| UNIV | 6 | 7 | 5 | 10 | 7 | 8 | 4 | 6 |
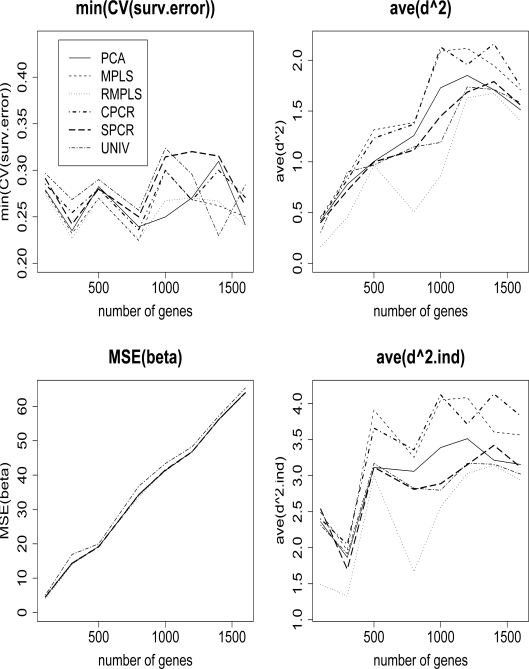
Once the CV is performed, we can use K with the simulated data as before, and obtain the mean square error for the β’s and the estimated survival function. Figure 9 compares the CV (surv.error), MSE(β), ave(d2) and ave(d2.ind) among PCA, MPLS, RMPLS, CPCR, SPCR and UNIV. RMPLS generally outperforms other methods in terms of CV (surv.error), ave(d2) and ave(d2.ind) for both cases when outliers are present and absent in the response. MPLS is affected by outliers, since the method performs worse than PCA in terms of ave(d2) and ave(d2.ind). In terms of MSE(β), PCA, MPLS, RMPLS, CPCR and SPCR perform relatively the same, and they all outperform UNIV. The standard errors (not shown) based on 5000 simulation runs of the four measures for RMPLS are small in magnitude, and are comparable to other methods. CPCR and UNIV have larger standard errors for MSE(β) compared to other methods. Using CV, RMPLS is also better variant of PLS than MPLS as in the case when the number of components, K, is fixed for all the methods.
Figure 9:
Cox model: 1/3 censored. K is chosen by CV. min(CV (surv.error)), MSE(β), ave(d2), and ave(d2.ind) comparing PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV based on 5000 simulations.
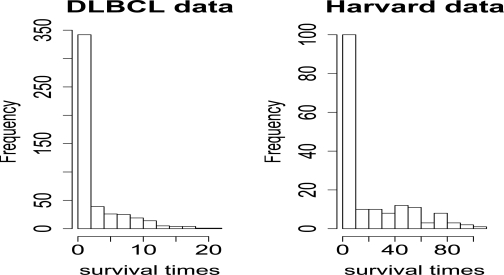
Real datasets: We also apply Cross-validation (CV) to two real datasets. The first dataset is the Diffuse Large-B-cell Lymphoma (DLBCL) data described in Rosenwald et al. (2002), and Bair and Tibshirani (2004). There are 240 patients, 7399 genes, and 42.5% of the patient survival times are censored. The second dataset is the Harvard lung carcinoma described in Bhattacharjee et al. (2001). There are 84 patients, 12625 genes, and 42.9% of the patient survival times are censored. Figure 10 shows the histograms of the survival times for the two datasets. The survival times of the Harvard dataset are heavily left-skewed, with few large observations which maybe outliers. We should observe that the survival times of the Harvard dataset have longer tail than those of the DLBCL dataset.
Figure 10:
Histograms of the survival times for stanford and harvard lung cancer datasets.
For the DLBCL data, we used a 9–fold CV with 25 samples in the test set, and 215 samples in the training set. For the Harvard data, we first screened out the genes with p – val > 0.5 using UNIV in a Cox model to retain 7189 genes. Then, we used 3–fold CV with 28 samples in the test set, and 56 samples in the training set. For both datasets, we repeat the CV 1000 times. Tables 2 and 3 show the minimized CV (surv.error) and the standard error of the 1000 repeated runs for the various methods. RMPLS outperforms all other methods for the Harvard data, in the presence of outliers in the response. Also, the method is comparable to MPLS and other methods for the DLBCL data in the absence of outliers.
Table 2:
Cox model: DLBCL data. K chosen by 9–fold CV for the different methods. The min(CV (surv.error)) and the standard error of the 1000 repeated runs are shown.
| K | PCA | MPLS | RMPLS | CPCR | SPCR | UNIV |
|---|---|---|---|---|---|---|
| 7 | 1 | 1 | 2 | 1 | 11 | |
| CV.surv.error | 0.1026 | 0.1074 | 0.1056 | 0.1014 | 0.1063 | 0.1221 |
| se.surv.error | 0.0336 | 0.0372 | 0.0354 | 0.0346 | 0.0353 | 0.0383 |
Table 3:
Cox model: Harvard data. K chosen by 3–fold CV for the different methods. The min(CV (surv.error)) and the standard error of the 1000 repeated runs are shown.
| K | PCA | MPLS | RMPLS | CPCR | SPCR | UNIV |
|---|---|---|---|---|---|---|
| 13 | 1 | 1 | 2 | 3 | 14 | |
| CV.surv.error | 0.1210 | 0.1304 | 0.1124 | 0.1402 | 0.1473 | 0.1663 |
| se.surv.error | 0.06 | 0.0654 | 0.0305 | 0.0727 | 0.0822 | 0.0863 |
For the DLBCL and Harvard datasets, we also explored the similarity between MPLS and RMPLS in the ranking of the significant genes based on the absolute value of the estimated weights on the genes (AEW), where AEW is defined as,
| (32) |
where W are the weights obtained from the dimension reduction step for MPLS or RMPLS using the whole datasets, and . Table 4 shows the number of top-ranked genes in common between MPLS and RMPLS out of K considered top-ranked genes for the two datasets using only the first component. We should observe that MPLS and RMPLS select many genes that are in common. Since the response of the Harvard dataset has outlying observations, the number of common genes selected by the two methods is generally less than that of the DLBCL dataset in the absence of outliers.
Table 4:
Cox model: Number of top-ranked genes in common between MPLS and RMPLS for DLBCL and Harvard datasets using the absolute of the estimated weights for the genes. The first row shows the number of considered top-ranked genes.
| K top-ranked genes | 25 | 50 | 100 | 250 | 500 | 1000 |
|---|---|---|---|---|---|---|
| DLBCL | 15 | 33 | 74 | 188 | 397 | 802 |
| HARVARD | 14 | 28 | 58 | 173 | 369 | 819 |
4. Conclusions and Discussion
In this paper, the simulation model of Nguyen and Rocke (2004) for gene expression data with censored response was adopted to assess the performance of several dimension reduction methods using a two-stage procedure employing the Cox regression model at the second stage. The dimension reduction methods considered in the simulations are: PCA, MPLS, RMPLS, SIR, UNIV, SPCR, and CPCR. The comparison of the different methods was based on five measures: 1) MSE(β), 2) ave(d2), 3) ave(d2.ind), 4) ave(bias), and 5) ave(bias.ind). Based on the simulation results, our conclusions are as follows.
Scenario 1: K is fixed
In the absence of outliers in the response, PCA, MPLS, RMPLS and SPCR perform relatively the same in terms of MSE(β), for all the considered TVPE (40%, 50%, 60%, and 70%). Also, all four methods outperform CPCR and UNIV. In terms of ave(d2) and ave(d2.ind), RMPLS is comparable to MPLS, and both methods substantially outperform other methods for low to moderate TVPE (40% and 50%). PCA and SPCR perform relatively the same, and both outperform CPCR. UNIV performs worst among the methods. In terms of ave(bias) and ave(bias.ind), RMPLS is comparable to MPLS for all deciles (q = .1, . . . , .9), and both RMPLS and MPLS outperform other methods for small to medium deciles (q = .1, . . . , .5). For large deciles (q = .6, . . . , .9), none of the methods dominates all others.
In the presence of outliers in the response, PCA, MPLS, RMPLS and SPCR perform relatively the same in terms of MSE(β), and all four methods outperform CPCR and UNIV. In terms of ave(d2) and ave(d2.ind), RMPLS outperforms all other methods. MPLS is affected by outliers in the response. SPCR generally outperforms MPLS, and UNIV surprisingly performs well compared to PCA. CPCR performs worst among the methods. In terms of ave(bias) and ave(bias.ind), RMPLS outperforms all other methods, including MPLS, for small to medium deciles (q = .1, . . . , .5).
Methods coupled with SIR (PCA and MPLS) do not improve their un-SIR counterparts based on the five measures.
As the TVPE increases, all methods improve.
Scenario 2: K is selected by cross-validation
RMPLS generally outperforms other methods in terms of CV (surv.error), ave(d2) and ave(d2.ind) for both cases when outliers are present and absent in the response.
MPLS is affected by outliers in terms of ave(d2) and ave(d2.ind).
MPLS, RMPLS, PCA, CPCR and SPCR perform relatively the same in terms of MSE(β), and they all outperform UNIV.
The covariance measure in the optimization criteria of PLS is influenced by outliers, and thus, the PLS method is sensitive to outliers. In this paper, we use the Spearman rank-based correlation, which is insensitive to outliers, in the optimization criteria of PLS. The simulation results indicate that RMPLS is a better dimension reduction method than MPLS in this case. Both approaches are variants of PLS that incorporate the censoring information.
When there are no outliers in the response, RMPLS yields similar results to MPLS, and both methods are superior to PCA. In these simulations, the response is generated as a function of the gene expressions to satisfy the proportional hazards assumption in the Cox model. Since PCA does not take into account the response in its construction of the components, the components selected for the Cox regression model are not necessarily predictive of the response. On the other hand, MPLS and RMPLS consider both response and predictors in their construction of the components.
One surprising result is that CPCR does not perform well in terms of mean squared error of the estimated coefficients for the genes nor in terms of mean squared error of the estimated survival function. In the dimension reduction stage, CPCR consists of two sequential steps: first use PCA to obtain all the PC’s, then apply UNIV to pick out K top-ranked PC’s. In the first step of CPCR, since PCA is used, the response is ignored. Thus, the PC’s selected in the second step of CPCR do not necessarily give better prediction than methods that incorporate the response such as MPLS or RMPLS. Furthermore, the PC’s selected for the final multivariate Cox model are not necessarily the first K PC’s, and thus, the TVPE of the selected PC’s can be much less than that of the first K PC’s.
There are some limitations to our simulation study. The regression model used is the Cox model, which models the hazard rate or survival probability, and not the actual survival times. An alternative is to use the Accelerated Failure Time (AFT) model. Preliminary results (not shown) indicate that RMPLS outperforms MPLS based on the five measures under the AFT model.
Extensions: In these simulations, the gene expression levels xij are taken to be , where the is composed of a linear combination of d underlying components, each normally distributed with a certain mean and variance, and an error component, normally distributed with a different mean and variance. We should observe that the linear combination of the d underlying components is also normally distributed, and thus, is only composed of an underlying component and an error component. By having d underlying components, we have to take into account the weights for these components, rki, for k = 1, . . . , d, and i = 1, . . . , N. The survival and censoring times depend on the gene expressions, which in turn depend on the rki. A poor choice of the weights would make some of the observed survival times Ti = min(yi, ci) outliers. For example, if we take rki ∼ Exp(10), then the response has outliers when p = 1000 as seen in Figure 11. Figure 12 compares the MSE(β), ave(d2), ave(d2.ind), ave(bias) for p = 100, and ave(bias.ind) for p = 100, for the case rki ∼ Exp(10) of PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV for censoring rate of 1/3. In terms of mean squared error of the estimated survival function (ave(d2) and ave(d2.ind)), RMPLS outperforms all other methods, including MPLS. Also, RMPLS outperforms all other methods for small to medium deciles (q = .1, . . . , .5) in terms of the bias of the estimated survival function (ave(bias) and ave(bias.ind)) in the case p = 100. Similar results were obtained for p = 300, 500, 800, 1000, 1200, 1400 and 1600. Also, a similar pattern is observed if rki ∼ Uniform(0, 0.5) or rki ∼ N(0, 0.252).
Figure 11:
rki ∼ Exp(10): 1/3 censoring with p = 100 and p = 1000 for one simulation run. The observed survival times Ti = min(yi, ci) are plotted against , where i = 1, . . . , N.
Figure 12:
Cox model: rki ∼ Exp(10), 1/3 censored. MSE(β), ave(d2), ave(d2.ind), ave(bias) for p = 100, and ave(bias.ind) for p = 100, for datasets with 50%, and 70% TVPE accounted by the first 3 PCs comparing PCA, MPLS, RMPLS, SPCR, CPCR, and UNIV based on 5000 simulations. Left panel: 50% TVPE, right panel: 70% TVPE.
Furthermore, the magnitude of the β’s, the coefficients for the genes, and hence, the survival times, are controlled by the variance . In these simulations, we fix σπ = 0.2, so that we have outliers in the response for large values of p. However, we can vary σπ as we increase p so that the survival times do not have outliers. The results (not included in this paper) indicate that the performance of the dimension reduction methods for large values of p are similar to that in the case p = 100 in the absence of outliers for rki ∼ Unif(–0.2, 0.2).
Acknowledgments
We thank two anonymous referees and the associate editor for their helpful suggestions and comments in bringing this paper to its present form. Research for this article was partially supported by NSF Grant SES-0532346, NSA RUSIS Grant H98230-06-1-0099, NSF REU Grant MS-0552590, and NCI Grant T32CA96520.
References
- Aalen OO. Nonparametric estimation of partial transition probabilities in multiple decrement models. Ann Statistics. 1978;6:701–726. doi: 10.1214/aos/1176344247. [DOI] [Google Scholar]
- Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biology. 2004;2:511–522. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair E, Hastie T, Paul D, Tibshirani R. Prediction by supervised principal components. Journal of American Statistical Association. 2006;101:119–137. doi: 10.1198/016214505000000628. [DOI] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. PNAS. 2001;98.24:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulesteix A. Statistical Applications in Genetics and Molecular Biology. 3.1.33. Berkeley Electronic Press; 2004. PLS dimension reduction for classification with microarray data. [DOI] [PubMed] [Google Scholar]
- Boulesteix A, Strimmer K. Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Briefings in Bioinformatics. 2006;8.1:32–44. doi: 10.1093/bib/bbl016. [DOI] [PubMed] [Google Scholar]
- Bovelstad HM, Nygard S, Storvold HL, Aldrin M, Borgan O, Frigessi A, Lingjaerde OC. Predicting survival from microarray data - a comparative study. Bioinformatics Advanced Access. 2007 doi: 10.1093/bioinformatics/btm305. [DOI] [PubMed] [Google Scholar]
- Bura E, Pfeiffer RM. Graphical methods for class prediction using dimension reduction techniques on DNA microarray data. Bioinformatics. 2003;19:1252–1258. doi: 10.1093/bioinformatics/btg150. [DOI] [PubMed] [Google Scholar]
- Cox DR. Regression Models and life tables (with discussion) Journal of Royal Statistical Society Series. 1972;B34:187–220. [Google Scholar]
- Dai JJ, Lieu L, Rocke DM. Dimension reduction for classification with gene expression microarray data. Statistical Applications in Genetics and Molecular Biology. 2006;5.1.6 doi: 10.2202/1544-6115.1147. http://www.bepress.com/sagmb/vol5/iss1/art6, [DOI] [PubMed] [Google Scholar]
- Datta S. Exploring relationships in gene expressions: a partial least squares approach. Gene Expressions. 2001;9:257–268. doi: 10.3727/000000001783992498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong S, Phatak A. Partial Least Squares Regression. In: van Huffel Sabine., editor. Recent Advances in Total Least Squares Techniques and Errors-in-Variables Modeling. Leuven, Belgium: 1996. pp. 25–36. [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Annals of Statistics. 2004;32:407–499. doi: 10.1214/009053604000000067. [DOI] [Google Scholar]
- Engler DA, Li Y. Survival analysis with large dimensional covariates: an application in microarray studies. Harvard University Biostatistics Working Paper Series. 2007;68 [Google Scholar]
- Frank IE, Friedman JH. A statistical view of some chemometrics regression tools (with discussion) Technometrics. 1993;35:109–148. doi: 10.2307/1269656. [DOI] [Google Scholar]
- Hoskuldsson A. PLS regression methods. Journal of Chemometrics. 1988;2:211–228. doi: 10.1002/cem.1180020306. [DOI] [Google Scholar]
- Geladi P. Wold, Herman: The father of PLS. Chemometrics and Intelligent Laboratory Systems. 1992;15.1:R7–R8. [Google Scholar]
- Gui J, Li H. Partial Cox regression analysis for high dimensional microarray gene expression data. Bioinformatics. 2004;20:208–215. doi: 10.1093/bioinformatics/bth900. [DOI] [PubMed] [Google Scholar]
- Gui J, Li H. Threshold gradient descent method for censored data regression, with applications in pharmacogenomics. Pacific Symposium on Biocomputing. 2005a;10:272–283. doi: 10.1142/9789812702456_0026. [DOI] [PubMed] [Google Scholar]
- Gui J, Li H. Penalized Cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinformatics. 2005b;21:3001–3008. doi: 10.1093/bioinformatics/bti422. [DOI] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of American Statistics Association. 1958;53:467–481. doi: 10.2307/2281868. [DOI] [Google Scholar]
- Klein JP, Moeschberger ML. Survival Analysis: techniques for censored and truncated data. second edition. Springer; New York: 2003. [Google Scholar]
- Li KC. Sliced inverse regression for dimension reduction. Journal of American Statistical Association. 1991;86:316–327. doi: 10.2307/2290563. [DOI] [Google Scholar]
- Li KC, Wang JL, Chen CH. Dimension reduction for censored regression data. The Annals of Statistics. 1999;27:1–23. doi: 10.1214/aos/1018031098. [DOI] [Google Scholar]
- Li L, Li H.Dimension reduction methods for microarrays with application to censored survival data Center for Bioinformatics and Molecular Biostatistics Paper surv2, 2004 [DOI] [PubMed]
- Li H, Luan Y. Kernel Cox regression models for linking gene expression profiles to censored survival data. Pacific Symposium of Biocomputing. 2003;8:65–76. [PubMed] [Google Scholar]
- Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. Academic Press; 2003. [Google Scholar]
- Martens H, Naes T. Multivariate calibration. Wiley; New York: 1989. [Google Scholar]
- Naik P, Tsai C. Partial least squares for single-index models. Journal of the Royal Stat. Soc., Series B. 2000;62.4:763–771. doi: 10.1111/1467-9868.00262. [DOI] [Google Scholar]
- Nguyen DV, Rocke DM. Partial least squares proportional hazard regression for application to DNA microarray survival data. Bioinformatics. 2002;18.1625 doi: 10.1093/bioinformatics/18.12.1625. [DOI] [PubMed] [Google Scholar]
- Nguyen DV, Rocke DM. On partial least squares dimension reduction for microarray-based classification: a simulation study. Computational Statistics and Data Analysis. 2004;46:407–425. doi: 10.1016/j.csda.2003.08.001. [DOI] [Google Scholar]
- Nguyen DV. Partial least squares dimension reduction for microarray gene expression data with a censored response. Mathematical Biosciences. 2005;193:119–137. doi: 10.1016/j.mbs.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Park PJ, Tian L, Kohane IS. Linking gene expression data with patient survival times using partial least squares. Bioinformatics. 2002;20:208–215. doi: 10.1093/bioinformatics/18.suppl_1.s120. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large Bcell lymphoma New England Journal of Medicine bf 3461937–1947.2002 10.1056/NEJMoa012914 [DOI] [PubMed] [Google Scholar]
- Rosipal R, Kramer N.Overview and recent advances in Partial Least Squares Springer-Verlag; Berlin Heidelberg: Saunders C, et al.SLSFS 2005, LNCS 394034–51.2006 [Google Scholar]
- Sun J. Correlation principal component regression analysis of NIR data. Journal of Chemometrics. 1995;9:21–29. doi: 10.1002/cem.1180090104. [DOI] [Google Scholar]
- VanWieringen WN, Kun D, Hampel R, Boulesteix A.Survival prediction using gene expression data: a review and comparison www.slcmsr.net/boulesteix/papers/survival.pdf, Preprint submitted to Elsevier; May2008 [Google Scholar]
- Wold H. Estimation of principal components and related models by iterative least squares. In: Krishnaiah P, editor. Multivariate Analysis. Academic Press; New York: 1966. pp. 391–420. [Google Scholar]
- Zhao Q, Sun J. Statistical Applications in Genetics and Molecular Biology. 6.1.16. Berkeley Electronic Press; 2007. Cox survival analysis of microarray gene expression data using correlation principal component regression. [DOI] [PubMed] [Google Scholar]