SUMMARY
Post-translational modifications of the carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) specify a molecular recognition code that is deciphered by proteins involved in RNA biogenesis. The CTD is comprised of a repeating heptapeptide (Y1S2P3T4S5P6S7). Recently, phosphorylation of Serine7 was shown to be important for co-transcriptional processing of two snRNAs in mammalian cells. Here, we report that Kin28/Cdk7, a subunit of the evolutionarily conserved TFIIH complex, is a Ser7 kinase. The ability of Kin28/Cdk7 to phosphorylate Ser7 is particularly surprising because this kinase functions at promoters of protein-coding genes, rather than being restricted to promoter-distal regions of snRNA genes. Kin28/Cdk7 is also known to phosphorylate Ser5 residues of the CTD at gene promoters. Taken together, our results implicate the TFIIH kinase in placing bivalent Ser5 and Ser7 marks early in gene transcription. These bivalent CTD marks, in concert with cues within nascent transcripts, specify the co-transcriptional engagement of the relevant RNA processing machinery.
INTRODUCTION
The CTD is reversibly modified by several enzymes, including kinases, phosphatases, prolyl isomerases and glycosylases (Phatnani and Greenleaf, 2006; Sims et al., 2004). Differentially modified CTD heptapeptides are selectively bound by different protein complexes that participate in RNA biogenesis (Buratowski, 2003; Corden, 2007; Egloff and Murphy, 2008; Hirose and Manley, 2000; Phatnani and Greenleaf, 2006). A hypo-phosphorylated CTD associates with the Mediator complex at gene promoters (Lee and Young, 2000; Myers and Kornberg, 2000; Ptashne and Gann, 2002). Phosphorylation of Ser5, primarily by the TFIIH associated kinase Kin28/Cdk7, enhances the association of the CTD with the m7G RNA capping machinery (Komarnitsky et al., 2000; Schroeder et al., 2000). The subsequent phosphorylation of Ser2 by Ctk1/Cdk9, leads to the recruitment of 3′ end processing complexes (Ahn et al., 2004; Licatalosi et al., 2002; Proudfoot et al, 2002). The sequential patterns of CTD modifications are postulated to define a spatio-temporal code that instructs ordered engagement with different functional complexes at various stages of the transcription cycle (Buratowski, 2003; Corden, 2007; Egloff and Murphy, 2008; Phatnani and Greenleaf, 2006).
Ser7 phosphorylation was recently detected in mammalian cells (Chapman et al., 2007; Egloff et al., 2007). This mark was most evident in polymerases engaged in transcription elongation, well within the coding regions of genes (Chapman et al., 2007). While this CTD modification is detected at protein-coding genes, it is functionally important for processing of two spliceosomal snRNAs that are transcribed by Pol II (Egloff et al., 2007). Substituting Ser7 with an alanine residue in tissue culture cells dramatically reduced transcription and processing of snRNA genes, but did not alter the transcription or processing of protein-coding genes. In agreement with this gene-specific role, Ser7 phosphorylation of the CTD was necessary for the stable association of the Integrator complex with the transcribing polymerase (Egloff et al., 2007). The multi-subunit Integrator complex is critical for 3′ processing of snRNA and does not participate in processing transcripts of protein-coding genes (Baillat et al., 2005). Thus, the Ser7 phosphorylation mark (Ser7-P) and the Integrator complex are functionally restricted to the transcription of a specific gene class. This gene-specific role for Ser7-P raises the key question whether the kinase that performs this task is uniquely recruited to snRNA genes. Furthermore, the enrichment of Ser7-P marks deep within the transcribed region suggests that the Ser7 kinase may associate with the elongating Pol II, consistent with its role in preparing Pol II for association with the 3′ snRNA processing machinery. Identifying the Ser7 kinase is therefore critical for understanding the mechanisms that underlie the ability of the CTD to orchestrate different processes during transcription.
The tandem heptapeptide (Y1S2P3T4S5P6S7) repeats of the CTD are conserved from protozoa to humans. The number of heptapeptide repeats increases in proportion to the complexity of the organism, with protozoa containing 15 repeats, budding yeast 26, and humans 52 (Chapman et al., 2008; Corden, 1990) However, nearly 35 repeats were required for efficient phosphorylation of Ser7 in mammalian cells (Chapman et al., 2007). Thus, organisms containing fewer than 35 heptapeptide repeats might lack Ser7-P marks. The restriction of Ser7-P function to higher eukaryotes was further suggested by the fact that most subunits of the Integrator complex have clearly conserved homologues in other metazoan organisms but are largely missing in budding yeast. Furthermore, DNA induced protein kinase, DNA-PK, the only kinase known to phosphorylate Ser7, has no orthologs in budding yeast (Trigon et al., 1998). Thus, it was not clear if the Ser7 residue of the heptapeptide is phosphorylated in single-celled eukaryotes or if this modification is restricted to higher eukaryotes.
RESULTS
Ser7 is phosphorylated in budding yeast
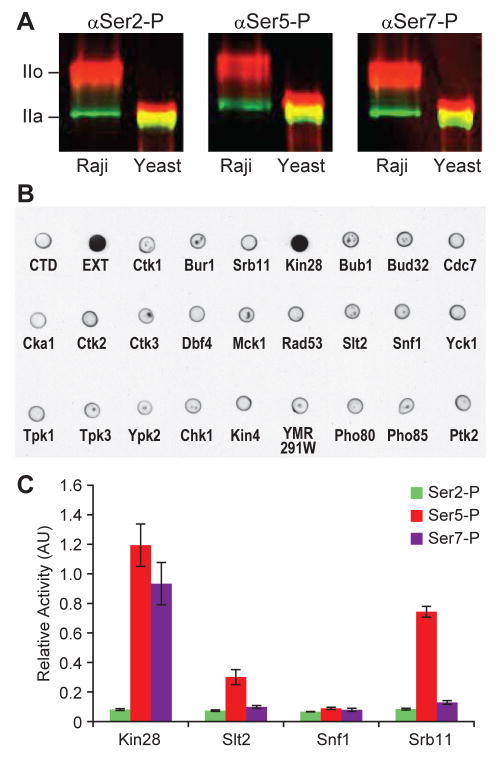
To examine if the Ser7-P mark occurred in simpler eukaryotes we used a monoclonal αSer7-P antibody (Chapman et al., 2007) to detect this modification on Pol II in whole cell extracts of the budding yeast, Saccharomyces cerevisiae. The data unambiguously show that this new modification occurs in budding yeast (Figure 1A). Given the conservation from yeast to humans of the Ser7-P mark and the CTD kinases that act on Ser5 and Ser2, it seemed reasonable that the Ser7 kinase could be identified by proteomic and chemical-genomic analyses in yeast (Bishop et al., 2001; Zhu et al., 2001). We therefore focused on 26 yeast kinases that are present in the nuclear compartment under log-phase growth in rich medium (Kumar et al., 2002). Twenty-three of these kinases were previously genetically tagged with the Tandem Affinity Purification (TAP) module (Open Biosystems). The TAP module is chromosomally integrated at the 3′ end of the kinase-coding region and the fusion protein is expressed at physiological levels (Ghaemmaghami et al., 2003, Puig et al, 2001). In the case of Srb10/Cdk8 kinase we used the TAP fusion to its cyclin (Srb11) to purify the functional cyclin-kinase complex. Because cyclins often direct substrates to their associated kinases (Nigg, 1996; Schulman et al., 1998) we also purified the cyclin Ctk2 that associates with Ctk1/Cdk9 and Pho80 that associates with Pho85 to examine if the co-precipitated kinases phosphorylate Ser7. The enzymes were purified from yeast, and their ability to phosphorylate a recombinant CTD was examined (Ansari et al., 2005). In this assay, only Kin28 efficiently phosphorylates Ser7 (Figure 1B). In control experiments, we readily detect the appearance of Ser5-P and Ser2-P marks with the four CTD kinases known to perform those reactions (Figure S1 A and B). Parallel immunoblots also show that all 23 fusions are expressed (Figure S1C) and the fused kinases phosphorylate an orthologous universal substrate, Ptk2 (Figure S1D). Finally, to quantify the extent of Ser5 and Ser7 phosphorylation by Kin28, we performed enzyme-linked immunoassays (ELISA). The results suggest that Kin28 phosphorylates Ser7 as efficiently as Ser5 (Figure 1C). In these assays Srb10, the other Ser5 kinase that associates with Pol II, does not phosphorylate Ser7.
Figure 1. Kin28 phosphorylates the Ser7 of Pol II CTD.
(A) Western blot of protein extracted from human T-cell line (Raji cells) and yeast. Dual labeling was performed with antibody to Rpb1 (green) and phospho-CTD (red). mAbs αSer2-P, αSer5-P and αSer7-P recognize the phosphorylated Ser2, Ser5 and Ser7 of the CTD.
(B) Dot blot probing Ser7-P marks on GST-CTD phosphorylated by individually purified yeast nuclear kinases. GST-CTD unphosphorylated (−) and phosphorylated by yeast cell extract (EXT) are used as negative and positive controls respectively.
(C) ELISA of synthetic CTD peptide phosphorylated by purified yeast Kin28, Slt2, Snf1 and Srb11 probed with αSer2-P (green), αSer5-P (red) and αSer7-P (purple) antibodies. The measurements were taken in triplicate and the error bars in this and all the subsequent figures correspond to SD.
TFIIH kinase phosphorylates Serine 7
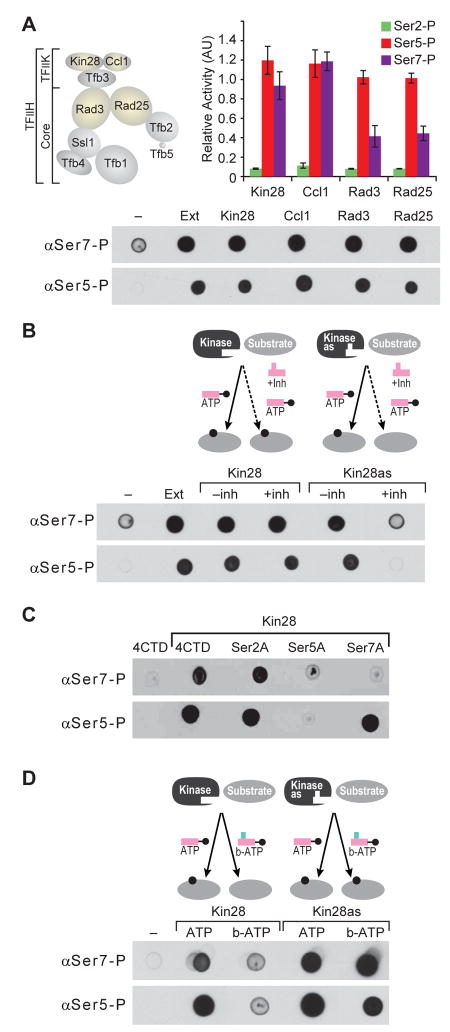
Cyclin-dependent kinases (Cdk) are promiscuous enzymes and their substrate choices are governed by cyclins that recruit specific target proteins (Nigg, 1996; Schulman et al., 1998). The substrate preference of Cdk7, the human homolog of Kin28, is specified by other subunits of the TFIIH complex (Feaver et al., 1994; Svejstrup et al., 1994; Watanabe et al., 2000). Thus, Kin28 within the TFIIH complex could display altered substrate selectivity and might not phosphorylate Ser7. We therefore examined if the TFIIH-associated Kin28 would phosphorylate Ser7 residue of the CTD. Kin28 associates with the cyclin Ccl1 and a third partner (Tfb3) to form an active TFIIK complex that phosphorylates Ser5 of the CTD (Feaver et al., 1994, Feaver et al., 1997). TFIIK further associates with the “core” seven-subunit complex to form the TFIIH complex (Figure 2A) (Feaver et al., 1994; Svejstrup et al., 1994; Ranish et al., 2004). As part of the TFIIH complex, the kinase shows a high degree of substrate specificity and selectively phosphorylates the CTD, two subunits of transcriptional machinery (Med4 and Rgr1), and the transcription activator Gal4 (Feaver et al., 1994; Watanabe et al., 2000; Guidi et al., 2004; Hirst et al., 1999; Liu et al., 2004; Muratani et al., 2005; Sadowski et al., 1991). We purified TFIIH using TAP-fusions to different subunits of the complex. In each case, the associated Kin28 kinase robustly phosphorylates Ser7 as well as Ser5 (Figure 2A). These results indicate that TFIIH-associated Kin28, despite its increased substrate-specificity, phosphorylates Ser7.
Figure 2. TFIIH-associated Kin28 phosphorylates the Ser7 of Pol II CTD.
(A) The schematic diagram (top left) shows the different subunits of TFIIK and the core TFIIH complex. Dot blot and ELISA of GST-CTD phosphorylated by TFIIH purified using TAP-tagged Kin28, Ccl1, Rad3 or Rad25.
(B) The diagram (top) illustrates the docking of ATP into the catalytic pocket of wild type kinase. Dot blot (bottom) of GST-CTD phosphorylated by Kin28 and its analog sensitive mutant Kin28as (Kin28-L83G) in the presence and absence of an inhibitor was probed with αSer5-P and αSer7-P antibodies.
(C) Dot blot of GST-CTD and its various mutants at positions 2, 5, and 7 phosphorylated by purified Kin28 and probed with αSer5-P and αSer7-P antibodies.
(D) The diagram (top) illustrates the ability of Kin28as to use of the ATP analog (N6-benzyl ATP) as a co-factor. Dot blot (bottom) of GST-CTD phosphorylated by Kin28 and Kin28as in presence and absence of the N6- benzyl ATP was probed with αSer5-P and αSer7-P antibodies.
TFIIH complex itself associates with numerous protein partners and even with the U1-snRNA (Esnault et al., 2008; Roeder, 2005; Kwek et al., 2002; Zurita and Merino, 2003). Hence, rather than Kin28, it is possible that an unrelated kinase that sub-stoichiometrically associates with TFIIH is responsible for the Ser7 modification. To address this concern we engineered the TAP-tagged Kin28 to accept a bulky analog of a kinase inhibitor (Figure S2). This bulky analog is incapable of docking into ATP binding pockets of unmodified kinases (Bishop et al., 2000; Bishop et al., 2001). In contrast, the engineered Kin28as (analog sensitized) is inhibited at micro-molar concentrations of the designed inhibitor, 1-NAPP1 (Kanin et al., 2007; Liu et al., 2004). The TAP-tagged Kin28as (Kin28-L83G) enzyme is unable to phosphorylate Ser5 or Ser7 in the presence of the inhibitor but is as active as the wild type kinase in the presence of ATP (Figure 2B). As expected, due to the inability of the inhibitor to dock in an unmodified ATP binding pocket, the activity of wild-type Kin28 is not detectably perturbed by the inhibitor. These results suggest that Kin28 directly phosphorylates Ser7. However, Ser7 phosphorylation might be dependent on prior phosphorylation of Ser5 by Kin28 (Chapman et al., 2007). We therefore examined the ability of Kin28 to phosphorylate recombinant CTD substrates wherein Ser2, 5 or 7 have been substituted by an alanine residue (Fig. 2C). The data show that an alanine at positions 2 or 7 does not detectably affect the ability of Kin28 to phosphorylate Ser5. In contrast, replacing Ser5 with an alanine dramatically reduces the ability of Kin28 to phosphorylate Ser7. In other words, inhibiting Ser5 phosphorylation by chemical inhibition could block subsequent phosphorylation of Ser7 by a different kinase. To directly test if Kin28 can phosphorylate both Ser5 and Ser7, we used N6-benzyl-ATP as a phospho-donor in kinase reactions with Kin28 or Kin28as (Figure 2D and Figure S2). This modified ATP docks into the expanded ATP binding pocket of Kin28as but is not used efficiently by unmodified kinases. The enlargement of the ATP binding site does not alter substrate specificity of the kinase (Liu et al., 1998; Bishop et al., 2000; Bishop et al., 2001; Liu et al., 2004). In this context, the appearance of Ser7-P mark with N6-benzyl-ATP therefore confirms that Kin28as phosphorylates both Ser7 and Ser5 residues of the CTD heptad (Figure 2D).
Chemical genomics evidence for Ser7 phosphorylation by Kin28 in vivo
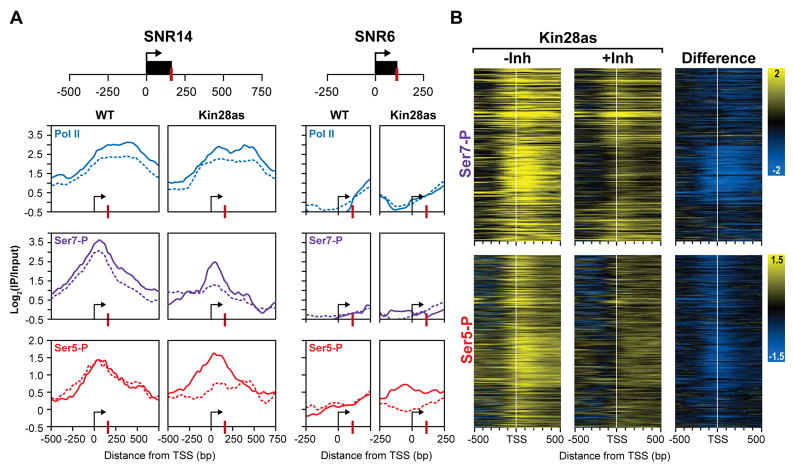
We examined the consequences of chemically inhibiting Kin28 on Ser7 phosphorylation patterns across the genome. The growth of cells bearing the Kin28as allele is greatly attenuated by micro-molar concentrations of the cell-permeable inhibitor (Figure S3) (Kanin et al., 2007; Liu et al., 2004). Chromatin immunoprecipitation (ChIP) assays show that within 20 minutes of inhibition, the Ser5-P marks of CTD are dramatically reduced in Kin28as strains (Kanin et al., 2007; Liu et al., 2004). Using high-resolution genome tiling microarrays, we examined the genome-wide profiles of Ser5-P and Ser7-P CTD modifications (Figure 3). We focused on the snRNA genes that code for spliceosomal transcripts because of the key role of Ser7 phosphorylation in the expression of these genes (Figure 3A). At all five snRNA genes the profiles of Ser7-P closely mirror those of Ser5-P (Figure 3 and Figure S4). SNR6 (U6), which is transcribed by Pol III, serves as negative control with background levels of Pol II and Ser5/7 phosphorylation profiles. Strikingly, inhibition of Kin28 eliminates both Ser7-P and Ser5-P peaks. A similar remodeling of the profiles is also seen at protein-coding genes (Figure 3B). We focused on protein-coding genes with uniformly robust Pol II occupancy across the entire open reading frame (Steinmetz et al., 2006). Of these 415 genes, 308 with well-defined transcription start sites were clustered based on the CTD modification profiles. Upon Kin28 inhibition, the attenuation of Ser7-P and Ser5-P profiles is clearly evident (Figure 3B). The results across the genome reflect this loss of Ser7-P phosphorylation at the promoter regions and are consistent with the ability of Kin28 to phosphorylate Ser7. The results strongly implicate Kin28 in the phosphorylation of Ser7 and Ser5 residues of the CTD in vivo.
Figure 3. In vivo inhibition of Kin28 affects the Ser7 phosphorylation pattern.
(A) ChIP-chip profiles for two snRNAs (remaining three are in the supplemental figures). The occupancy profiles of Pol II (blue), Ser7-P (purple) and Ser5-P (red) at SNR14 and SNR6 (control) are shown. Uninhibited profiles are shown as solid lines and profiles in chemically inhibited cells are shown as dashed lines. TSS and 3′ processing sites are marked by an arrow and a red bar respectively. All x-axes are shown as the distance in base pairs relative to the TSS and y-axes are shown on a log2 scale for ChIP-chip enrichment (IP/input).
(B) Heat maps of 308 protein-coding genes showing Ser7-P and Ser5-P occupancy profiles over a 1kb region centered over the TSS (white bar) for the Kin28as strain with and without inhibition. The change in profiles upon inhibition is shown on the right. Yellow profiles indicate enrichment and blue profiles indicate depletion of the phosphorylated CTD.
Human homolog, Cdk7 phosphorylates Ser 7 residue
TFIIH and its associated kinase are highly conserved from yeast to humans (Chapman et al., 2008; Coin and Egly, 1998; Egloff and Murphy, 2008; Zurita and Merino, 2003). To determine if the substrate specificity for Ser7 was conserved in the human ortholog we examined the ability of the Cdk7/cyclinH complex, as well as other purified Cdks, to phosphorylate Ser7. The results demonstrate that the human ortholog of Kin28 phosphorylates both Ser7 as well as the expected Ser5 residue of the CTD heptapeptide (Figure 4). Moreover, Cdk8/CycC like its yeast ortholog Srb10/Srb11 phosphorylates Ser5 but not Ser7. Thus, Kin28 and Cdk7 show identical substrate specificity for Ser5 and Ser7 of the CTD.
Figure 4. Mammalian Cdk7 phosphorylates the Ser7 of Pol II CTD.
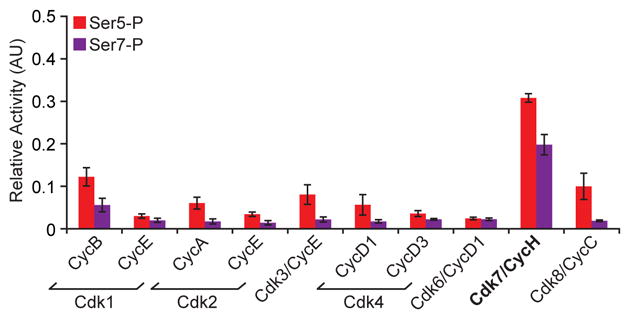
ELISA of a synthetic CTD peptide (four repeats) phosphorylated by purified mammalian kinases and their corresponding cyclins were probed with αSer5-P (red) and αSer7-P (purple) antibodies.
DISCUSSION
The identification of TFIIH-associated kinase as the Ser7 kinase is unexpected for multiple reasons. First, it belongs to the cyclin-dependent kinase (Cdk) family. Members of this family prefer Ser-Pro dipeptide cores in its substrates whereas Ser7 of the CTD is flanked by a tyrosine residue (Songyang et al., 1996). Second, Kin28/Cdk7 as a subunit of TFIIH associates with Pol II at gene promoters and not deep within transcribed regions where the Ser7-P mark was first found to be enriched (Chapman et al., 2007). Third, this kinase associates with most, if not all, Pol II transcribed genes and is not known to play a specific role in snRNA gene expression. Fourth, biochemical analyses suggest that TFIIH is not associated with Pol II and the transcriptional machinery at snRNA promoters (Hernandez, 2001). Finally, Kin28/Cdk7 is known to specifically phosphorylate Ser5 residues of the CTD and does not display an ability to phosphorylate the adjacent Ser2 residues appreciably.
In contrast to expectations, our data strongly implicate TFIIH in the placement of Ser7 marks at promoters of Pol II-transcribed genes and reveal the involvement of TFIIH in the transcription of snRNA in vivo. We anticipate that the Integrator complex (or its functional analog in budding yeast) is recruited, in part, by Ser7 phosphorylation at snRNA gene promoters. This recruitment is a consequence not only of Ser7-P marks, but also the promoter elements and the specialized pre-initiation complex that binds those elements. After promoter-escape, the RNA processing complex travels with the elongating polymerase, and in response to the 3′-end box at the end of the snRNA transcription unit, the processing complex associates with the nascent transcript and engages in co-transcriptional processing. At protein-coding genes, the promoter-bound factors facilitate the association of the CPSF complex (Dantonel et al., 1997; Glover-Cutter et al., 2007) or the Nrd1 complex (Vasiljeva et al., 2008; Gudipati et al., 2008) with Pol II. Each of these complexes, Integrator, Nrd1 or CPSF responds to different processing sequences within nascent transcripts.
Our data suggest that bivalent Ser5-P+Ser7-P marks are placed by the TFIIH-associated kinase during early stages of gene transcription. It is likely that this bivalent mark, rather than Ser5-P alone, is recognized by Bur1 or pTEFb/Cdk9, enzymes that act next on the CTD (Viladevall et al., 2009; Qiu et al., 2009). Furthermore, given the universality of the Ser7-P mark, we propose that information relayed by promoter-specific complexes, differential CTD modifications, and RNA-borne signals is integrated to engage the relevant protein complex to process different classes of transcripts. These observations force a re-evaluation of the basic recognition determinants and mechanisms used by partnering proteins to decipher the CTD code.
EXPERIMENTAL PROCEDURES
Strains and kinases
TAP-tagged Kin28as (Kin28-L83G) was a gift from Prof. Steven Hahn (Seattle, Washington). Mammalian kinases were purchased from Proqinase/Biomol. TAP tagged yeast kinases were purchased from Open biosystems and purified as described previously (Liu et al., 2004; Puig et al., 2001).
In vitro kinase assay detected by dot blot
The substrate, GST-CTD3 (three repeats of YSPTSPS attached to GST), GST-CTD16, GST-CTD4 (and its respective alanine mutants, 2A,5A and 7A) purifications and enzymatic assays were carried out as described previously (Ansari et al., 2005; Patturajan et al., 1998). For inhibition assays, the kinase was pre-incubated for 5 minutes with an inhibitor (6μM of 1-NA-PP1) or synthetic analog of ATP (0.6mM of N6-benzyl ATP) prior to the reaction. For the assay, 1:200 dilution of primary rat IgG (αSer2-P or αSer5-P or αSer7-P) and 1:10000 dilution of secondary HRP anti-rat IgG (Southern Biotech) antibodies were used.
In vitro kinase assay detected by ELISA
Recombinant kinase (100ng) was incubated with excess CTD-peptide linked to 96-well plates and phosphorylation of CTD was quantitated via ELISA experiments after incubation with αSer2-P, αSer5-P and αSer7-P specific antibodies (for details see supplementary materials).
ChIP-chip analysis
The Chromain immunoprecipitation (ChIP) followed by identification on genome-tiled microarray (chip) analysis performed as described previously (Kanin et al., 2007; Steinmetz et al., 2006). In brief, for ChIP the yeast whole cell extract was incubated over night at 4°C with the either αSer7-P (4E12) or αSer5-P (H14) or αRpb3 (Neoclone) antibodies. Amplified DNA was hybridized to custom designed NimbleGen microarrays containing 379,876 features tiling the yeast genome (Tietjan and Ansari, unpublished). Microarray data was analyzed using standard techniques as described previously (Steinmetz et al., 2006).
Supplementary Material
Acknowledgments
We are grateful to J. Rodriguez-Molina and S. Boeing for helpful discussions, A. Flatley and E. Kremmer for technical support and to A. Gasch, A. Kumar, J. Corden, K. Shokat, S. Hahn, P. Fisher and D. Barilla for strains and plasmids. This work was funded by the NSF-CAREER (MCB07147), the W.M. Keck, Shaw Scholar and Vilas associate awards to A. Z. Ansari and grant from the Deutsche Forschungsgemeinschaft (SFB/TR5 and SFB684) to D. Eick. M. Heidemann was funded by Boehringer Ingelheim Fonds travel grant and J. Tietjen was supported by the NHGRI training grant of the Genome Sciences Training program (5T32HG002760).
Footnotes
SUPPLEMENTAL DATA
The supplemental data include four figures, figure legends, supplemental references and detailed experimental procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA Polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Ansari AZ, Ogirala A, Ptashne M. Transcriptional activating regions target attached substrates to a cyclin-dependent kinase. Proc Natl Acad Sci USA. 2005;102:2346–2349. doi: 10.1073/pnas.0409671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA Polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Buzko O, Shokat KM. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–172. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Coin F, Egly JM. Ten years of TFIIH. Cold Spring Harb Symp Quant Biol. 1998;63:105–110. doi: 10.1101/sqb.1998.63.105. [DOI] [PubMed] [Google Scholar]
- Corden JL. Transcription: seven ups the code. Science. 2007;318:1735–1736. doi: 10.1126/science.1152624. [DOI] [PubMed] [Google Scholar]
- Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA Polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1110. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Feaver WJ, Henry NL, Wang Z, Wu X, Svejstrup JQ, Bushnell DA, Friedberg EC, Kornberg RD. Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH. J Biol Chem. 1997;272:19319–19327. doi: 10.1074/jbc.272.31.19319. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2007;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- Guidi BW, Bjornsdottir G, Hopkins DC, Lacomis L, Erdjument-Bromage H, Tempst P, Myers LC. Mutual targeting of Mediator and the TFIIH kinase Kin28. J Biol Chem. 2004;279:29114–29120. doi: 10.1074/jbc.M404426200. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J Biol Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme–associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci USA. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Shokat KM. Features of Selective Kinase Inhibitors. Chem Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas BO, Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast Pre-mRNA 3′ end processing factors with RNA Polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shah K, Yang F, Witucki L, Shokat K. Engineering Src family protein kinases with unnatural nucleotide specificity. Chem Biol. 1998;5:91–101. doi: 10.1016/s1074-5521(98)90143-0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol Cell Biol. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Kung C, Shokat KM, Tansey WP. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and co-transcriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Myers LC, Kornberg RD. Mediator of transcription regulation. Annu Rev Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent kinase 7: at the cross-roads of transcription, DNA repair and cell cycle control. Curr Opin Cell Biol. 1996;8:312–317. doi: 10.1016/s0955-0674(96)80003-2. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA Polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Genes & Signals. Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by kin28 enhances Bur1/Bur2 recruitment and ser2 CTD phosphorylation near promoters. Mol Cell. 2009;33:752–7662. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish JA, Hahn S, Lu Y, Yi EC, Li XJ, Eng J, Aebersold R. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat Genet. 2004;36:707–713. doi: 10.1038/ng1385. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Niedbala D, Wood K, Ptashne M. GAL4 is phosphorylated as a consequence of transcriptional activation. Proc Natl Acad Sci USA. 1991;88:10510–10514. doi: 10.1073/pnas.88.23.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CW, Kuehner JN, Panbehi B, Ansari AZ, Brow D. Genome-wide distribution of yeast RNA Polymerase II and Its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ, Feaver WJ, LaPointe J, Kornberg RD. RNA polymerase transcription factor IIH holoenzyme from yeast. J Biol Chem. 1994;269:28044–28048. [PubMed] [Google Scholar]
- Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different c-terminal domain kinases. J Biol Chem. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3- Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladevall L, St Amour CV, Rosebrock A, Schneider S, Zhang C, Allen JJ, Shokat KM, Schwer B, Leatherwood JK, Fisher RP. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol Cell. 2009;33:738–751. doi: 10.1016/j.molcel.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Fujimoto H, Watanabe T, Maekawa T, Masutani C, Hanaoka F, Ohkuma Y. Modulation of TFIIH-associated kinase activity by complex formation and its relationship with CTD phosphorylation of RNA polymerase II. Genes Cells. 2000;5:407–423. doi: 10.1046/j.1365-2443.2000.00336.x. [DOI] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- Zurita M, Merino C. The transcriptional complexity of the TFIIH complex. Trends Genet. 2003;19:578–584. doi: 10.1016/j.tig.2003.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.