Abstract
The epicardium plays an essential role in coronary artery formation and myocardial development, but signals controlling the development and differentiation of this tissue are not well understood. To investigate the role of platelet derived growth factor receptor β (PDGFRβ) in development of epicardial-derived vascular smooth muscle cells (VSMC), we examined PDGFRβ-/- and PDGFRβ epicardial-mutant hearts. We found that PDGFRβ-/- hearts failed to form dominant coronary vessels on the ventral heart surface, had a thinned myocardium, and completely lacked coronary VSMC (cVSMC). This constellation of defects was consistent with a primary defect in the epicardium. To verify that these defects were specific to epicardial derivatives, we generated mice with an epicardial deletion of PDGFRβ that resulted in reduced cVSMC distal to the aorta. The regional absence of cVSMC suggested that cVSMC could arise from two sources, epicardial and non-epicardial, and that both were dependent on PDGFRβ. In the absence of PDGFRβ signaling, epicardial cells adopted an irregular actin cytoskeleton leading to aberrant migration of epicardial cells into the myocardium in vivo. In addition, PDGF receptor stimulation promoted epicardial cell migration, and PDGFRβ-driven phosphoinositide 3' kinase (PI3K) signaling was critical for this process. Our data demonstrate that PDGFRβ is required for the formation of two distinct cVSMC populations and that loss of PDGFRβ-PI3K signaling disrupts epicardial cell migration.
Keywords: epicardium, PDGF, coronary vascular smooth muscle, migration, PI3K
Introduction
Vascular smooth muscle cells (VSMC) arise from a diverse range of tissues including neural crest, somites, splanchnic mesoderm, gut mesothelium, and epicardium (reviewed by Majesky1), and past data has demonstrated that many coronary VSMC (cVSMC) are derived from the embryonic epicardium2, 3. While several genes have been identified that are essential for the formation, attachment, and spreading of the epicardium, few genes have been identified that are essential during epithelial to mesenchymal transition (EMT) and subsequent differentiation into cVSMC and cardiac fibroblasts.
Platelet derived growth factor receptor (PDGFR) tyrosine kinases are one family of signaling proteins that are potentially involved in epicardial cell function. Analyses in the mouse have shown that PDGFRβ signaling promotes proliferation and migration of VSMC in multiple vascular beds including the heart4-8. Therefore, we investigated the function of PDGFRβ signaling during epicardial development. We have examined PDGFRβ-/-, epicardial-specific PDGFRβ mutant, and PDGFRβ signaling deficient embryos. We discovered that epicardial deletion resulted in the absence of cVSMC distal to the aorta and that PDGFRβ signaling through PI3K was required for proper cytoskeletal organization in epicardial cells. Our results designate PDGF receptor signaling as another growth factor system involved in epicardial development.
Materials and methods
Experimental animals
Mice were maintained on a mixed C57Bl/6 X 129SV background. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Texas Southwestern Medical Center and performed according to the Guide for the Care and Use of Laboratory Animals published by the NIH. Dr. Eric Olson kindly provided capsulinLacZ/+ mice17, and Dr. Pilar Ruiz-Lozano kindly provided GataCreTg/0 mice9. Online materials and methods provide additional strain and breeding information.
Migration
E12.5 hearts were isolated from indicated genotypes and incubated with GFP-expressing adenovirus. Hearts were cultured in 10% FBS 1:1 DMEM:M199 supplemented with bFGF (2ng/ml, Sigma) for 48h with vehicle, 50ng/ml PDGFBB, 50ng/ml PDGFBB + LY294002 (2μM, Cell Signaling), hTGFβ1 (20ng/ml, R&D Systems), or bFGF (25ng/ml, Sigma). Hearts were isolated, fixed, and frozen embedded as performed in the CCFSE experiments. Ten-micron sections were DAPI stained and mounted in 1:1 PBS glycerol. Migration was quantified by counting the number of GFP-positive cells beneath the epicardium in a 40x field of view from five non-consecutive sections. Percentage of migrated cells was determined by dividing the average number of GFP-positive cells for each sample by the average of wild type GFP-positive cells that had migrated in response to PDGFBB stimulation and multiplying by 100.
Standard protocols were used for histology, immunohistochemistry, western blotting, and primary epicardial cultures. Detailed procedures can be found in the online materials and methods.
Results
Loss of PDGFRβ results in coronary blood vessel and myocardial defects
Although PDGFRβ signaling is considered mitogenic for VSMC, only select populations of VSMC are affected by the loss of PDGFRβ or PDGFB4. Because cVSMC arise from a unique mesothelial origin, we wanted to determine how PDGFRβ signaling regulates this cell population. The Gata5CreTg mouse line expresses Cre recombinase in the proepicardial organ (PEO; the embryonic source of epicardial cells) and the epicardium9. Crossing this transgenic line to mice with a PDGFRβ loxP-flanked allele, generated animals lacking PDGFRβ in the epicardium (PDGFRβEKO). Using mice that possessed either a PDGFRβ null allele or this epicardial deletion of PDGFRβ, we examined coronary vessel formation.
We began our analysis by investigating the myocardial thickness in these PDGFRβ deficient hearts. In comparison to the myocardial defects reported in PDGFRβ-/- hearts10 and data not shown, the myocardial compact zone in PDGFRβEKO hearts was relatively normal. PDGFRβEKO compact zones at E18.5 were 88 ± 4% the thickness of controls (n=4 controls and n=4 PDGFRβEKO). Therefore, the myocardial defects observed in PDGFRβ-/- hearts may be secondary to systemic vascular defects rather than epicardial loss of PDGFRβ as the epicardial mutant hearts had minimal myocardial disruption.
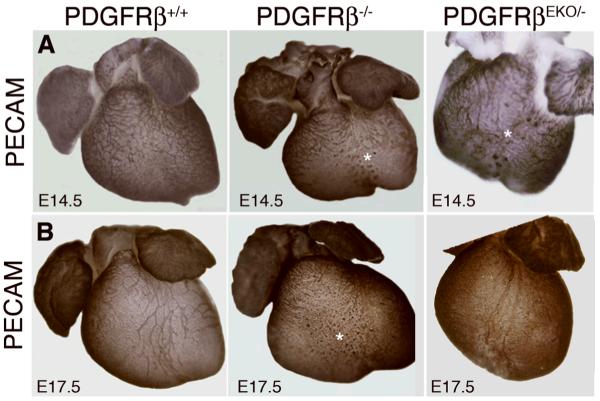
We next examined coronary artery development in these hearts. At E14.5, hearts of all genotypes possessed a capillary plexus, but PDGFRβ-/- and PDGFRβEKO hearts possessed abnormal clusters of endothelial cells (Figure 1A). In PDGFRβ-/- hearts these defects persisted and dominant coronary vessels failed to form on the ventral surface. In contrast, at E17.5 endothelial vessel formation in PDGFRβEKO hearts was similar to wild type (Figure 1B). These results indicated that coronary vessel disruption occurs in PDGFRβ-deficient hearts, but in the case of the epicardial deletion, vessel remodeling is later recovered.
Figure 1. PDGFRβ expression is required for proper coronary artery development.
Whole mount (A) E14.5 and (B) E17.5 hearts stained for PECAM (endothelial cells) by immunohistochemistry. Asterisks indicate sinusoidal PECAM staining.
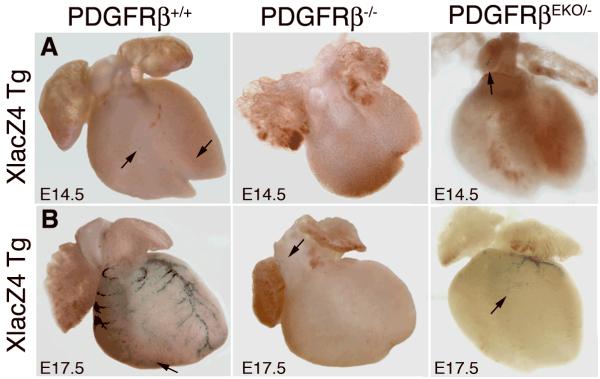
To determine if the endothelial defects were accompanied by similar VSMC defects, we generated PDGFRβ deficient animals that possessed the XlacZ4 transgene. XlacZ4 mice express a nuclear-localized β-galactosidase protein in VSMC11, including those derived from the epicardium (unpublished observation CS and MT, 2007). In wild type hearts, a few β-galactosidase+ cells were observed as early as E14.5. A majority of cells were present in close proximity to nascent coronary vessels, while single cells were found scattered on the surface of (Figure 2A). In comparison, PDGFRβ-/- and PDGFRβEKO hearts contained no β-galactosidase+ cells (Figure 2A). At E17.5 VSMC were present in wild type hearts both along the major coronary arteries as well as scattered along smaller vessels within the ventricles. Remarkably, E17.5 PDGFRβ-/- hearts did not possess β-galactosidase+ cells along the heart surface or within the ventricles (Figure 2B and data not shown), although a few β-galactosidase+ cells were observed in the aortic arch. In contrast, epicardial loss of PDGFRβ caused loss of a majority of cVSMC, but β-galactosidase+ cells were still present along the vessels descending from the aortic root (Figure 2B). The absence of cVSMC in PDGFRβ-/- hearts and reduction in PDGFRβEKO hearts was confirmed by staining for smooth muscle myosin heavy chain (smMHC) and α smooth muscle actin (αSMA) (data not shown). Therefore, the requirement for PDGFRβ in the formation of cVSMC is different than the requirement for VSMC in other tissues that have been investigated. The complete loss of this VSMC population in the PDGFRβ-/- heart demonstrates a greater dependence on PDGFRβ signaling in cVSMC development and suggests that PDGFRβ may function beyond the role of a mitogen or chemotactic factor.
Figure 2. Loss of cVSMC in the absence of PDGFRβ.
Whole mount (A) E14.5 and (B) E17.5 hearts stained for β-galactosidase activity (cVSMC) of the XlacZ4 transgene. Arrows indicate scattered β-galactosidase+ cells.
Expression of PDGF receptors during heart development
To identify when PDGFRβ was expressed in the heart, we examined wild type embryos at various stages of coronary vascular development. Consistent with chick expression studies12, PDGFRβ was expressed as early as E9.5 in the proepicardium (data not shown). At E13.5, we observed PDGFRβ in the cells of the epicardium. At E15.5 PDGFRβ expression remained in the epicardium but was also observed in cells surrounding vessels within the myocardium. By E17.5, PDGFRβ expression was reduced in the epicardium, and the majority of positive cells were presumably VSMC adjacent to endothelial vessels (Online Figure II A).
Recent reports have also demonstrated that PDGFRα is expressed by epicardial cells13. Using an allele of the PDGFRα that expresses a nuclear localized green fluorescent protein from the PDGFRα locus14, we investigated the spatial and temporal expression of this receptor. Similar to PDGFRβ, PDGFRα is expressed by epicardial cells (Online Figure II B). PDGFRα continues to be expressed by epicardial cells after they have migrated into the myocardium. At E17.5 very few cells co-express both receptors within the myocardium (Online Figure II C).
Non-epicardial source for cVSMC
We next investigated the source of the persisting cVSMC present in PDGFRβEKO hearts. From the whole mount images, we found that cVSMC were present at the aortic root. We then quantified the number of cVSMC and found that PDGFRβEKO hearts contained similar numbers of cVSMC in the base of the heart but exhibited a significant reduction of these cells within the heart apex (Online Figure III).
This region-specific loss of cVSMC suggested that these cVSMC arise from an origin separate from the epicardium or that recombination of the PDGFRβ locus was inefficient or too late. To test these possibilities, we examined PDGFRβ expression by generating E11.5 epicardial cells. Real-time PCR showed that expression of PDGFRβ transcripts was significantly reduced (Online Figure IV A). We also examined PDGFRβ protein expression in the epicardium at E12.5 and found that PDGFRβ was absent from the epicardium (podoplanin+ cells15) of PDGFRβEKO hearts including cells surrounding the conotruncal region (Online Figure B-C). At E13.5 we found that PDGFβ was absent from all epicardial cells but was still present in other smooth muscle populations and cardiac valve primordial (Online Figure IV D-E). We next determined the cell populations within the heart that have had Cre activity using ROSA26 reporter mice. As shown in Online Figure IV F, Cre recombination could be detected throughout the epicardium by E12.5, including the epicardium surrounding the conotruncal region and atria. Finally, to determine if an independent epicardial Cre-deletion strain yielded similar results, we used WT1CreGFP mice. In this mouse line Cre is expressed very early during epicardial development and has been used to trace epicardial derived cells16. Using the ROSA26 reporter to follow Cre activity, we observed that a substantial number of these cVSMC were not epicardial derivatives (Online Figure IV G-H). Thus, it appears that there are two distinct cell populations for generating coronary artery VSMC, and PDGFRβ function is required in both populations.
Epicardium and epicardial derivative formation in the PDGFRβ-/- hearts
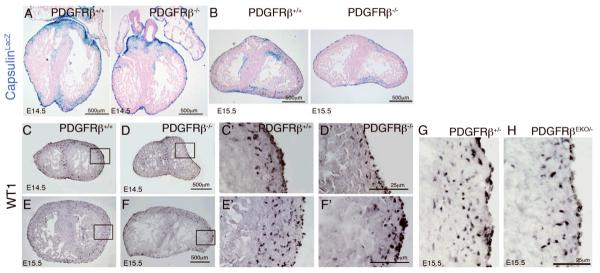
Because we observed a complete absence of epicardial-derived cVSMC we determined if the epicardium developed normally in our mutant hearts. To identify the proepicardium and epicardium we used a mouse line that expresses β-galactosidase from the capsulin locus (capsulinLacZ/+)17, which is expressed by proepicardial and epicardial cells. Online Figure V A-B demonstrates that at E9.5 both wild type and PDGFRβ-/- embryos formed a proepicardium and have clusters of cells that have attached to the dorsal aspect of the heart. From E13.5- E15.5, capsulin+ epicardial cells have spread over the surface of the heart (Online Figure V C-H). Although the PDGFRβ-/-; capsulinLacZ+ epicardial cell pattern appeared less uniform and exhibited small gaps between cells compared to the controls, we still concluded that epicardial cell attachment and spreading over the myocardium occurred in the absence of PDGFRβ signaling.
After epicardial cells attach to and spread over the heart, a subset undergoes EMT and subsequently migrate into the myocardium. Using β-galactosidase expression from capsulinLacZ/+ mice to trace epicardial cell presence in the myocardium, we found that PDGFRβ-/- hearts had reduced numbers of β-galactosidase+ cells within the myocardium at both E14.5 and E15.5 (Figure 3A-B). To verify this reduction of epicardial cell derivatives in the myocardium, we used an independent marker for undifferentiated epicardial cells, Wilms tumor 1 (WT1)18, 19. We observed a marked decrease in WT1+ cells in the myocardium of PDGFRβ-/- and PDGFRβEKO hearts compared to wild type controls, although WT1 expressing cells are clearly found in the epicardium (Figure 3C-H). The reduction in epicardial-derived cells within the myocardium could be caused by either reduced proliferation, increased cell death, or decreased migration into the heart. We analyzed PDGFRβ-/- hearts for proliferation and apoptosis and found no difference between control and PDGFRβ-/- hearts for either of these parameters at E13.5 and E14.5 (Online Figure VI and Online Table I). Taken together these data suggest that the reduction of epicardial cells within the myocardium was caused by a failure of epicardial cells to exit the epicardium.
Figure 3. Decreased presence of epicardial derivatives in PDGFRβ-/- hearts.
(A, B) Cells of the epicardium were identified using capsulinlacZ+ driven-expression of β-galactosidase (blue). (C-H) Sections stained for WT1 of E14.5 (C,D) and E15.5 (E-H) hearts. Figures in C'-F' are higher magnification views of boxed areas in C-F.
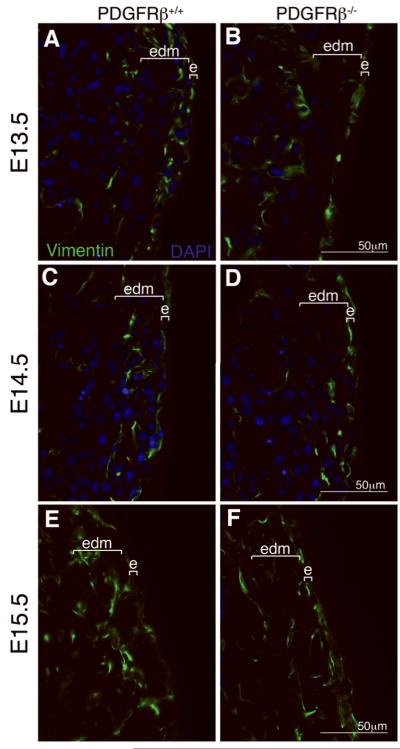
Because failure of epicardial cell migration into the myocardium could also be a result of EMT disruption, we examined the ability of PDGFRβ-/- epicardial cells to upregulate transcription of a mesenchymal marker, vimentin20. At E13.5 abundant vimentin+ cells were observed in the myocardium of control hearts (Figure 4A). At E14.5 vimentin-expressing cells progressed further into the myocardium (Figure 4C), and at E15.5 the vimentin+ cells were distributed throughout the myocardium (Figure 4E). In contrast, fewer vimentin+ cells were observed within the myocardium of PDGFRβ-/- hearts at each developmental stage, even though vimentin was expressed in PDGFRβ-/- epicardium at each stage (Figure 4B,D,F). Quantification of epicardial derived mesenchyme demonstrated a 52% reduction in vimentin+ cells within the heart, similar to what we observe with WT1 staining (%vimentin+ area excluding epicardium: control 11.39% ± 2.56 (n=5) and PDGFRβ-/- 5.88% ± 1.44 (n=5); p<0.005). In addition, there appears to be more cells in the PDGFRβ-/- epicardium that retain vimentin expression at E14.5 and E15.5 than in wild type epicardium. This leads to the possibility that although the cells initiate the transcription program for EMT, they lack the appropriate signals for migration. All results thus far are consistent with a role for PDGFRβ signaling in instructing epicardial cell invasion into the myocardium.
Figure 4. PDGFRβ-/- hearts express the mesenchymal marker, vimentin.
(A-F) Vimentin expression in the indicated genotypes. e; epicardium, and edm; epicardial-derived mesenchyme.
A role for PDGFRβ in epicardial migration
The reduction in capsulin+ and WT1+ cells within the myocardium of PDGFRβ-deficient hearts caused us to investigate epicardial cell migration in greater detail. To identify the signaling pathway responsible for epicardial cell migration, we examined hearts of mice that fail to activate specific signaling pathways downstream of the PDGFRβ alongside PDGFRβ-/- and PDGFRβEKO hearts. We first quantified the number of WT1+ cells in the myocardium at E15.5. We found that both PDGFRβ-/- and PDGFRβEKO hearts possessed roughly half the WT1+ cells of littermate controls in both the left and right ventricles of the heart (Figure 5A). We next investigated WT1+ cell migration in mice bearing signaling point mutants of PDGFRβ. Previous analyses have shown that cVSMC development occurs less efficiently in these animals21. The PDGFRβ mutant receptors lacked PDGFRβ-induced PI3K (PDGFRβF2/F2); PI3K, RasGAP, Shp2, and PLCγ (PDGFRβF5/F5); or Src, Grb2, PI3K, RasGAP, Shp2 and PLCγ (PDGFRβF7/F7) pathways. All three PDGFRβ mutant strains exhibited a 40-50% reduction in the number of WT1+ cells within the myocardium (Figure 5A). Because PI3K is the only signaling pathway disrupted in all three mutant strains, we infer that the PI3K pathway may be the essential and predominant signaling pathway for initiating PDGFRβ-driven epicardial cell migration into the heart. This observation agrees with the known role of PI3K signaling in actin reorganization and cell migration22.
Figure 5. PDGFRβ stimulation induces epicardial cell migration.
(A) Quantification of WT1+ cells in right and left lateral ventricles of E15.5 hearts from the indicated genotypes. Numbers in parentheses indicate the number of independent hearts quantified. All mutant alleles exhibited a reduction in WT1+ cells within the myocardium compared to those present in wild type myocardium. (One- way ANOVA, *p<0.001). See text for allele description. (B-E) Migration of epicardial cells by CCFSE dye tracing. Inset in B shows a time zero E13.5 heart labeled with CCFSE. Arrowheads mark the epicardial boundary. F. Quantification of adeno-viral GFP-labeled epicardial cells that have migrated into the myocardium. Numbers in parentheses indicate the number of hearts assayed. LY=LY294002. All samples were compared to wild type cell migration. (Two-way ANOVA, *p<0.001)
To further evaluate epicardial cell responses to PDGF ligand stimulation, we used a wound closure assay on epicardial monolayers. We saw that stimulation with PDGFBB and PDGFDD enhanced the rate of wound closure in wild type cultures when compared to unstimulated cultures but had no effect on PDGFRβ-/- culture wound closure (Online Table II). The migration of PDGF-stimulated wild type epicardial cells was similar to the migration induced by FGF, which also has a demonstrated role in migration of epicardial cells23.
To directly determine if PDGFRβ stimulation could accelerate the number of cells entering the myocardium, we used an ex vivo system of migration. It has been demonstrated previously that the epicardial surface of hearts can be specifically labeled with the intravital, fluorescent dye, carboxyfluorescein (CCFSE), to follow the epicardial cells as they undergo EMT23 (Figure 5B-E). We cultured hearts from PDGFRβ-/- and wild type embryos in media containing either PDGFBB or serum. When wild type hearts were cultured in 10% FBS, dye-labeled cells were observed in the myocardium. In contrast, when PDGFRβ-/- hearts were cultured under the same conditions, few cells were observed entering the heart. These data suggested that in the absence of PDGFRβ signaling epicardial cells migrated inefficiently. When wild type hearts were stimulated with PDGFBB, numerous labeled cells migrated into the myocardium compared to hearts stimulated by vehicle alone. These results demonstrated that PDGFRβ is necessary and sufficient for epicardial cell migration.
To quantify the effect of PDGFRβ signaling in epicardial cell migration and examine migration in the signaling point mutants, we performed a similar ex vivo assay using adenoviral GFP transduction of the epicardium24. When wild type hearts were cultured in the presence of PDGFBB, GFP+ cells that migrated from the epicardium were observed in the myocardium. The response was similar to known migratory factors, TGFβ1 and bFGF. However, when PDGFRβ (F2/F2, F5/F5, F7/F7) mutant hearts were cultured with PDGFBB, a significant reduction of GFP+ cells within the myocardium was observed. Additionally, wild type hearts treated with a PI3K inhibitor (LY294002) and stimulated with PDGFBB showed a reduction in migration similar to PDGFRβF2/F2 signaling mutants (Figure 5F). Consistent with our previous data, these data suggest the PI3K pathway is essential for PDGFRβ-driven epicardial cell migration and is the dominant signaling pathway involved in this response.
Cellular morphology and cytoskeletal organization in PDGFRβ-/- epicardial cells
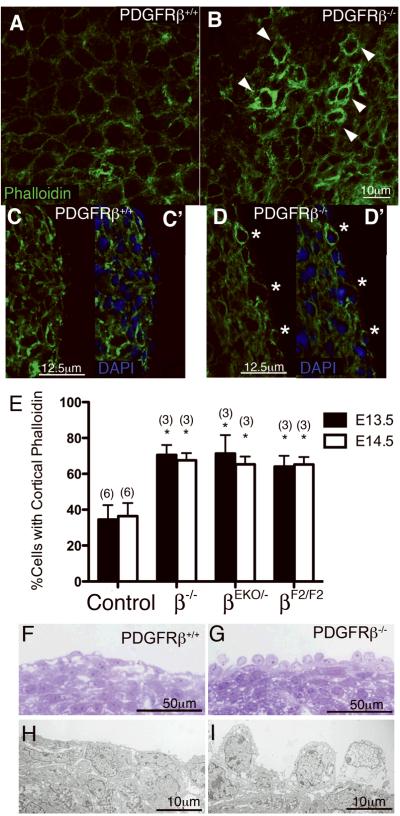
Because proper organization of the cytoskeleton is essential for cell migration we examined actin localization in epicardial cells. Using phalloidin staining we imaged actin organization of hearts in whole mount and in sections of E14.5 PDGFRβ+/+ and PDGFRβ-/-hearts (Figure 6A-D). In wild type hearts, actin was consistently localized to the basal surface of the epicardial cells (Figure 6C). However, PDGFRβ-/- and PDGFRβEKO/- hearts showed an increase in cells that exhibited a subcortical actin distribution (Figure 6C-E). Consistent with a role for PI3K in actin localization, PDGFRβF2/F2 hearts also demonstrated an increase in cortical actin localization (Figure 6E).
Figure 6. Aberrant actin organization and epicardial cell migration in PDGFRβ signaling mutant hearts.
(A-D) Actin localization in control and PDGFRβ mutant hearts using fluorescent-phalloidin. (A,B) Confocal imaging of whole mount actin localization of E14.5 hearts. Arrowheads indicate cells with cortical actin distribution. (C, D) Confocal imaging of E14.5 sectioned hearts. Figures indicated by the prime have DAPI staining to permit identification of epicardial cells on the exterior of the heart. Asterisks indicate cells that have cortical actin localization. (E) Quantification of cells possessing cortical actin at the specified ages in the indicated genotype compared to controls at the same age. (Two-way ANOVA *p<0.001) (F, G) Thick sections of E14.5 hearts processed for TEM and imaged on a light microscope. (H,I) Representative TEM images of thin sections.
We examined the ultrastructure of epicardial cells at E14.5 (Figure 6F-I). TEM revealed that epicardial cells in PDGFRβ-/- hearts lacked the epithelial morphology exhibited by wild type epicardial cells. Occasionally, a few rounded cells were observed in wild type hearts but these were rare in occurrence compared to the number of those observed in PDGFRβ-/-. These results further support a role for PDGFRβ in promoting the cellular processes involved in actin reorganization and migration of epicardial cells.
PI3K signaling is required for PDGFRβ-dependent cortactin localization
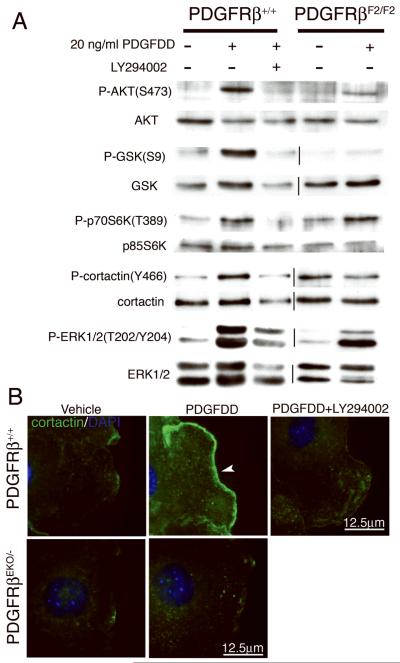
We next examined the phosphorylation status of proteins downstream of the PI3K pathway (AKT, p70S6K, GSK3β) and involved in actin polymerization (cortactin) in LY294002-treated and PDGFRβF2/F2 epicardial cultures. PDGFDD stimulation of wild type epicardial cells induced phosphorylation of AKT, GSK3β, p70S6K, cortactin and ERK1/2 (Figure 7A), but when PI3K signaling was disrupted, phosphorylation of all of these proteins was significantly reduced or absent, with the exception of ERK1/2. Residual AKT and p70S6K activation was observed in PDGFRβF2/F2 epicardial cells. This was likely due to the fact that epicardial cells express both PDGFRα and PDGFRβ, and PDGFDD weakly induces PDGFRα/β heterodimers25.
Figure 7. Disruption of PDGF signaling results in failure to activate and localize cortactin.
(A) Western blot analysis of primary epicardial cell lysates from the indicated genotypes and treatments were probed for phosphorylated (top panel) and total protein (bottom panel). (B) Cortactin localization on wounded epicardial cells. Cells were stimulated with 50ng/ml PDGFDD. Arrowhead indicates abundant cortactin localization to lamellipodia.
Because activation of many PI3K downstream components is associated with actin polymerization, we next evaluated the ability of epicardial cells to form lamellipodia and localize proteins to these cellular areas (Figure 7B and Online Figure VII). Stimulation with PDGFDD increased cortactin localization to lamellipodia in wild type cells. However, PDGFRβ stimulation was unable to direct cortactin to lamellipodia in PDGFRβEKO/- or LY294002-treated cells. These data suggest PI3K is required downstream of PDGF signaling for activating proteins involved in actin reorganization and localizing cortactin to the leading edge of a migrating cell.
Discussion
The epicardium retains stem cell-like properties and when given the appropriate cues, it can differentiate into multiple cardiac cell types26-28. In a zebrafish cardiac injury model, the epicardium is an essential component of the regeneration process29. In addition to being a source of cardiac16, 26 and vascular cells2, 3, the epicardium also secretes growth factors essential for myocardial development30, 31. Therefore, a better understanding of the epicardium during development may help define the signals essential for reactivation of these differentiation processes to improve outcomes of human heart disease.
We show that PDGFRβ provides essential cues for efficient epicardial migration, cVSMC formation, and coronary vessel maturation. Previous expression studies have demonstrated ligands and receptors within the epicardium foreshadowing a requirement for PDGF signal transduction12, 32. Explant studies in rat and chick proepicardial and epicardial cells have demonstrated that stimulation with PDGF ligands leads to filamentous actin formation and expression of smooth muscle cell markers33, 34. We now provide in vivo illustration of the role for PDGFRβ in epicardial function and show that disruption of this signaling impacts more than just VSMC proliferation. The heart is the first tissue to demonstrate an absolute requirement for PDGFRβ signaling to promote VSMC differentiation. In most other tissues VSMC differentiation occurs but expansion is disrupted in the absence of PDGFRβ4.
It is an established fact that VSMC are a heterogeneous population and that they come from a vast range of embryonic origins. In the chick, lineage-tracing analysis has demonstrated that the majority of cVSMC are derived from the proepicardium3, 35, 36. Consistent with recent lineage tracing studies16, 26, we have shown that in the mouse, a majority of VSMC also arise from the epicardium. However, one question concerning the heterogeneity of cVSMC is the origin of the residual cells that are present around the coronary arteries in PDGFRβ epicardial mutant hearts. From the current experiments it is difficult to determine if these cVSMC arise ectopically due to the absence of epicardially-derived VSMC, or if they are a normal subpopulation of cells contributing to the coronary arteries. Both neural crest-derived cells and cells from the secondary heart field can contribute to VSMC in the outflow tract and the coronary arteries37-39. The possibility that cVSMC are heterogeneous in origin is an important consideration because the two cell populations are likely to express different genes and respond differently under pathological conditions or in response to pharmacologic intervention.
Based on our analysis of the formation of the coronary vessels in PDGFRβ mutant hearts, we propose that PDGFRβ signaling can indirectly help shape the mature coronary vasculature. The observation that the coronary arteries defects in PDGFRβEKO but not PDGFRβ-/- hearts improve over time suggests that cVSMC may be involved in the coronary vessel remodeling process. We have shown that cVSMC formation was completely disrupted, while epicardial cell migration was reduced but not abrogated in PDGFRβ-/- hearts. The most likely explanation for this observation is that presence of PDGFRα can compensate for loss of PDGFRβ in the epicardium but not cVSMC. These two receptors signal through very similar pathways and can bind some but not all of the same ligands40. We have recently shown that PDGF receptor function in neural crest cells is also partially redundant, therefore ligand availability may be a key factor in determining the contribution of each receptor. The exquisite expression of the PDGFDD ligand in the epicardium32, which predominantly activates PDGFRβ41, could favor signaling through the PDGFRβ. Therefore, it is possible that PDGFDD may provide an autocrine signal that induces cytoskeletal rearrangements necessary for EMT. Recently, PDGFDD overexpression has been demonstrated to induce an EMT-like transformation in prostate cancer cells42.
Our current data cannot determine the temporal requirement for PDGFRβ signaling. The migration defect we observe may predominantly affect only the cells destined to become cVSMC. In this scenario, cVSMC progenitors might not reach their final destination to receive the differentiation cues from myocardium or endothelium. In support of this possibility, clonal analysis of VSMC in the chick has shown that specification of VSMC occurs prior to the formation of the epicardium and that some VSMC markers are expressed in the proepicardium3. Because the requirement for PDGFRβ signaling in many tissues is to promote VSMC proliferation6, 7, another way to explain our results is that PDGFRβ may be required at two stages: within the epicardium for migration and within the epicardial-derived mesenchyme to promote VSMC differentiation or expansion.
A role for PDGFRβ in directing cytoskeletal rearrangements via PI3K has been established in multiple cell types (reviewed by Heldin43). Here, we have demonstrated that PDGF stimulation of PI3K is also required for cortactin localization to lamellipodia. The failure of cortactin localization is likely due to the loss of PI3K-induced Rac activation44. The epicardial cells are incapable of controlling directed actin filament growth, and directed migration into the myocardium is inefficient.
In conclusion, we have demonstrated a novel role for the PDGFRβ in epicardial development in vivo and identified PI3K signaling as one of the pathways associated with this process. In the absence of this signaling, the epicardium fails to adopt a motile phenotype leading to a reduction in cVSMC and abnormal coronary vessels. Overall, these findings suggest that PDGF signaling is acting to promote epicardial migration and that modulation of PDGF receptor signaling should be considered when exploring options for therapeutic applications of epicardial-derived cells.
Acknowledgments
Sources of Funding Supported by NHLBI grant R01HL074257 (MDT), AHA SDG 0330351 (MDT), Harvard Stem Cell Institute (WP), and AHA postdoctoral fellowship (BZ).
Footnotes
Disclosures None.
References
- 1.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27:1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- 2.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- 3.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 4.Hellstrom M, Kalen M, Lindahl P, Abramsson A, Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development. 1999;126:3047–3055. doi: 10.1242/dev.126.14.3047. [DOI] [PubMed] [Google Scholar]
- 5.Leveen P, Pekny M, Gebre-Medhin S, Swolin B, Larsson E, Betsholtz C. Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev. 1994;8:1875–1887. doi: 10.1101/gad.8.16.1875. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl P, Hellstrom M, Kalen M, Karlsson L, Pekny M, Pekna M, Soriano P, Betsholtz C. Paracrine PDGF-B/PDGF-Rbeta signaling controls mesangial cell development in kidney glomeruli. Development. 1998;125:3313–3322. doi: 10.1242/dev.125.17.3313. [DOI] [PubMed] [Google Scholar]
- 7.Lindahl P, Johansson BR, Leveen P, Betsholtz C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science. 1997;277:242–245. doi: 10.1126/science.277.5323.242. [DOI] [PubMed] [Google Scholar]
- 8.Soriano P. Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 1994;8:1888–1896. doi: 10.1101/gad.8.16.1888. [DOI] [PubMed] [Google Scholar]
- 9.Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Nat Acad Sci. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van den Akker NM, Winkel LC, Nisancioglu MH, Maas S, Wisse LJ, Armulik A, Poelmann RE, Lie-Venema H, Betsholtz C, Gittenberger-de Groot AC. PDGF-B signaling is important for murine cardiac development: its role in developing atrioventricular valves, coronaries, and cardiac innervation. Dev Dyn. 2008;237:494–503. doi: 10.1002/dvdy.21436. [DOI] [PubMed] [Google Scholar]
- 11.Tidhar A, Reichenstein M, Cohen D, Faerman A, Copeland NG, Gilbert DJ, Jenkins NA, Shani M. A novel transgenic marker for migrating limb muscle precursors and for vascular smooth muscle cells. Dev Dyn. 2001;220:60–73. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1089>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 12.Van Den Akker NM, Lie-Venema H, Maas S, Eralp I, DeRuiter MC, Poelmann RE, Gittenberger-De Groot AC. Platelet-derived growth factors in the developing avian heart and maturating coronary vasculature. Dev Dyn. 2005;233:1579–1588. doi: 10.1002/dvdy.20476. [DOI] [PubMed] [Google Scholar]
- 13.Kang J, Gu Y, Li P, Johnson BL, Sucov HM, Thomas PS. PDGF-A as an epicardial mitogen during heart development. Dev Dyn. 2008;237:692–701. doi: 10.1002/dvdy.21469. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahtab EA, Wijffels MC, Van Den Akker NM, Hahurij ND, Lie-Venema H, Wisse LJ, Deruiter MC, Uhrin P, Zaujec J, Binder BR, Schalij MJ, Poelmann RE, Gittenberger-De Groot AC. Cardiac malformations and myocardial abnormalities in podoplanin knockout mouse embryos: Correlation with abnormal epicardial development. Dev Dyn. 2008;237:847–857. doi: 10.1002/dvdy.21463. [DOI] [PubMed] [Google Scholar]
- 16.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–13. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, Chang P, Richardson JA, Gan L, Weiler H, Olson EN. The basic helix-loop-helix transcription factor capsulin controls spleen organogenesis. Proc Nat Acad Sci. 2000;97:9525–9530. doi: 10.1073/pnas.97.17.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- 19.Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Nat Acad Sci. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Pomares JM, Macias D, Garcia-Garrido L, Munoz-Chapuli R. Contribution of the primitive epicardium to the subepicardial mesenchyme in hamster and chick embryos. Dev Dyn. 1997;210:96–105. doi: 10.1002/(SICI)1097-0177(199710)210:2<96::AID-AJA3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Tallquist MD, French WJ, Soriano P. Additive effects of PDGF receptor beta signaling pathways in vascular smooth muscle cell development. PLoS Biol. 2003;1:E52. doi: 10.1371/journal.pbio.0000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki AT, Firtel RA. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur J Cell Biol. 2006;85:873–895. doi: 10.1016/j.ejcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Morabito CJ, Dettman RW, Kattan J, Collier JM, Bristow J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev Biol. 2001;234:204–215. doi: 10.1006/dbio.2001.0254. [DOI] [PubMed] [Google Scholar]
- 24.Compton LA, Potash DA, Mundell NA, Barnett JV. Transforming growth factor-beta induces loss of epithelial character and smooth muscle cell differentiation in epicardial cells. Dev Dyn. 2006;235:82–93. doi: 10.1002/dvdy.20629. [DOI] [PubMed] [Google Scholar]
- 25.LaRochelle WJ, Jeffers M, McDonald WF, Chillakuru RA, Giese NA, Lokker NA, Sullivan C, Boldog FL, Yang M, Vernet C, Burgess CE, Fernandes E, Deegler LL, Rittman B, Shimkets J, Shimkets RA, Rothberg JM, Lichenstein HS. PDGF-D, a new protease-activated growth factor. Nature Cell Biol. 2001;3:517–521. doi: 10.1038/35074593. [DOI] [PubMed] [Google Scholar]
- 26.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR, Riley PR. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 28.van Tuyn J, Atsma DE, Winter EM, van der Velde-van Dijke I, Pijnappels DA, Bax NA, Knaan-Shanzer S, Gittenberger-de Groot AC, Poelmann RE, van der Laarse A, van der Wall EE, Schalij MJ, de Vries AA. Epicardial Cells of Human Adults Can Undergo an Epithelial-to-Mesenchymal Transition and Obtain Characteristics of Smooth Muscle Cells In Vitro. Stem Cells. 2007;25:271–278. doi: 10.1634/stemcells.2006-0366. [DOI] [PubMed] [Google Scholar]
- 29.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 30.Chen TH, Chang TC, Kang JO, Choudhary B, Makita T, Tran CM, Burch JB, Eid H, Sucov HM. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acidinducible trophic factor. Dev Biol. 2002;250:198–207. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- 31.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Ponten A, Folestad EB, Pietras K, Eriksson U. Platelet-derived growth factor D induces cardiac fibrosis and proliferation of vascular smooth muscle cells in heart-specific transgenic mice. Circ Res. 2005;97:1036–1045. doi: 10.1161/01.RES.0000190590.31545.d4. [DOI] [PubMed] [Google Scholar]
- 33.Landerholm TE, Dong XR, Lu J, Belaguli NS, Schwartz RJ, Majesky MW. A role for serum response factor in coronary smooth muscle differentiation from proepicardial cells. Development. 1999;126:2053–2062. doi: 10.1242/dev.126.10.2053. [DOI] [PubMed] [Google Scholar]
- 34.Wada AM, Smith TK, Osler ME, Reese DE, Bader DM. Epicardial/Mesothelial cell line retains vasculogenic potential of embryonic epicardium. Circ Res. 2003;92:525–531. doi: 10.1161/01.RES.0000060484.11032.0B. [DOI] [PubMed] [Google Scholar]
- 35.Dettman RW, Denetclaw W, Jr., Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Pomares JM, Macias D, Garcia-Garrido L, Munoz-Chapuli R. The origin of the subepicardial mesenchyme in the avian embryo: an immunohistochemical and quail-chick chimera study. Dev Biol. 1998;200(1):57–68. doi: 10.1006/dbio.1998.8949. [DOI] [PubMed] [Google Scholar]
- 37.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304(1):286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 40.Fredriksson L, Li H, Eriksson U. The PDGF family: four gene products form five dimeric isoforms. Cytokine & Growth Factor Rev. 2004;15:197–204. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Bergsten E, Uutela M, Li X, Pietras K, Ostman A, Heldin CH, Alitalo K, Eriksson U. PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor. Nature Cell Biol. 2001;3:512–516. doi: 10.1038/35074588. [DOI] [PubMed] [Google Scholar]
- 42.Kong D, Wang Z, Sarkar SH, Li Y, Banerjee S, Saliganan A, Kim HR, Cher ML, Sarkar FH. Platelet-derived growth factor-D overexpression contributes to epithelialmesenchymal transition of PC3 prostate cancer cells. Stem Cells. 2008;26:1425–1435. doi: 10.1634/stemcells.2007-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heldin CH, Ostman A, Ronnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim et Biophys Acta. 1998;1378:F79–113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 44.Head JA, Jiang D, Li M, Zorn LJ, Schaefer EM, Parsons JT, Weed SA. Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol Biol Cell. 2003;14:3216–3229. doi: 10.1091/mbc.E02-11-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]