Abstract
DevS is one of the two sensing kinases responsible for DevR activation and the subsequent entry of Mycobacterium tuberculosis into dormancy. Full length wild-type DevS forms a stable oxy-ferrous complex. The DevS autooxidation rates are extremely low (half-lives > 24 h) in the presence of cations such as K+, Na+, Mg2+, and Ca2+. At relatively high concentrations (100 µM), Fe3+ mildly increases the autooxidation rate (six-fold increase) while Cu2+ accelerates autooxidation more than 1500-fold. Contrary to expectations, removal of the key hydrogen bond between the iron-coordinated oxygen and Tyr171 in the Y171F mutant provides a protein of comparable stability to autooxidation and similar oxygen dissociation rate. This correlates with our earlier finding that the Y171F mutant and wild-type kinase activities are similarly regulated by the binding of oxygen: namely, the ferrous 5c complex is active whereas the oxy ferrous 6c species is inactive. Our results indicate that DevS is a gas sensor in vivo rather than a redox sensor and that the stability of its ferrous-oxy complex is enhanced by inter-domain interactions.
Tuberculosis remains a health concern in the 21st century, as it is a leading cause of mortality among infectious diseases and results in the death of 2–3 million people each year (1). Active tuberculosis cases are recruited from an immense reservoir of about 2 billion people latently infected with the bacillus worldwide (1, 2). Despite the magnitude of this health problem, no new drug has been introduced for tuberculosis therapy during the past 30 years. The current tuberculosis treatment requires at least six months, which makes it costly and reduces patient compliance. The available medications can only be used with limited success in infections caused by the rapidly emerging resistant tuberculosis strains, particularly multi-drug resistant and extensively drug resistant tuberculosis (3). Latent tuberculosis is itself very difficult to treat, since dormant Mycobacterium tuberculosis displays a diminished susceptibility to drugs.
The mechanism of Mycobacterium tuberculosis entrance into the dormant phase still needs to be unraveled (4); this is a key step in the development of new and more effective approaches to the therapy of this disease. When Mycobacterium tuberculosis enters dormancy, it alters its metabolism in response to unfavorable environmental stimuli and undergoes striking morphological changes (5, 6). Dormant Mycobacterium tuberculosis bacilli have a thicker modified cell wall (7) that has lost its acid-fastedness and is Ziehl-Neelsen negative (8, 9). HspX, (Rv2031) an α-crystallin-like heat shock protein upregulated in this phase (10, 11), is, in fact, one of the most abundant proteins in dormant bacilli (11). Rv2031 belongs to the “dormancy regulon” (12, 13), a set of 48 genes whose expression is induced upon Mycobacterium tuberculosis entry into the dormant state. α-Crystallin performs a key function in Mycobacterium tuberculosis: it maintains proteins in their functional form, and thus reduces the need for the de novo biosynthesis of proteins required for bacterial survival (10, 11).
Induction of the dormancy regulon is accomplished exclusively via the DevS/DosT/DevR two component system. DevS and DosT are the only sensing kinases within the Mycobacterium tuberculosis genome that are capable of activating DevR (14). Both kinases are heme proteins that bind oxygen, nitric oxide and carbon monoxide, the very stimuli that determine whether or not Mycobacterium tuberculosis will enter the dormant state. The identification of DevR protein in Mycobacterium tuberculosis inside in vitro infected human monocytes (15) suggests that the DevS/DosT/DevR transduction pathway is operational in vivo and is clinically relevant. Interestingly, antibodies against HspX are frequently identified in tuberculosis patients (16), providing further support for the relevance of a functional DevS/DosT/DevR system in human tuberculosis infections.
So far, DevS and DosT appear to be redundant in their function, suggesting that Mycobacterium tuberculosis needs multiple genes to ensure that this important transduction system is functional. A difference that has been noted, however, is differential induction of protein expression in response to hypoxia: DevS expression is induced by hypoxia whereas DosT expression is not (17). Recent reports have also suggested that the two proteins may display different stabilities of the oxy complexes (18–20). DevS and DosT are highly homologous and both possess a heme moiety anchored to the N-terminal GAF domain. The facile oxidation of oxy DevS as compared to DosT in the hands of Kumar et al. (19) and Cho et al. (20) led the authors to propose that DevS is a redox sensor while DosT is a gas sensor. In contrast, the two proteins have also been reported to have similar low autooxidation rates (18). A discrepancy thus exists in the reported autooxidation rate for DevS, and consequently in our understanding of its physiological function.
We have performed a detailed study of full length wild-type DevS autooxidation in an effort to understand the role of DevS in vivo as well as the factors that may contribute to its stability/lability to autooxidation. We compare full length DevS to its truncated versions and to the full length Y171F DevS mutant. Tyr171 is hypothesized to assist in signal transduction and to stabilize the oxy complex by donating a hydrogen bond to the oxygen molecule that coordinates to the ferrous heme iron.
MATERIALS AND METHODS
Chemicals (99.999+% pure metal salts) were purchased from Sigma Aldrich. Ampicillin was purchased from Fisher Biotech while chloramphenicol was obtained from Roche, hemin from Fluka Biochemica, dithionite from JT Baker, and IPTG from Promega. Lysozyme and phenylmethylsulfonyl fluoride were from Sigma. Protease inhibitors (antipain, leupeptin and pepstatin), were obtained from Roche.
Protein Expression and Purification
Genetic manipulations were performed and recombinant proteins were expressed and purified as described in our previous publications (21–23).
Autooxidation Reactions: General Procedure
The protein sample was reduced inside a glove box with excess dithionite (100 mM solution) and then desalted on a PD10 column. All tubes and glassware were first rinsed with 5 mM dithionite solution and then 5 times with anaerobic buffer. The protein sample (600 µL) was removed from the glove box in an anaerobic cuvette and the starting spectrum was recorded. Then oxygenated buffer was added (250 – 600 µL) and UV-visible spectra were typically recorded for 1–3 days at 15 min time intervals. The data obtained were analyzed with Specfit and fitted to the single exponential model reaction A converted to B. The starting and ending spectra of DevS were used as limits for the fitting procedure. Two types of buffers were used: either 20 mM Hepes pH 7.5, pretreated with Chelex 100 resin, or 50 mM phosphate buffer (pH 7.5) containing 200 mM NaCl and 1 mM EDTA. If Hepes buffer treated with Chelex was used, the protein sample was treated with EDTA before the desalting step. Reagents of high purity (99.999+%) were used to determine the effect of metal ions on autooxidation rates. Autooxidation rate measurements were carried out at 25 °C in duplicate.
Reduction of Full Length DevS
Ferric full length DevS was reduced by ferredoxin (Rv0763c) and ferredoxin reductase (Rv0688) under anaerobic conditions to the ferrous form. These reductase proteins were provided by Hugues Ouellet (UCSF). Full length DevS was fully oxidized inside the glove box using ferricyanide to the ferric form and it was desalted on a PD10 column to transfer it to 20 mM Hepes buffer containing 1 mM EDTA. The final protein concentration was 2.5 µM. Ferredoxin was then added to a final concentration of 5 µM and ferredoxin reductase was present at 1 µM concentration. The final NADH concentration was 100 µM. Glucose oxidase (2 U/mL) glucose (10 mM) and catalase (100 µg/mL) were also added to ensure complete removal of oxygen traces. The ferredoxin/ferredoxin reductase combination was added to the DevS protein sample, the solution was mixed quickly in the 500 µL cuvette and then spectra were recorded every 30 s for 30 min. The data were fit to the first order rate equation by monitoring the absorbance changes at the two wavelengths (405 and 433 nm) where changes were maximal, as established from the difference spectrum between the ferric and oxy ferrous DevS complex. The data from 150 s onwards were fit to the single exponential equation to obtain the reduction reaction rates. This experiment was carried out three times and the results were averaged.
Stopped flow experiments and determination of kinetic constants of ligand binding
Kinetic parameters were measured using a Hi-Tech KinetAsyst Stopped Flow System (SF-61DX2) in diode array mode, with some experiments repeated in photomultiplier mode at the wavelength of maximal spectral change. CO binding constants were determined at 4 °C, while oxygen dissociation rates were measured at 25 °C. Datasets were analyzed using Specfit software (version 3.0.36 for 32 bit Windows systems). Each data point represents the average of 5–6 independent determinations. All kinetic experiments were performed at least three times.
Phosphate buffer (50 mM), pH 7.5 containing 200 mM NaCl and EDTA 1 mM was used for all solutions. Heme protein concentrations were in the range 3–7 µM. Protein samples were typically reduced with excess dithionite inside the anaerobic glovebox and desalted on PD10 columns. The oxy complexes were prepared by mixing the reduced protein sample with aerated buffer, while CO complexes were formed by adding CO saturated buffer to the ferrous species to a final concentration of 50 µM. Ligand solutions were prepared in gas tight syringes under anaerobic conditions inside the glovebox from saturated CO and NO buffers.
For the determination of CO association constants, ligand concentrations ranged from 50 µM to 250 µM. Generally the kobs (s−1) was determined at 7 different concentrations and plotted as a function of ligand concentration (µM); the slope provided the CO association constant.
The CO dissociation constant was measured using wild-type-DevS CO complex and air saturated buffer. The result was confirmed using 80% air saturated buffer, NO 100% and 80% saturated solutions. The CO dissociation constant for DevSY171F mutant was determined using NO solutions (80% and 100% saturated.)
Oxygen dissociation rates were measured using CO saturated solutions for the wild-type-DevS protein constructs, while dithionite solutions (5 and 6 mM) were used in the case of Y171FDevS.
Laser flash photolysis experiments
Oxygen association rates were measured using laser flash photolysis with a pulsed dye laser system fitted with a Quantel Brillian B Nd:YAG laser. Solutions were irradiated at 540 nm with a pulse of 10 ns and monitored at 434 nm. Formation of the complex was confirmed by optical absorption spectra recorded before and after laser flash photolysis measurements. Both, in the case of wild-type DevS and the Y171F mutant, the ferric heme was first reduced with sodium dithionite in deoxygenated buffer. Then, the excess dithionite was removed by loading the sample onto a column of Sephadex G-25 and the protein sample was eluted with buffer containing known oxygen concentrations. The oxygen stock solution was prepared by equilibrating the buffer with 1 atm of pure oxygen gas at room temperature. All of the kinetic measurements were carried out at pH 7.5 in phosphate buffer (20 mM) containing 200 mM NaCl and 1 mM EDTA.
RESULTS
The autoxidation rates of ferrous oxygen-bound complexes are widely considered a key parameter that reflects the stability of heme proteins (24, 25). In the present paper, we have examined the autooxidation of full length DevS over extended periods of time at room temperature in the presence and absence of metal ions. In addition, we have investigated the autooxidation behavior of the isolated GAF A domain, GAF A/B domains, and Y171F full-length mutant to investigate the influence of the protein context on the autooxidation rates.
The heme proteins used in this study were co-expressed with the GroEL/ES chaperones as previously reported in order to promote protein folding (21). This is a key step, as it is the protein matrix that protects the heme from oxidation (26). Minimal changes in the distal pocket that affect the size and hydrophobicity of the heme environment have been shown to modulate the stability of the corresponding oxy-complexes (26–28). To this end, site directed mutagenesis experiments, mainly done with myoglobins, have demonstrated that the shape, size and lipophilicity of the heme pocket modulate resistance to autooxidation. These earlier results show that the protein architecture is of prime importance and may account for the discrepancy in the previously published results on DevS autooxidation (18, 19).
The degradation of the full-length DevS oxy complex to the met form upon exposure to air was monitored by UV-vis spectroscopy. In buffers free of transition metals, DevS proved to be very stable to autooxidation (Table 1). The conversion of the oxy-complex to the ferric form is extremely slow in phosphate buffer in the presence of 1 mM EDTA or in 20 mM hepes buffer pretreated with Chelex, with t1/2 values >30 h. Removal of transition metal ions either by using a cation exchange resin (Chelex 100) or by sequestration as an inert EDTA chelate, provides an environment in which oxy-DevS is highly stable. Mg2+ and Ca2+ ions are commonly used by kinases and DevS requires Mg2+ for autophosphorylation (17, 29). Neither Ca2+ nor Mg2+ at 0.1 mM or 1 mM concentration, respectively, accelerated the autooxidation of full-length DevS. We next examined the stability of DevS in the presence of 200 mM KCl or 200 mM NaCl. Again DevS was very resistant to autooxidation. We then selected two ions found in biological fluids at low concentrations, Fe3+ and Cu2+. In the presence of 0.1 mM Fe3+ the autooxidation of DevS was mildly accelerated (half life of 7.8 h.) Interestingly, addition of 0.1 mM Cu2+ resulted in a large enhancement of the autooxidation rate from more than 60 h to less than 2 min. We conclude that DevS displays a very high stability to autooxidation in the presence of physiologically relevant cations such as Na+, K+, Mg2+, and Ca2+, but Cu2+ at relatively high concentrations is able to act as an efficient electron acceptor and oxidizes oxy-DevS at high rates, k = 25 h−1. The stability of oxy-DevS to autooxidation suggests that DevS functions as a gas sensor in vivo. Autooxidation is very slow, characterized by half-lives greater than one day in the absence of Fe3+ and Cu2+. The physiological concentrations of iron and copper are very low compared to the levels tested in this study and therefore are unlikely to cause a significant acceleration of DevS autooxidation. We note that a tenfold decrease in Cu2+ concentration (from 100 to 10 µM) decreased the DevS autooxidation rate over 300 fold and correspondingly increased the autooxidation half life from less than 2 min. to 11h. We conclude that oxy-DevS is very stable in the mycobacterial cell.
TABLE 1.
Autooxidation rates of full-length wild-type DevS in the presence of common metal ions. Rates were measured at 25 °C. Buffer conditions:
| conditions | k (h−1) | t1/2 (h) |
|---|---|---|
| KCl 200 mMa | 0.013 | 78 |
| CaCl2 0.1 mMa | 0.0049 | 170 |
| MgCl2 1 mMa | 0.012 | 59 |
| NaCl 200 mMa | 0.010 | 96 |
| hepes 20 mMa | 0.0033 | 210 |
| Phosphate EDTAb | 0.019 | 36 |
| CuCl2 0.1 mMa | 25 | 0.03 |
| CuCl2 10 µMa | 0.062 | 11 |
| FeCl3 0.1 mMa,c | 0.089 | 7.8 |
hepes buffer (20 mM), pH 7.5, pretreated with Chelex 100 resin. The cations indicated were added as salts of high purity (>99.999+%)
phosphate buffer (50 mM), pH 7.5, 1 mM EDTA and 200 mM NaCl.
The nominal FeCl3 concentration is indicated.
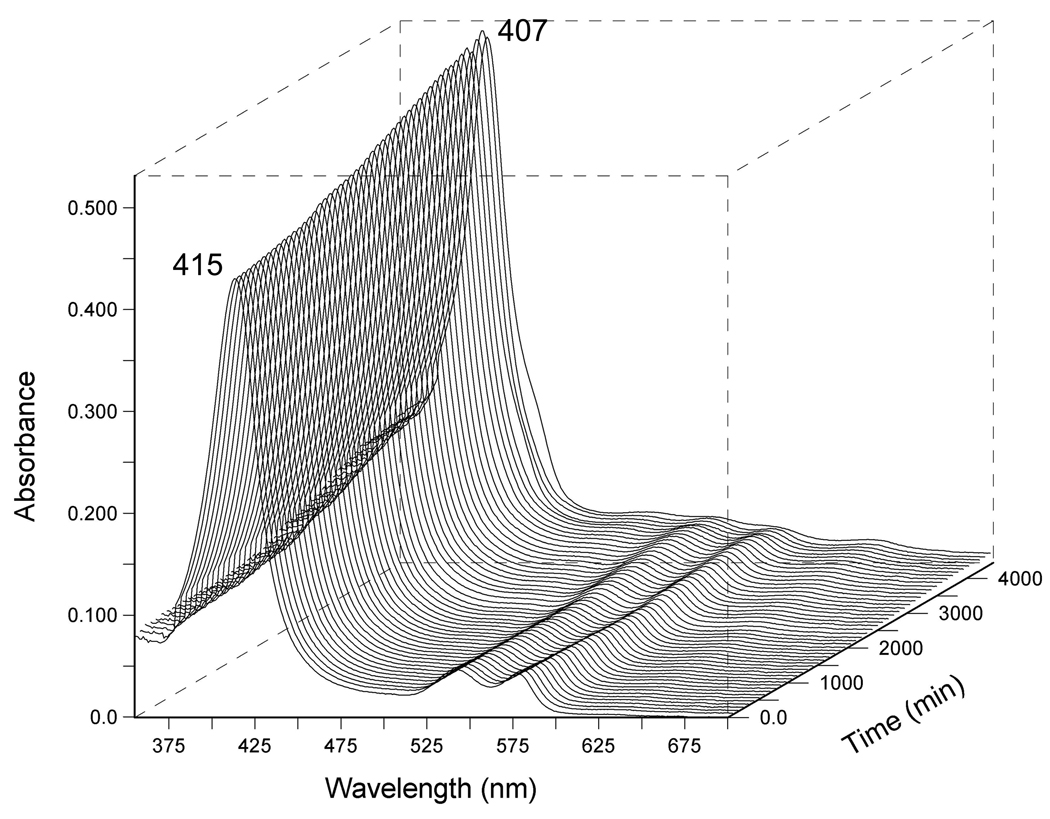
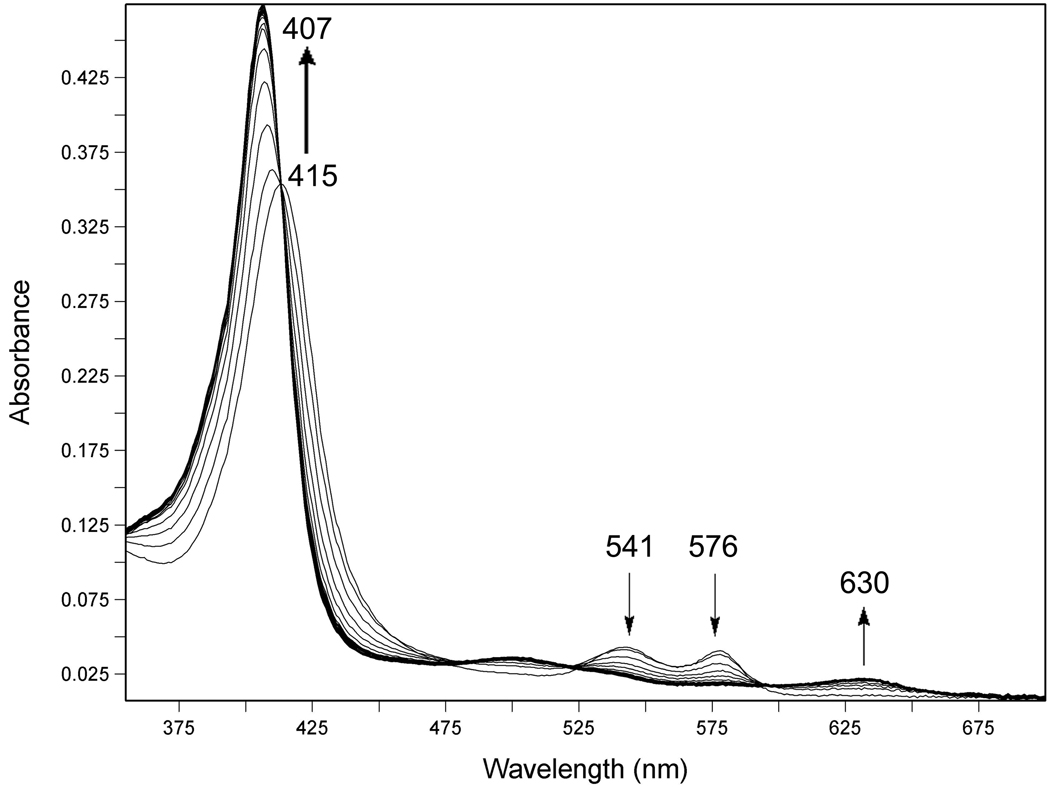
Representative traces of autooxidation experiments are shown in Figure 1 and Figure 2. In Figure 1, a three dimensional plot of UV-visible spectra collected over time, shows that oxy-DevS is very slowly converted to ferric DevS in the presence of 200 mM NaCl over the course of roughly three days. In Figure 2, DevS quickly undergoes autooxidation from the oxy-form to the ferric complex in the presence of Cu2+. In this experiment, spectroscopic traces were collected every 30 s for a total of 8 h (only traces from the first 30 min of the experiment are shown). Clear isosbestic points at 413, 524, 594, and 648 nm indicate that a smooth conversion occurs from the oxy complex reactant into the product met form under these conditions.
FIGURE 1.
Autooxidation of wild-type full-length DevS in the presence of 200 mM NaCl, 20 mM Hepes buffer, pH 7.5 at 25 °C. The reaction was monitored every 15 min for 3 days. Other metal ions were excluded using a Chelex 100 anion exchange resin.
FIGURE 2.
Autooxidation of wild-type full-length DevS in the presence of 100 µM CuCl2, 20 mM Hepes buffer, pH 7.5 at 25 °C. The reaction was monitored every 30 s for 8 h. Only 50 spectra recorded during the first half hour of the experiment are shown. Other metal ions were excluded using a Chelex 100 anion exchange resin.
Next we compared full-length wild-type DevS to the truncated proteins GAF A DevS and GAF A/B DevS in phosphate buffer containing EDTA (Table 2). The first of these proteins consists only of the heme-containing GAF A domain, and the second of the GAF A and B domains. All the proteins are fairly stable to autooxidation under these conditions. GAFA DevS is the least stable (half life 16.5 h) followed by GAF A/B DevS (half life 26.0 h). Full length wild-type DevS (half life 73.6 h) is the most stable in this series.
TABLE 2.
Autooxidation rates of the truncated proteins, DevS GAFA and DevS GAFA/B, and of the the full-length Y171F DevS mutant. Rates were measured at 25 °C in phosphate buffer (50 mM) containing 1 mM EDTA and 200 mM NaCl.
| Protein | k(h−1) | t1/2 (h) |
|---|---|---|
| DevS GAFA |
0.046 | 16.5 |
| DevS GAFA/B |
0.027 | 26.0 |
| full-length Y171F DevS |
0.012 | 73.6 |
The autooxidation behavior of Y171F is similar to that of wild-type full-length DevS (Table 2).
Heme proteins used for oxygen storage and transportation are physiologically active only in the ferrous form and autooxidize spontaneously to the inactive met form. Reduction systems are normally present to reduce the ferric forms back to the ferrous complexes that are capable of binding oxygen. For this reason we have examined the reduction of full-length DevS by an endogenous Mycobacterium tuberculosis ferredoxin reductase/ferredoxin system, the Rv0688 and Rv0763c gene products, respectively. As shown in Supplemental Figure 1 and Supplemental Figure 2, ferric full-length DevS was quickly reduced by this system to the ferrous form, at a rate of 0.0027 s−1, as monitored both at 405 nm and 433 nm (t1/2 4.4 min). We propose that DevS, if oxidized to met-DevS, can be reduced inside Mycobacterium tuberculosis to the physiologically relevant ferrous DevS, the active kinase.
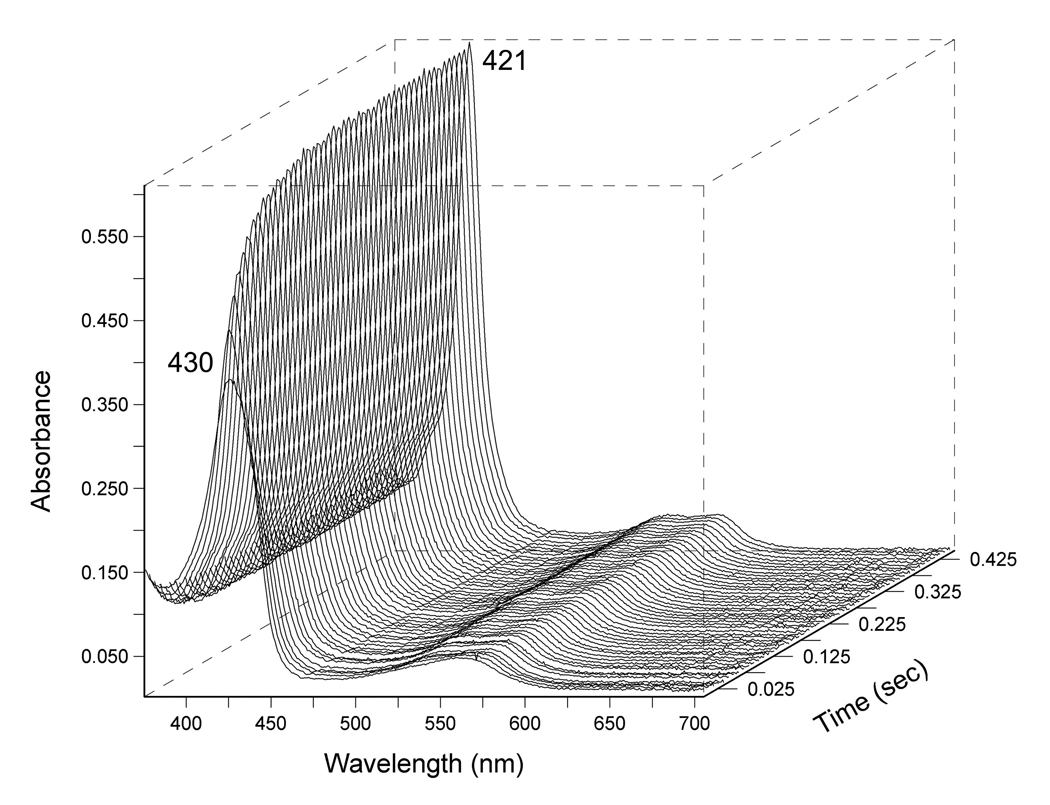
Kinetic CO and O2 binding parameters were determined for wild-type DevS and Y171F DevS (Figure 3). Additionally CO association and dissociation constants were measured for the truncated DevS constructs. While the differences in CO on-rates are small, ranging from 0.18 to 0.73 µM−1s−1, a clear trend is established; CO association constants increase in the order: DevSGAFA < DevSGAFA/B < DevS < Y171F DevS. Representative traces of CO binding to Y171F DevS are shown in Figure 3. The CO kon for wild-type DevS proteins is higher than for FixL gas sensors including BjFixL (0.005 µM−1s−1) (30) and comparable to the CO association rate of the AxPHEA heme domain (31).
FIGURE 3.
Representative traces of the CO binding reaction to Y171F DevS monitored in diode array at 4 °C at 50 µM CO concentration in the stopped-flow instrument.
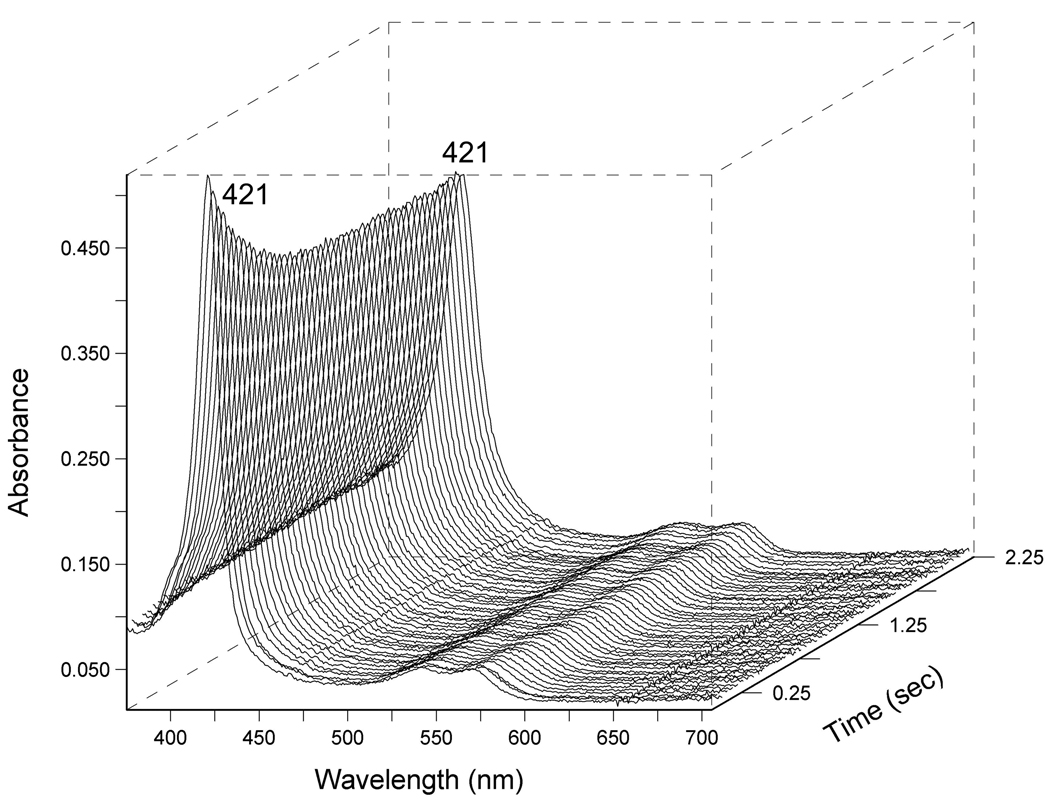
CO off-rates are virtually constant in the DevS protein series at around 2.5 s−1, and notably higher than CO off values for FixLs. UV-visible spectra of the displacement reaction between Y171F Fe2+ CO and NO are included in Figure 4. These trends result in an increase in KDCO in the following order: Y171F DevS < DevS < DevSGAFA/B < DevSGAFA. Wild-type full length DevS has a KDCO value that is quite close to the BjFixL constant (KDCO 9 µM) (30).
FIGURE 4.
Representative traces of the CO displacement reaction of Y171F DevS Fe(II)CO with NO saturated buffer, monitored in diode array at 4 °C in the stopped-flow instrument.
The oxygen association constant for the Y171F mutant is about three times larger than that of the wild-type (Table 3). The wild-type DevS value is lower, while the mutant O2 association constant is higher than the corresponding constants of FixLs (BjFixL and RmFixL), EcDosH (32) and AxPDEAH (31). The O2 association constants for all gas sensors, DevS and FixLs included, are considerably lower than for the globin family of proteins, represented by myoglobin and hemoglobin. O2 dissociation rates for the wild-type DevS and Y171FDevS are comparable. In both cases, O2 dissociation rates are notably lower than for FixLs and globins. Only RmFixLH provided a similar koffO2 value (6.8 s−1) (30). Taken together, these data suggest that DevS proteins are more effective stores of O2 than CO. The Y171F mutant has slightly increased affinity for both gases, roughly threefold, as compared to wild-type DevS.
TABLE 3.
Summary of kinetic constants.
| CO | |||
|---|---|---|---|
| Protein | kon (µM−1s−1) | koff (s−1) | Kd (µM) |
| Y171FdevS | 0.73 ± 0.071b | 2.43 ± 0.15b | 3.33 |
| DevS | 0.31 ± 0.031b | 2.57 ± 0.40b | 8.29 |
| DevSGAFA/B | 0.26 ± 0.024b | 2.45 ± 0.26 | 9.42 |
| DevSGAFA | 0.18 ± 0.007b | 2.74 ± 0.40 | 15.22 |
| EcDosH(32) | 0.0011 | 0.011 | 10 |
| AxPDEAH(31) | 0.21 | 0.058 | 0.28 |
| BjFixL(30) | 0.005 | 0.045 | 9 |
| RmFixLT(30) | 0.012 | ||
| RmFixLH(30) | 0.017 | 0.083 | 4.9 |
| O2 | |||
| Protein | kon (µM−1s−1) | koff (s−1) | Kd (µM) |
| Y171FDevS | 32.4a | 3.85 ± 0.69b | 0.12 |
| DevS | 11.8a | 6.79 ± 0.33b | 0.58 |
| EcDosH(32) | 0.0026 | 0.034 | 13 |
| AxPDEAH(31) | 6.6 | 77 | 12 |
| BjFixL(30) | 0.145 | 20 | 138 |
| RmFixLT(30) | 0.217 | 11 | 51 |
| RmFixLH(30) | 0.217 | 6.8 | 31 |
The oxygen association constants were determined in this study using laser flash photolysis.
All other rates were determined in this work using the stopped flow method.
DISCUSSION
Full length DevS, one of the two kinases that control DevR phosphorylation, plays a key role in allowing Mycobacterium tuberculosis to enter the dormant phase.
In our previous work, wild-type full length DevS was compared to Y171F DevS in terms of resonance Raman data, UV-visible spectra, and autophosphorylation behavior (Table 4 and Table 5). Functional assays performed with wild-type DevS revealed that the ferrous unligated form has intermediate kinase activity, and ligand binding either further activates the kinase (CO and NO) or completely inactivates it (O2) (23). Earlier work using the kinase domain of DevS by itself showed that the isolated kinase core is active (33). Therefore, the role of the GAFA and GAFB domains is to repress/enhance the activity of the DevS kinase as a function of ligand identity and the diatomic gas concentration. The Y171F mutant is only capable of discriminating between the presence and absence of ligands and is unable to distinguish among them. Y171F DevS is active in the ferrous unligated form (5c complex) and inactive in the ligand bound form (6c complex with CO, NO or O2) (23).
TABLE 4.
Summary of UV-visible spectra of full length Y171F DevS and wild-type DevS.
TABLE 5.
Resonance Raman vibrations of CO complexes, with the dominant complex shown in bold.
These data indicate that the Tyr 171 side-chain is critical for activation and repression of the kinase activity as well as for discrimination among ligands. Here we present the kinetic and autooxidation data of Y171F DevS and compare it to that of wild-type DevS in order to glean more insight into the role of this particular amino acid side-chain as well as the biological role of DevS in Mycobacterium tuberculosis infections.
Previous resonance Raman studies established that the CO complexes of wild-type DevS exist as two conformers: the hydrogen bonded conformer defined by the high correlation point at 1936 cm−1 and 524 cm−1 in the plot of νC-O versus νFe-CO and the non-hydrogen bonded conformer defined by the low correlation point at 1971 cm−1 and 490 cm−1 (Supplemental Table 2) (21, 22).
These conformers were identified in the truncated versions of DevS; however, their proportion was altered as a function of truncation. Briefly, while in DevS GAFA the two conformers were present in roughly equal concentrations, in full length DevS the non-hydrogen bonded conformer predominates (85%). In DevS GAFA/B the conformers are present in intermediate proportions (22).
Our kinetic analysis provides support for the existence of two conformers in the Fe2+ unligated complex of DevS, analogous to the two CO conformers detected by resonance Raman in the Fe-CO complex. In only one conformer, a hydrogen bond between the Tyr171 hydroxyl and a water molecule serves to position the water in the distal heme pocket in the proximity of the iron atom; however, not close enough to achieve coordination. Recently, the crystal structure of DevS GAFA was solved, confirming this conclusion (20). In the other conformer (for which structural or crystallographic data are lacking), no water molecule is fixed in the distal site at that position, resulting in unhindered access of the ligand to the heme iron. The absence of that “fixed” water molecule in the distal heme pocket can be interpreted as the result of the displacement of the Tyr 171 side-chain, its attachment point. Tyr 171 may adopt a different conformation than in the crystal structures (20, 34), where it serves as a gate that separates the immediate small distal heme pocket (10 Å3) from a larger cavity of 45 Å3; perhaps the phenol ring is flipped out of the way of the incoming ligand.
As the proportion of the hydrogen bonded conformer of the Fe2+ unligated complex increases, the CO and O2 association rates are expected to decrease. This trend was confirmed by the CO on-rates determined in this study: DevS > DevS GAFA/B > DevS GAFA. In the case of the Y171F DevS mutant, no such hydrogen bonded conformer can exist, and, in fact, CO and O2 association rates for Y171F DevS were roughly threefold higher than for full length wild-type DevS, providing further support for this trend. The gas association rates for our DevS proteins suggest that interdomain interactions preferentially stabilize the conformer lacking the hydrogen bonded water molecule.
We evaluated the stability of the oxy-complexes of DevS and Y171F DevS in terms of autooxidation rates. Three recent papers have provided conflicting data on the autooxidation properties of DevS. In 2007, Steyn et al. reported that their full length DevS protein, isolated as a SUMO fusion, exists in the ferric form; the oxygen ferrous complex could not be isolated by the authors and they concluded that it is completely unstable. On this basis alone, Steyn and co-workers concluded that DevS is a redox sensor (19). More recently, Cho et al. arrived at the same conclusion based on work done with the isolated GAF A domain of DevS (20). Yet another research group used a slow expressing vector to produce full length DevS and they reported an autooxidation half life of 4 h at 37 °C (18). Our examination of the full-length DevS autooxidation behavior reveals that this heme protein actually forms a very stable oxy complex. The DevS autooxidation rates are extremely low under a variety of conditions. In the presence of K+, Na+, Mg2+, and Ca2+, ions that are universally present in biological fluids, DevS retains its stability, with autooxidation rates between 0.005-0.02 h−1. Fe3+ increases the autooxidation rate to 0.09 h−1, while Cu2+ accelerates the autooxidation reaction by 3 orders of magnitude to 25 h−1, when present at the non-physiological concentration of 0.1 mM. The high stability of oxy-DevS establishes that this kinase acts as a gas rather than redox sensor. This is consistent with DevS kinase activity being turned off upon oxygen binding to the ferrous complex.
In fact, as shown in Table 6, DevS is one of the most stable heme proteins. DevS is much more stable than the oxygen sensors EcDos or FixL; the DevS autooxidation rate is between 30 and 240 times lower. In addition, DevS is more stable than the human α-and β-hemoglobin chains and sperm whale myoglobin (SWMb); its stability at the physiological pH is comparable to that of bovine myoglobin in basic conditions (63 h half-life at pH 9.)
TABLE 6.
Autooxidation rates of various heme proteins, where Mb is myoglobin; Hb is hemoglobin; EcDos is the Escherichia coli Dos. Conditions:
| Protein | k (h−1) | t1/2 (h) |
|---|---|---|
| EcDos-PAS(47)a | 0.3 | 2.3 |
| EcDos-PAS R97A(47) a | 570 | 0.0012 |
| EcDos-PAS R97E(47) a | 2760 | 0.00025 |
| EcDos(48) | 0.7 | 1 |
| AxPDEA1(31)b | >12 | |
| BjFixL(30) c | 2.8 | 0.25 |
| RmFixLT(30) c | 1.9 | 0.37 |
| RmFixLH(30) c | 2.3 | 0.3 |
| SWMb(49) c | 0.06 | 12 |
| SWMb H(E7)L(49) c | 10.4 | 0.07 |
| Bovine Mb(50) d | 0.01 | 63 |
| Human Hb chain alpha(51) e | 0.05 | 13 |
| human Hb chain beta(51) e | 0.09 | 7.8 |
pH 8, 25 °C;
pH 8, Tris buffer, 23 °C;
pH7, 37 °C;
pH 9, 25 °C;
pH 8, 35 °C.
In DevS the heme moiety is associated with a GAF domain, whereas in all the other oxygen sensors in Table 6, including the E. coli phophodiesterase EcDos and the FixL kinases, the heme group is anchored in a PAS domain. This comparison suggests that the GAF protein fold is better able to stabilize the oxygen-bound heme against oxidation than the PAS domain. From the previously characterized PAS oxygen sensors, only AxPDEA1 is fairly stable (autooxidation half life > 12 h.)
The previous data also suggest that the globin fold encountered in proteins used either for oxygen transport (hemoglobins) or oxygen storage (myoglobins) is more efficient in preventing autooxidation of the heme iron than are the PAS domains.
In this work, we show that in DevS the GAF domain effectively shelters the heme from autooxidation, leading to properties similar to myoglobins. The GAF domain was only recently shown to bind heme (21, 35). DosT GAF was the first heme binding GAF domain to be crystallized (34). DevS is thus similar to DosT, another protein possessing a GAF anchored heme moiety that has been shown to possess enhanced stability against autooxidation (18, 19) (unpublished results.)
In the case of the myoglobins, the presence of a distal pocket hydrogen bond donor that stabilizes oxygen in the ferrous species and a water molecule in the met form correlates with a γmet/γoxy>1 (the ratio of molar absorptivities at the wavelengths corresponding to the Soret peak). Typically the distal hydrogen bond donor is a histidine residue which confers these spectral characteristics, and its lack usually results in a lower γmet/γoxy<1 (36).
Replacement of this hydrogen bonding residue with a hydrophobic amino acid results in an altered γmet/γoxy<1 and in a blue shift of the Soret band for the met form. The same characteristic changes were observed in the case of DevS: mutation of Tyr 171 to Phe decreased the γmet/γoxy ratio from 1.37 for the wild-type to 0.97 for the mutant. This was accompanied by a blue-shifted Soret for the respective ferric species, as expected. The intensity and position of the Soret band is considered to be strongly influenced by the structure of the distal heme pocket and by the interaction between the heme group and the apoprotein (37). In our case the ferrous oxy complex of the mutant had a red-shifted Soret at 422 nm as compared to the wild-type at 414 nm (Table 5), suggesting that the oxygen molecule is coordinating to the heme iron in a different distal environment.
Mutation of other residues in the myoglobin distal pocket that increased lipophilicity resulted in decreased autooxidation rates (26). In our case, mutation of Tyr to Phe also increases the lipophilicity of the heme pocket, and, due to the absence of the Tyr hydroxyl in the mutant, favors exclusion of solvent/water molecules. This, in turn, increases the stability of the oxy-complex and accounts at least in part for its low autooxidation rate.
The full length protein demonstrated an increased stability to autooxidation as compared to the truncated GAFA DevS and GAFA/B DevS. The presence of the second GAF domain and, additionally, the kinase core decrease susceptibility to autooxidation. The full length Y171F mutant displays an autooxidation behavior similar to that of the wild-type.
Inter-domain interactions between the GAFA domain of DevS, GAFB and the kinase core and possibly inter-domain interactions resulting from DevS dimerization are hypothesized to decrease the flexibility of the GAFA domain and stabilize the conformation resistant to autooxidation. The hypothesis that inter-domain interactions enhance stability of heme proteins against autooxidation is not unprecedented. In fact, Kawano et al. studied the relationship between artificial multiple domain myoglobins and their tendency to undergo autooxidation. They found that as the number of domains increased, autooxidation rates decreased. The authors hypothesized that poly-domain formation stabilizes oxygen storage and reduces autooxidation rates, and they concluded that the structural flexibility of the globin domain is restricted in their unusual poly-domain myoglobins (25). In addition, naturally occurring polymeric hemoglobins from Barbatia lima demonstrate higher stability and reduced autooxidation rates as compared to the simple dimers of the hemoglobin chains (25). Human hemoglobin is much more resistant to autooxidation than the isolated α- and β- chains, and inter-domain contacts are hypothesized to be responsible for this differential stabilization of the human hemoglobin tetramer (38).
Heme proteins are typically maintained in their active reduced state by a reduction system. This is the case for both myoglobin (39) and hemoglobin (40). In fact, in the absence of a functional reduction system, myoglobin would exist entirely in its met form, which is physiologically useless. In erythrocytes, oxidizing compounds convert 1.5–3% per day of hemoglobin to methemoglobin, but in vivo the methemoglobin is present in less than 1% due to the activity of this reducing enzymatic system (41).
In the current work, we determined that DevS can be quickly reduced by a physiological reducing system, the gene products of Rv0688 and Rv0763c, an endogenous Mycobacterium tuberculosis ferredoxin reductase and ferredoxin pair. This system may provide reducing equivalents to DevS in Mycobacterium tuberculosis in vivo and thus help to maintain this kinase in its reduced active form. It is known that ferredoxins are highly similar proteins (42). FdxA is upregulated upon entrance into dormancy (10, 13). The expression of the Rv0763c gene was induced in the murine lungs during tuberculosis infection (43). Thus, either the gene product of Rv0763c or FdxA could support DevS reduction in vivo. In addition to the already hypothesized roles of FdxA in dormancy, FdxA may serve to maintain DevS in its functional state.
As mentioned earlier, the positioning of the 171 Tyr residue side-chain in the distal heme pocket determines the activation or repression of the kinase function in DevS. It has previously been shown that the hydrogen bond to the coordinated O2 is different from the hydrogen bond formed with CO or NO in the active site (22). The hydrogen bond to oxygen is not sensitive to protein truncations, suggesting two possibilities: 1) only one hydrogen bonded O2 conformer is present in all constructs (Figure 5), or 2) several hydrogen bonded O2 conformers are present but they are indistinguishable by resonance Raman. These data suggest that the DevS protein is able to accommodate hydrogen bond formation to the coordinating O2 more easily than in the case of NO or CO. This results in apparently relaxed steric constraints for the hydrogen bond to the oxygen molecule serving as the 6th heme ligand, with the Tyr171 side-chain positioned differently than in the CO and NO complexes. Thus oxygen binds to both wild-type DevS and Y171FDevS and forms complexes of comparable stability, as determined by O2 dissociation and autooxidation rates. The downstream effect is in both cases inactivation of the histidine kinase activity (23).
FIGURE 5.
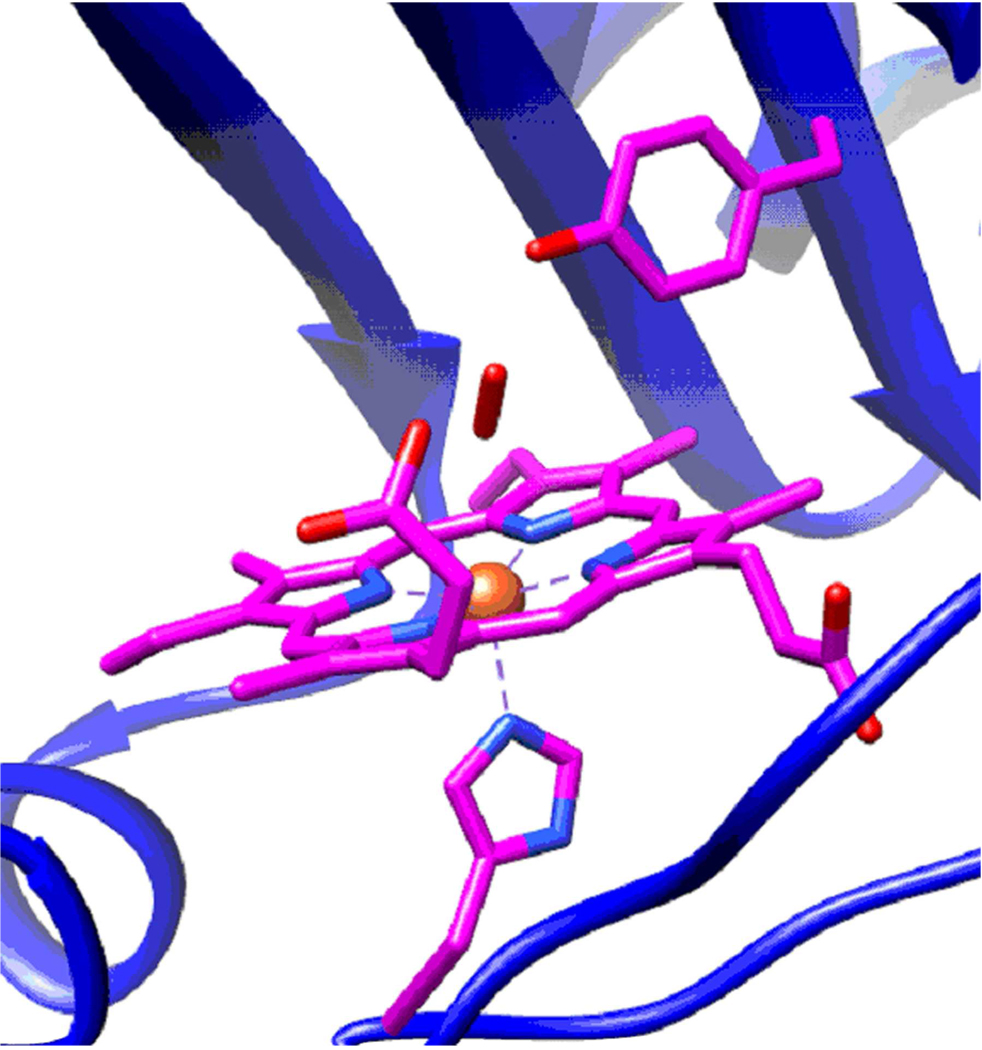
Model of oxygen binding to DevSGAFA based on the structure from the crystallized DevS GAF A (PDB ID 2W3F)(20) with the oxygen ligand positioned according to the DosT GAFA crystal structure (PDB ID 2VZW)(34). The porphyrin ring, His 149, and Tyr 171 are shown in purple as capped sticks, while the protein backbone is represented as a blue ribbon. The oxygen molecule coordinating to the iron in the distal pocket is shown in red.
We hypothesize that a different mechanism is at work in Y171F DevS in order to stabilize the oxy complex of the mutant. This mechanism that substitutes for the stabilizing effect of the O2 hydrogen bond in the wild-type is most likely the interaction between the heme bound oxygen molecule and the phenyl edge of F171. A large body of crystallographic data provides support for the favorable interaction between oxygen atoms bearing partial or integral negative charges and the edge of phenyl rings (44). Oxygen atoms from carbonyl groups (44) and negatively charged carboxylate oxygens (45) are frequently positioned at the edge of phenyl rings in proteins. This interaction is thought to be due in part to the electrostatic forces between the negative charge on the oxygen atom and the positive charge at the edge of the phenyl ring, which acts as a multipole having negative charge density below and above its surface. Another, perhaps more important, contribution is the polarization effect of the aromatic ring. In our case, the phenyl ring of F171 is hypothesized to interact with the partial negative charge on the oxygen atom. Heme bound oxygen molecules are known to display superoxide character. This effect may be responsible for the stabilization of the Y171F oxy complex and correlates with its high stability to autooxidation and the low oxygen dissociation rate. This is analogous to the F29 oxy-myoglobin mutant which displayed a higher stability to autooxidation and a lower oxygen dissociation rate than the wild-type protein. The crystal structure of F29 oxy-myoglobin revealed a short distance of 3.2 Å between the bound oxygen molecule and the nearest carbon atom from the F29 residue; this suggested the presence of a favorable interaction between the oxygen partial negative charge and the positive edge of the phenyl ring (28).
The presence of partial negative charges in the distal pocket in close proximity to the bound ligand have been shown to destabilize the oxy complex in heme proteins causing a decrease in the oxy complex stability of myoglobin mutants such as V68S and V68T (26). Partial negative charges present on the hydroxyl group from T68 in which the negative end of the T68 dipole points toward the coordinating ligand have also been shown to increase the νC-O frequency (46). The presence of the Y171 hydroxyl group in wild-type DevS may cause similar effects in terms of νC-O stretching frequencies. The Y171 hydroxyl moiety in wild type DevS is involved in hydrogen bonding to the ligand in one CO conformer (νC-O 1936 cm−1) while in the other the Tyr171 residue may adopt an entirely different conformation or its hydroxyl may orient itself with the negative end of its dipole towards the bound ligand giving the νC-O at 1971 cm−1. Removal of this partial negative charge in the Y171F DevS mutant results in a lower CO stretching frequency (νC-O 1965 cm−1) and, of course, the hydrogen bonded conformer is entirely absent in this DevS mutant.
As a result, the distal heme pocket of the mutant provides only one CO conformer according to resonance Raman data, νC-O 1965 cm−1 and νFe-CO 487 cm−1 (23) located close to the non hydrogen bonded conformer of DevS. In the DevS series we therefore see a trend in autooxidation rates that is opposite to the myoglobin or hemoglobin models where hydrogen bonding stabilization between the distal His and bound CO corresponds to enhanced stability to autooxidation and higher correlation points on the νC-O versus νFe-CO plots.
We conclude that the N-terminal GAF domain in full-length wild-type DevS effectively protects the ferrous oxygen-bound heme from oxidation under a variety of conditions. The presence of the most abundant cations, K+, Na+, Mg2+, and Ca2+ does not decrease the stability of DevS. Two transition metal ions, Fe3+ and Cu2+, enhance autooxidation 6-fold and over 1000-fold respectively when present at 100 µM concentrations. However, in biological fluids these ions are present at much lower levels.
The autooxidation properties of DevS clearly indicate that DevS functions as a gas sensor in vivo. Its stability to autooxidation is enhanced by inter-domain interactions and replacement of the key distal tyrosine with a phenyl alanine residue results in a protein equally resistant to autooxidation.
Supplementary Material
Supporting Information available: The time dependence of the reduction of DevS by an endogenous Mtb reducing system and the spectra before and after reduction of full length DevS are presented as supporting infromation. This material is available free of charge via Internet at http://pubs.acs.org.
ACKNOWLEDGMENTS
We thank Hughes Ouellet for providing the ferredoxin (Rv0763c) and ferredoxin reductase (Rv0688) proteins and Dr. Wytze van der Veer for assistance with the laser flash photolysis experiments.
Abbreviations
- IPTG
isopropyl-β-D-thiogalactopyranoside
- wt
wild-type
- GAF
domain, protein domain conserved in cyclic GMP-specific and stimulated phosphodiesterases, adenylate cyclases, and E. coli formate hydrogenlyase transcriptional activator (Pfam accession number PF01590)
Footnotes
This work was supported by grants AI074824 (P.R.O.M.) and GM42614 (T.L.P.) from the National Institutes of Health.
REFERENCES
- 1.World Health Organization. Global tuberculosis control - surveillance, planning, financing. 2006
- 2.Kochi A. The global tuberculosis situation and the new control strategy of the World Health Organization. Tubercle. 1991;72:1–6. doi: 10.1016/0041-3879(91)90017-m. [DOI] [PubMed] [Google Scholar]
- 3.Bloom BR, Murray CJ. Tuberculosis: Commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 4.Kendall SL, Rison SC, Movahedzadeh F, Frita R, Stoker NG. What do microarrays really tell us about M. tuberculosis? Trends Microbiol. 2004;12:537–544. doi: 10.1016/j.tim.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham AF, Spreadbury CL. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J. Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulrichs T, Kaufmann SH. New insights into the function of granulomas in human tuberculosis. J. Pathol. 2006;208:261–269. doi: 10.1002/path.1906. [DOI] [PubMed] [Google Scholar]
- 9.Seiler P, Ulrichs T, Bandermann S, Pradl L, Jorg S, Krenn V, Morawietz L, Kaufmann SH, Aichele P. Cell-wall alterations as an attribute of Mycobacterium tuberculosis in latent infection. J. Infect. Dis. 2003;188:1326–1331. doi: 10.1086/378563. [DOI] [PubMed] [Google Scholar]
- 10.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan Y, Crane DD, Barry CE., 3rd Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial alpha-crystallin homolog. J. Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roupie V, Romano M, Zhang L, Korf H, Lin MY, Franken KL, Ottenhoff TH, Klein MR, Huygen K. Immunogenicity of eight dormancy regulon-encoded proteins of Mycobacterium tuberculosis in DNA-vaccinated and tuberculosis-infected mice. Infect. Immun. 2007;75:941–949. doi: 10.1128/IAI.01137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts DM, Liao RP, Wisedchaisri G, Hol WG, Sherman DR. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 2004;279:23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasgupta N, Kapur V, Singh KK, Das TK, Sachdeva S, Jyothisri K, Tyagi JS. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 2000;80:141–159. doi: 10.1054/tuld.2000.0240. [DOI] [PubMed] [Google Scholar]
- 16.Lee BY, Hefta SA, Brennan PJ. Characterization of the major membrane protein of virulent Mycobacterium tuberculosis. Infect. Immun. 1992;60:2066–2074. doi: 10.1128/iai.60.5.2066-2074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini DK, Malhotra V, Tyagi JS. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 2004;565:75–80. doi: 10.1016/j.febslet.2004.02.092. [DOI] [PubMed] [Google Scholar]
- 18.Sousa EH, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 2007;16:1708–1719. doi: 10.1110/ps.072897707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr, Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho HY, Cho HJ, Kim YM, Oh JI, Kang BS. Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 2009 doi: 10.1074/jbc.M808905200. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ioanoviciu A, Yukl ET, Moenne-Loccoz P, Ortiz de Montellano PR. DevS, a heme-containing two-component oxygen sensor of Mycobacterium tuberculosis. Biochemistry. 2007;46:4250–4260. doi: 10.1021/bi602422p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yukl ET, Ioanoviciu A, Ortiz de Montellano PR, Moenne-Loccoz P. Interdomain interactions within the two-component heme-based sensor DevS from Mycobacterium tuberculosis. Biochemistry. 2007;46:9728–9736. doi: 10.1021/bi7008695. [DOI] [PubMed] [Google Scholar]
- 23.Yukl ET, Ioanoviciu A, Nakano MM, Ortiz de Montellano PR, Moenne-Loccoz P. A distal tyrosine residue is required for ligand discrimination in DevS from Mycobacterium tuberculosis. Biochemistry. 2008;47:12532–12539. doi: 10.1021/bi801234w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki T, Watanabe YH, Nagasawa M, Matsuoka A, Shikama K. Dual nature of the distal histidine residue in the autoxidation reaction of myoglobin and hemoglobin comparison of the H64 mutants. Eur. J. Biochem. 2000;267:6166–6174. doi: 10.1046/j.1432-1327.2000.01685.x. [DOI] [PubMed] [Google Scholar]
- 25.Kawano K, Uda K, Otsuki R, Suzuki T. Preparation of artificial 2-, 3-, 4- and 8-domain myoglobins and comparison of their autoxidation rates. FEBS Lett. 2004;574:203–207. doi: 10.1016/j.febslet.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 26.Brantley RE, Jr, Smerdon SJ, Wilkinson AJ, Singleton EW, Olson JS. The mechanism of autooxidation of myoglobin. J. Biol. Chem. 1993;268:6995–7010. [PubMed] [Google Scholar]
- 27.Wallace WJ, Houtchens RA, Maxwell JC, Caughey WS. Mechanism of autooxidation for hemoglobins and myoglobins. Promotion of superoxide production by protons and anions. J. Biol. Chem. 1982;257:4966–4977. [PubMed] [Google Scholar]
- 28.Carver TE, Brantley RE, Jr, Singleton EW, Arduini RM, Quillin ML, Phillips GN, Olson JS., Jr A novel site-directed mutant of myoglobin with an unusually high O2 affinity and low autooxidation rate. J. Biol. Chem. 1992;267:14443–14450. [PubMed] [Google Scholar]
- 29.Saini DK, Malhotra V, Dey D, Pant N, Das TK, Tyagi JS. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology. 2004;150:865–875. doi: 10.1099/mic.0.26218-0. [DOI] [PubMed] [Google Scholar]
- 30.Gilles-Gonzalez MA, Gonzalez G, Perutz MF, Kiger L, Marden MC, Poyart C. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry. 1994;33:8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- 31.Chang AL, Tuckerman JR, Gonzalez G, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Gilles-Gonzalez MA. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry. 2001;40:3420–3426. doi: 10.1021/bi0100236. [DOI] [PubMed] [Google Scholar]
- 32.Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry. 2000;39:2685–2691. doi: 10.1021/bi991911s. [DOI] [PubMed] [Google Scholar]
- 33.Saini DK, Tyagi JS. High-throughput microplate phosphorylation assays based on DevR-DevS/Rv2027c 2-component signal transduction pathway to screen for novel antitubercular compounds. J. Biomol. Screen. 2005;10:215–224. doi: 10.1177/1087057104272090. [DOI] [PubMed] [Google Scholar]
- 34.Podust LM, Ioanoviciu A, Ortiz de Montellano PR. 2.3 A X-ray structure of the heme-bound GAF domain of sensory histidine kinase DosT of Mycobacterium tuberculosis. Biochemistry. 2008;47:12523–12531. doi: 10.1021/bi8012356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sardiwal S, Kendall SL, Movahedzadeh F, Rison SC, Stoker NG, Djordjevic S. A GAF domain in the hypoxia/NO-inducible Mycobacterium tuberculosis DosS protein binds haem. J. Mol. Biol. 2005;353:929–936. doi: 10.1016/j.jmb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Korenaga S, Igarashi J, Matsuoka A, Shikama K. A primitive myoglobin from Tetrahymena pyriformis: Its heme environment, autoxidizability, and genomic DNA structure. Biochim. Biophys. Acta. 2000;1543:131–145. doi: 10.1016/s0167-4838(00)00187-4. [DOI] [PubMed] [Google Scholar]
- 37.Puett D. The equilibrium unfolding parameters of horse and sperm whale myoglobin. Effects of guanidine hydrochloride, urea, and acid. J. Biol. Chem. 1973;248:4623–4634. [PubMed] [Google Scholar]
- 38.Tsuruga M, Matsuoka A, Hachimori A, Sugawara Y, Shikama K. The molecular mechanism of autoxidation for human oxyhemoglobin. Tilting of the distal histidine causes nonequivalent oxidation in the beta chain. J. Biol. Chem. 1998;273:8607–8615. doi: 10.1074/jbc.273.15.8607. [DOI] [PubMed] [Google Scholar]
- 39.Hagler L, Coppes RI, Jr, Herman RH. Metmyoglobin reductase. Identification and purification of a reduced nicotinamide adenine dinucleotide-dependent enzyme from bovine heart which reduces metmyoglobin. J. Biol. Chem. 1979;254:6505–6514. [PubMed] [Google Scholar]
- 40.Ogasawara Y, Funakoshi M, Ishii K. Glucose metabolism is accelerated by exposure to t-butylhydroperoxide during NADH consumption in human erythrocytes. Blood Cells Mol. Dis. 2008;41:237–243. doi: 10.1016/j.bcmd.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Bodansky O. Methemoglobinemia and methemoglobin-producing compounds. Pharmacol. Rev. 1951;3:144–196. [PubMed] [Google Scholar]
- 42.Ricagno S, de Rosa M, Aliverti A, Zanetti G, Bolognesi M. The crystal structure of FdxA, a 7Fe ferredoxin from Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 2007;360:97–102. doi: 10.1016/j.bbrc.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 43.Talaat AM, Ward SK, Wu CW, Rondon E, Tavano C, Bannantine JP, Lyons R, Johnston SA. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J. Bacteriol. 2007;189:4265–4274. doi: 10.1128/JB.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas KA, Smith GM, Thomas TB, Feldmann RJ. Electronic distributions within protein phenylalanine aromatic rings are reflected by the three-dimensional oxygen atom environments. Proc. Natl. Acad. Sci. U. S. A. 1982;79:4843–4847. doi: 10.1073/pnas.79.16.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson MR, Beahm R, Duvvuru S, Narasimhan C, Wu J, Wang HN, Philip VM, Hinde RJ, Howell EE. A preference for edgewise interactions between aromatic rings and carboxylate anions: the biological relevance of anion-quadrupole interactions. J. Phys. Chem. B. 2007;111:8242–8249. doi: 10.1021/jp0661995. [DOI] [PubMed] [Google Scholar]
- 46.Li T, Quillin ML, Phillips GN, Jr, Olson JS. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 1994;33:1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- 47.Ishitsuka Y, Araki Y, Tanaka A, Igarashi J, Ito O, Shimizu T. Arg97 at the heme-distal side of the isolated heme-bound PAS domain of a heme-based oxygen sensor from Escherichia coli (Ec DOS) plays critical roles in autoxidation and binding to gases, particularly O2. Biochemistry. 2008;47:8874–8884. doi: 10.1021/bi800248c. [DOI] [PubMed] [Google Scholar]
- 48.Sasakura Y, Yoshimura-Suzuki T, Kurokawa H, Shimizu T. Structure-function relationships of EcDOS, a heme-regulated phosphodiesterase from Escherichia coli. Acc. Chem. Res. 2006;39:37–43. doi: 10.1021/ar0501525. [DOI] [PubMed] [Google Scholar]
- 49.Quillin ML, Arduini RM, Olson JS, Phillips GN., Jr High-resolution crystal structures of distal histidine mutants of sperm whale myoglobin. J. Mol. Biol. 1993;234:140–155. doi: 10.1006/jmbi.1993.1569. [DOI] [PubMed] [Google Scholar]
- 50.Shikama K, Sugawara Y. Autoxidation of native oxymyoglobin. Kinetic analysis of the pH profile. Eur. J. Biochem. 1978;91:407–413. doi: 10.1111/j.1432-1033.1978.tb12693.x. [DOI] [PubMed] [Google Scholar]
- 51.Poli AL, Moreira LM, Hidalgo AA, Imasato H. Autoxidation studies of extracellular hemoglobin of Glossoscolex paulistus at pH 9: cyanide and hydroxyl effect. Biophys. Chem. 2005;114:253–260. doi: 10.1016/j.bpc.2004.12.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information available: The time dependence of the reduction of DevS by an endogenous Mtb reducing system and the spectra before and after reduction of full length DevS are presented as supporting infromation. This material is available free of charge via Internet at http://pubs.acs.org.