Abstract
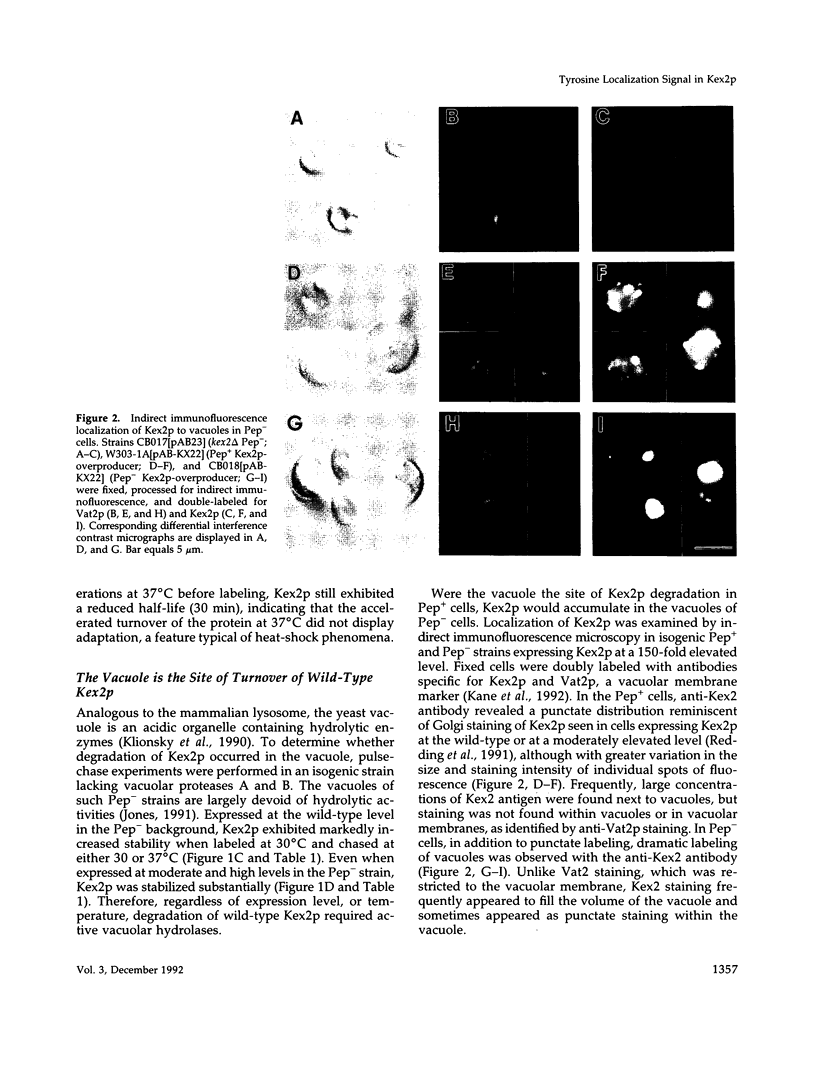
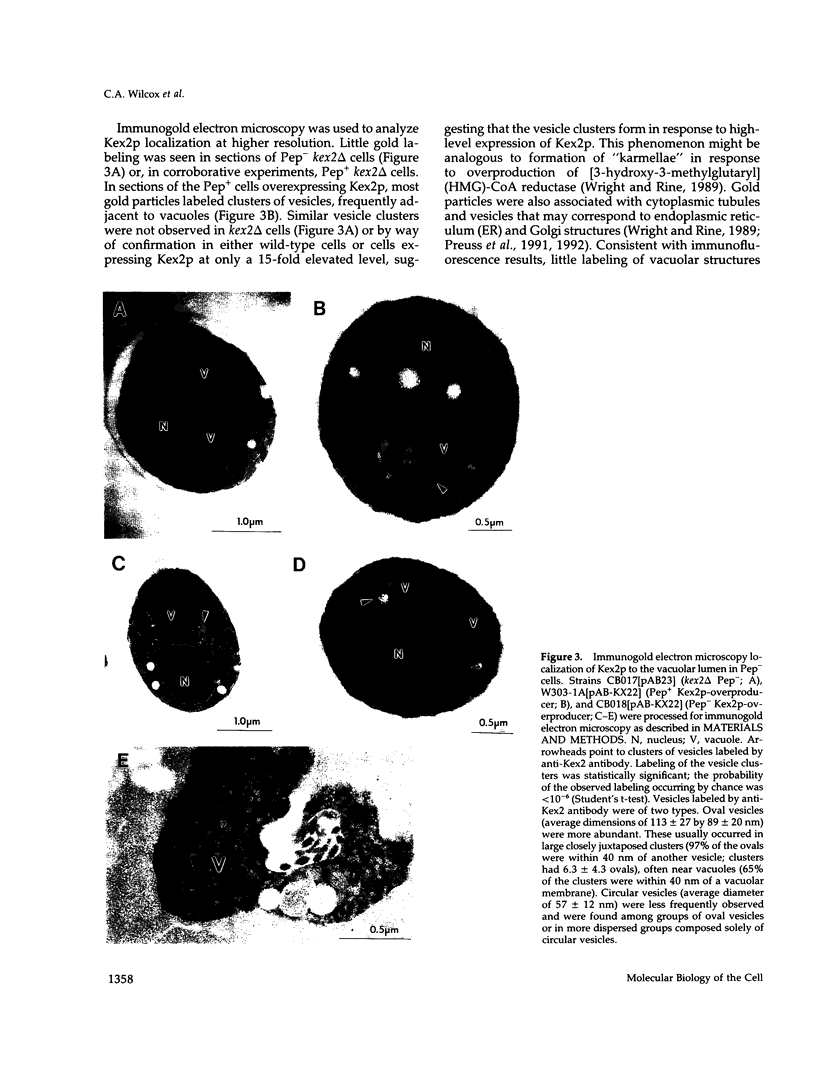
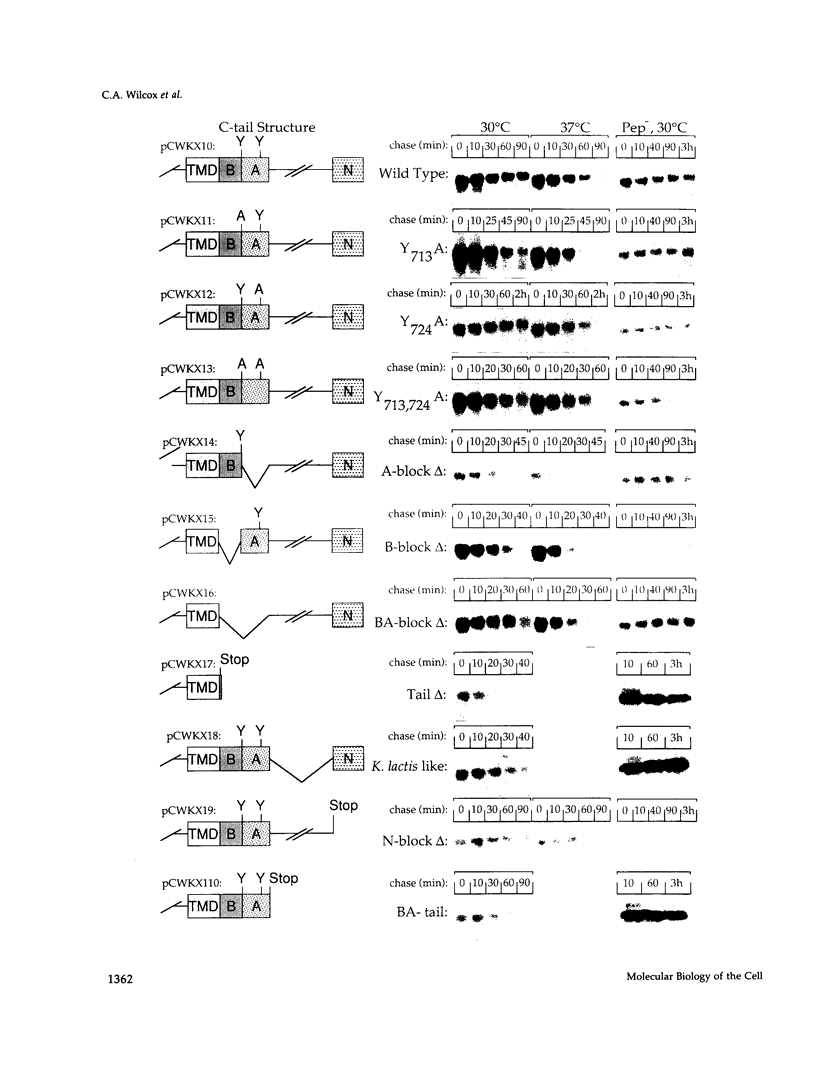
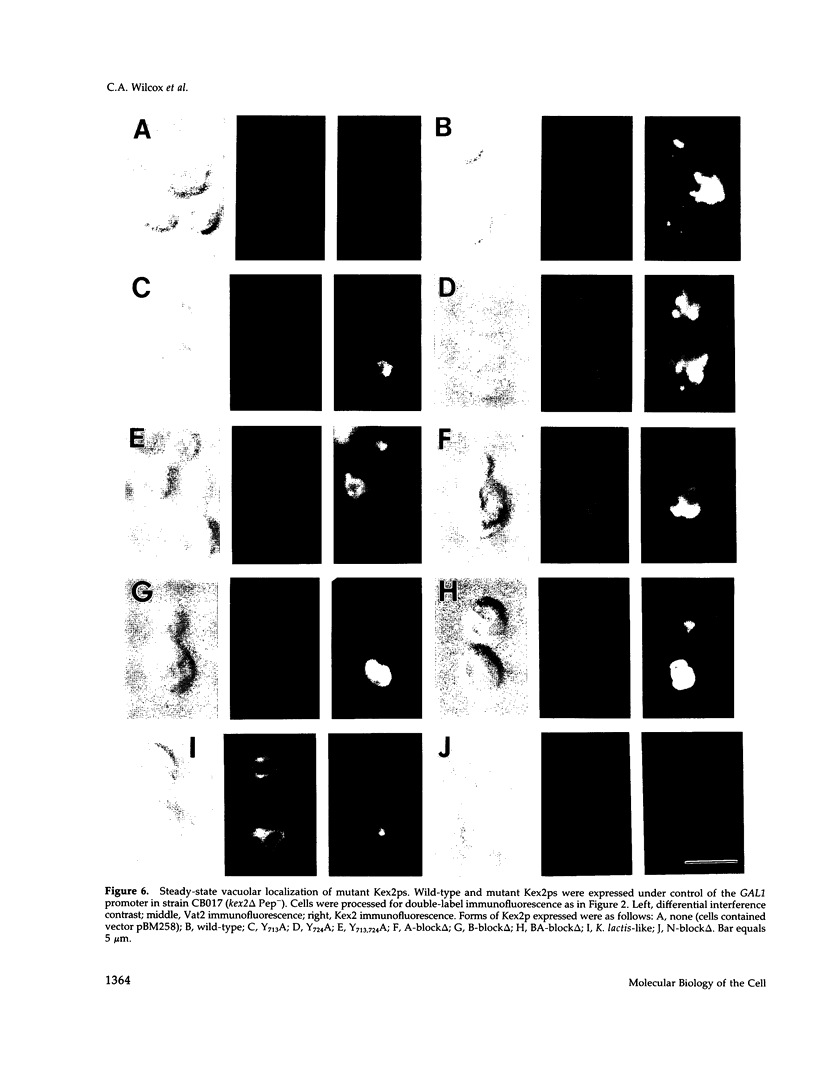
Kex2 protease processes pro-alpha-factor in a late Golgi compartment in Saccharomyces cerevisiae. The first approximately 30 residues of the 115 amino acid CO2H-terminal cytosolic tail (C-tail) of the Kex2 protein (Kex2p) contain a Golgi retention signal that resembles coated-pit localization signals in mammalian cell surface receptors. Mutation of one (Tyr713) of two tyrosine residues in the C-tail or deletion of sequences adjacent to Tyr713 results in loss of normal Golgi localization. Surprisingly, loss of the Golgi retention signal resulted in transport of C-tail mutant Kex2p to the vacuole (yeast lysosome), as judged by kinetics of degradation and by indirect immunofluorescence. Analysis of the loss of Kex2 function in vivo after shutting off expression of wild-type or mutant forms proved that mutations that cause rapid vacuolar turnover do so by increasing the rate of exit of the enzyme from the pro-alpha-factor processing compartment. The most likely explanation for these results is that mutation of the Golgi retention signal in the C-tail results in transport of Kex2p to the vacuole by default. Wild-type Kex2p also was transported to the vacuole at an increased rate when overproduced, although apparently not due to saturation of a Golgi-retention mechanism. Instead, the wild-type and C-tail mutant forms of Kex2p may follow distinct paths to the vacuole.
Full text
PDF







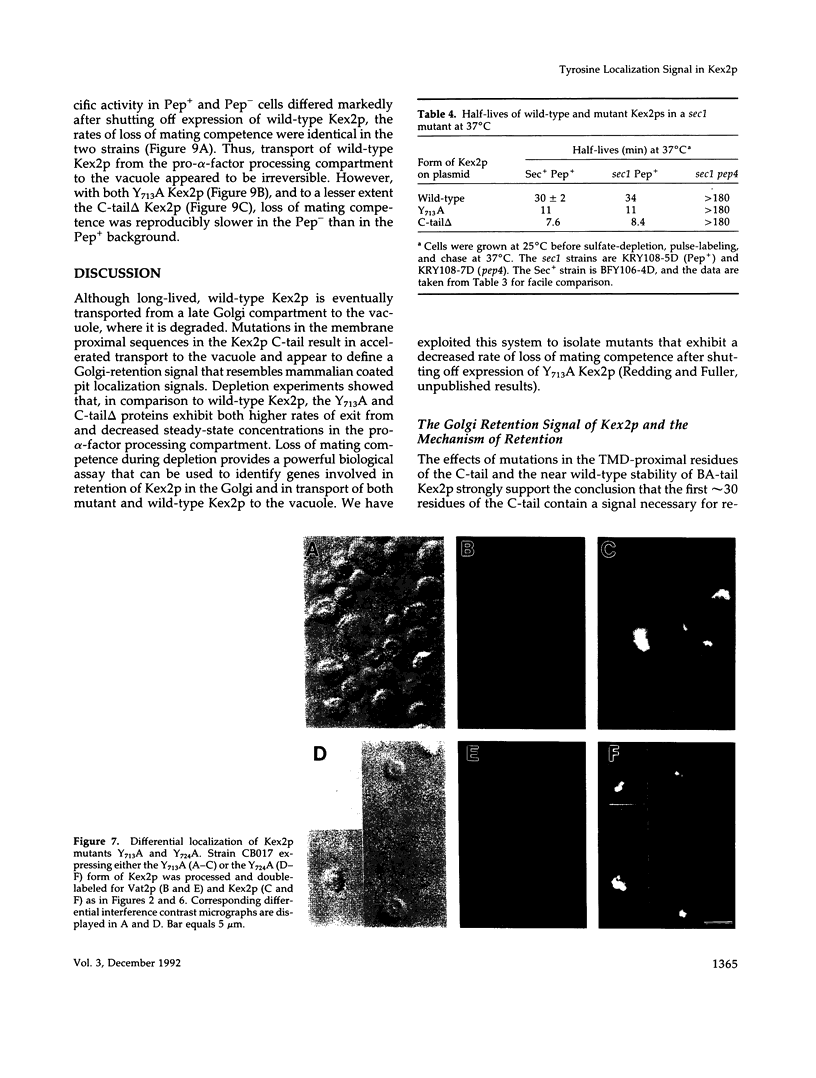
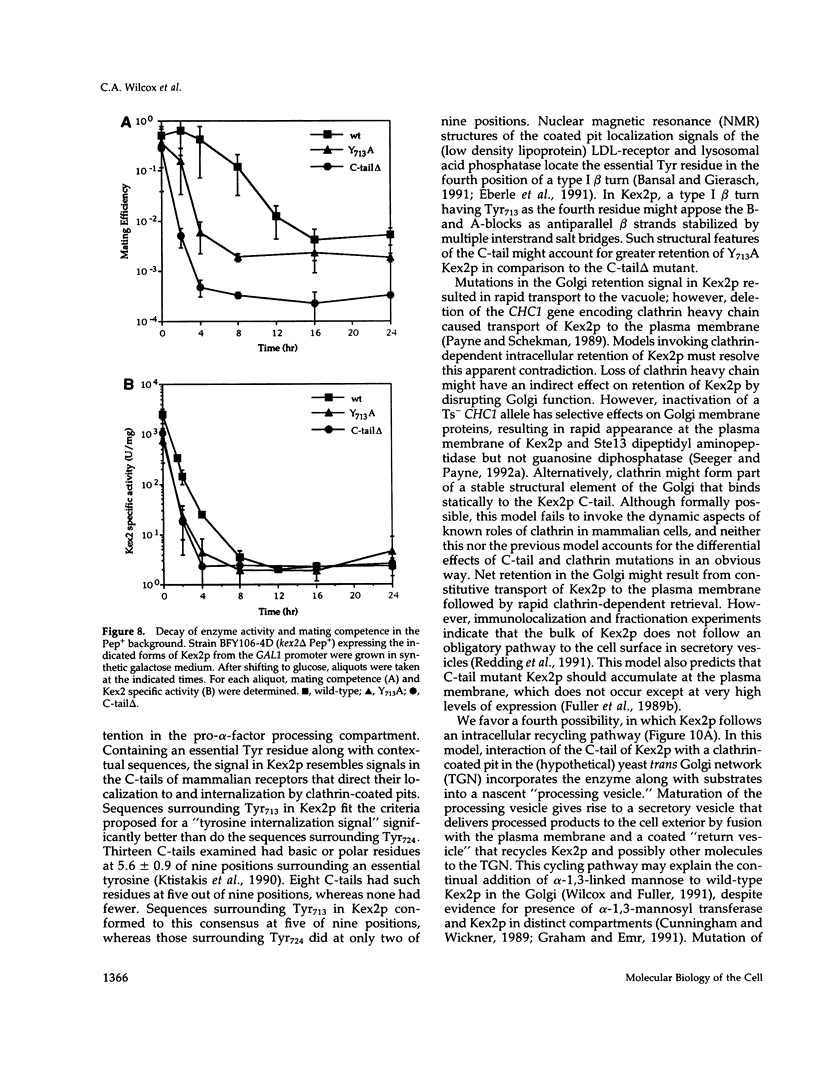
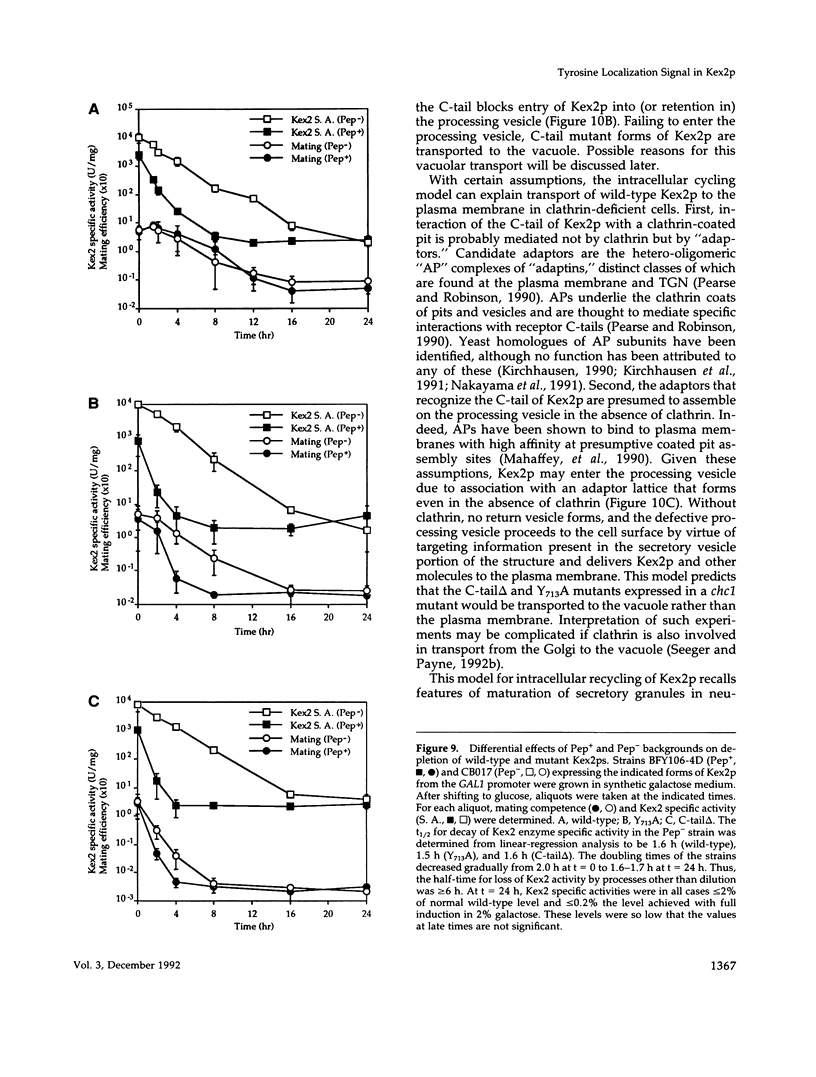
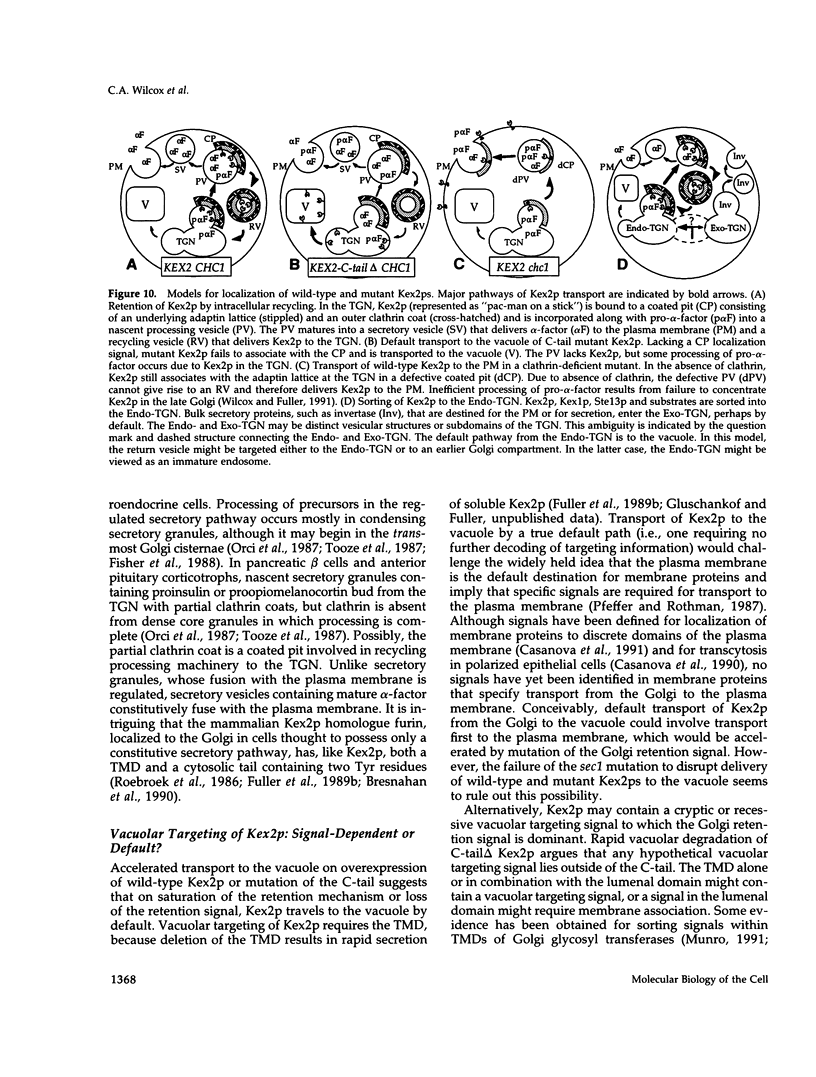



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bansal A., Gierasch L. M. The NPXY internalization signal of the LDL receptor adopts a reverse-turn conformation. Cell. 1991 Dec 20;67(6):1195–1201. doi: 10.1016/0092-8674(91)90295-a. [DOI] [PubMed] [Google Scholar]
- Barr P. J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991 Jul 12;66(1):1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Brenner C., Fuller R. S. Structural and enzymatic characterization of a purified prohormone-processing enzyme: secreted, soluble Kex2 protease. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):922–926. doi: 10.1073/pnas.89.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnahan P. A., Leduc R., Thomas L., Thorner J., Gibson H. L., Brake A. J., Barr P. J., Thomas G. Human fur gene encodes a yeast KEX2-like endoprotease that cleaves pro-beta-NGF in vivo. J Cell Biol. 1990 Dec;111(6 Pt 2):2851–2859. doi: 10.1083/jcb.111.6.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J. E., Apodaca G., Mostov K. E. An autonomous signal for basolateral sorting in the cytoplasmic domain of the polymeric immunoglobulin receptor. Cell. 1991 Jul 12;66(1):65–75. doi: 10.1016/0092-8674(91)90139-p. [DOI] [PubMed] [Google Scholar]
- Casanova J. E., Breitfeld P. P., Ross S. A., Mostov K. E. Phosphorylation of the polymeric immunoglobulin receptor required for its efficient transcytosis. Science. 1990 May 11;248(4956):742–745. doi: 10.1126/science.2110383. [DOI] [PubMed] [Google Scholar]
- Cleves A. E., McGee T. P., Whitters E. A., Champion K. M., Aitken J. R., Dowhan W., Goebl M., Bankaitis V. A. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991 Feb 22;64(4):789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland C. S., Zimmer K. P., Wagner K. R., Healey G. A., Mellman I., Helenius A. Folding, trimerization, and transport are sequential events in the biogenesis of influenza virus hemagglutinin. Cell. 1988 Apr 22;53(2):197–209. doi: 10.1016/0092-8674(88)90381-9. [DOI] [PubMed] [Google Scholar]
- Cunningham K. W., Wickner W. T. Yeast KEX2 protease and mannosyltransferase I are localized to distinct compartments of the secretory pathway. Yeast. 1989 Jan-Feb;5(1):25–33. doi: 10.1002/yea.320050105. [DOI] [PubMed] [Google Scholar]
- Davis C. G., van Driel I. R., Russell D. W., Brown M. S., Goldstein J. L. The low density lipoprotein receptor. Identification of amino acids in cytoplasmic domain required for rapid endocytosis. J Biol Chem. 1987 Mar 25;262(9):4075–4082. [PubMed] [Google Scholar]
- Doms R. W., Ruusala A., Machamer C., Helenius J., Helenius A., Rose J. K. Differential effects of mutations in three domains on folding, quaternary structure, and intracellular transport of vesicular stomatitis virus G protein. J Cell Biol. 1988 Jul;107(1):89–99. doi: 10.1083/jcb.107.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle W., Sander C., Klaus W., Schmidt B., von Figura K., Peters C. The essential tyrosine of the internalization signal in lysosomal acid phosphatase is part of a beta turn. Cell. 1991 Dec 20;67(6):1203–1209. doi: 10.1016/0092-8674(91)90296-b. [DOI] [PubMed] [Google Scholar]
- Fisher J. M., Sossin W., Newcomb R., Scheller R. H. Multiple neuropeptides derived from a common precursor are differentially packaged and transported. Cell. 1988 Sep 9;54(6):813–822. doi: 10.1016/s0092-8674(88)91131-2. [DOI] [PubMed] [Google Scholar]
- Franzusoff A., Redding K., Crosby J., Fuller R. S., Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991 Jan;112(1):27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A., Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989 Sep;8(9):2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Brake A. J., Thorner J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science. 1989 Oct 27;246(4929):482–486. doi: 10.1126/science.2683070. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Brake A., Thorner J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1434–1438. doi: 10.1073/pnas.86.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Sterne R. E., Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- Gething M. J., McCammon K., Sambrook J. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell. 1986 Sep 12;46(6):939–950. doi: 10.1016/0092-8674(86)90076-0. [DOI] [PubMed] [Google Scholar]
- Graham T. R., Emr S. D. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991 Jul;114(2):207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980 Jun;85(3):811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasilik A., Tanner W. Biosynthesis of the vacuolar yeast glycoprotein carboxypeptidase Y. Conversion of precursor into the enzyme. Eur J Biochem. 1978 Apr 17;85(2):599–608. doi: 10.1111/j.1432-1033.1978.tb12275.x. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Three proteolytic systems in the yeast saccharomyces cerevisiae. J Biol Chem. 1991 May 5;266(13):7963–7966. [PubMed] [Google Scholar]
- Julius D., Brake A., Blair L., Kunisawa R., Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984 Jul;37(3):1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Julius D., Schekman R., Thorner J. Glycosylation and processing of prepro-alpha-factor through the yeast secretory pathway. Cell. 1984 Feb;36(2):309–318. doi: 10.1016/0092-8674(84)90224-1. [DOI] [PubMed] [Google Scholar]
- Kane P. M., Kuehn M. C., Howald-Stevenson I., Stevens T. H. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992 Jan 5;267(1):447–454. [PubMed] [Google Scholar]
- Kirchhausen T., Davis A. C., Frucht S., Greco B. O., Payne G. S., Tubb B. AP17 and AP19, the mammalian small chains of the clathrin-associated protein complexes show homology to Yap17p, their putative homolog in yeast. J Biol Chem. 1991 Jun 15;266(17):11153–11157. [PubMed] [Google Scholar]
- Kirchhausen T. Identification of a putative yeast homolog of the mammalian beta chains of the clathrin-associated protein complexes. Mol Cell Biol. 1990 Nov;10(11):6089–6090. doi: 10.1128/mcb.10.11.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Emr S. D. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989 Aug;8(8):2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Herman P. K., Emr S. D. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990 Sep;54(3):266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis T. E., Lodish H. F. Oligomerization is essential for transport of vesicular stomatitis viral glycoprotein to the cell surface. Cell. 1986 Sep 12;46(6):929–937. doi: 10.1016/0092-8674(86)90075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ktistakis N. T., Thomas D., Roth M. G. Characteristics of the tyrosine recognition signal for internalization of transmembrane surface glycoproteins. J Cell Biol. 1990 Oct;111(4):1393–1407. doi: 10.1083/jcb.111.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lazarovits J., Roth M. A single amino acid change in the cytoplasmic domain allows the influenza virus hemagglutinin to be endocytosed through coated pits. Cell. 1988 Jun 3;53(5):743–752. doi: 10.1016/0092-8674(88)90092-x. [DOI] [PubMed] [Google Scholar]
- Leibowitz M. J., Wickner R. B. A chromosomal gene required for killer plasmid expression, mating, and spore maturation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2061–2065. doi: 10.1073/pnas.73.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey D. T., Peeler J. S., Brodsky F. M., Anderson R. G. Clathrin-coated pits contain an integral membrane protein that binds the AP-2 subunit with high affinity. J Biol Chem. 1990 Sep 25;265(27):16514–16520. [PubMed] [Google Scholar]
- Munro S. Sequences within and adjacent to the transmembrane segment of alpha-2,6-sialyltransferase specify Golgi retention. EMBO J. 1991 Dec;10(12):3577–3588. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N., Miyajima A., Arai K. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985 Oct;4(10):2643–2648. doi: 10.1002/j.1460-2075.1985.tb03982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y., Goebl M., O'Brine Greco B., Lemmon S., Pingchang Chow E., Kirchhausen T. The medium chains of the mammalian clathrin-associated proteins have a homolog in yeast. Eur J Biochem. 1991 Dec 5;202(2):569–574. doi: 10.1111/j.1432-1033.1991.tb16409.x. [DOI] [PubMed] [Google Scholar]
- Nilsson T., Lucocq J. M., Mackay D., Warren G. The membrane spanning domain of beta-1,4-galactosyltransferase specifies trans Golgi localization. EMBO J. 1991 Dec;10(12):3567–3575. doi: 10.1002/j.1460-2075.1991.tb04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L., Ravazzola M., Storch M. J., Anderson R. G., Vassalli J. D., Perrelet A. Proteolytic maturation of insulin is a post-Golgi event which occurs in acidifying clathrin-coated secretory vesicles. Cell. 1987 Jun 19;49(6):865–868. doi: 10.1016/0092-8674(87)90624-6. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989 Sep 22;245(4924):1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- Pearse B. M., Robinson M. S. Clathrin, adaptors, and sorting. Annu Rev Cell Biol. 1990;6:151–171. doi: 10.1146/annurev.cb.06.110190.001055. [DOI] [PubMed] [Google Scholar]
- Pfeffer S. R., Rothman J. E. Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Annu Rev Biochem. 1987;56:829–852. doi: 10.1146/annurev.bi.56.070187.004145. [DOI] [PubMed] [Google Scholar]
- Preuss D., Mulholland J., Franzusoff A., Segev N., Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992 Jul;3(7):789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Mulholland J., Kaiser C. A., Orlean P., Albright C., Rose M. D., Robbins P. W., Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991 Dec;7(9):891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- Redding K., Holcomb C., Fuller R. S. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991 May;113(3):527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. J., Nothwehr S. F., Stevens T. H. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992 Oct;119(1):69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebroek A. J., Schalken J. A., Leunissen J. A., Onnekink C., Bloemers H. P., Van de Ven W. J. Evolutionary conserved close linkage of the c-fes/fps proto-oncogene and genetic sequences encoding a receptor-like protein. EMBO J. 1986 Sep;5(9):2197–2202. doi: 10.1002/j.1460-2075.1986.tb04484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rüther U., Müller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2(10):1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Payne G. S. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J. 1992 Aug;11(8):2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Payne G. S. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992 Aug;118(3):531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguy-Rougeau C., Wésolowski-Louvel M., Fukuhara H. The Kluyveromyces lactis KEX1 gene encodes a subtilisin-type serine proteinase. FEBS Lett. 1988 Jul 18;234(2):464–470. doi: 10.1016/0014-5793(88)80139-x. [DOI] [PubMed] [Google Scholar]
- Tooze J., Hollinshead M., Frank R., Burke B. An antibody specific for an endoproteolytic cleavage site provides evidence that pro-opiomelanocortin is packaged into secretory granules in AtT20 cells before its cleavage. J Cell Biol. 1987 Jul;105(1):155–162. doi: 10.1083/jcb.105.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurushita N., Maki H., Korn L. J. Site-directed mutagenesis with Escherichia coli DNA polymerase III holoenzyme. Gene. 1988;62(1):135–139. doi: 10.1016/0378-1119(88)90587-2. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wilcox C. A., Fuller R. S. Posttranslational processing of the prohormone-cleaving Kex2 protease in the Saccharomyces cerevisiae secretory pathway. J Cell Biol. 1991 Oct;115(2):297–307. doi: 10.1083/jcb.115.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. H., Low S. H., Hong W. The 17-residue transmembrane domain of beta-galactoside alpha 2,6-sialyltransferase is sufficient for Golgi retention. J Cell Biol. 1992 Apr;117(2):245–258. doi: 10.1083/jcb.117.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R., Rine J. Transmission electron microscopy and immunocytochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy. Methods Cell Biol. 1989;31:473–512. doi: 10.1016/s0091-679x(08)61624-6. [DOI] [PubMed] [Google Scholar]









