Summary
The transcriptional control of CNS myelin gene expression is poorly understood. Here we identify gene model 98, which we have named Myelin-gene Regulatory Factor (MRF), as a transcriptional regulator required for CNS myelination. Within the CNS, MRF is specifically expressed by postmitotic oligodendrocytes. MRF is a nuclear protein containing an evolutionarily conserved DNA binding domain homologous to a yeast transcription factor. Knockdown of MRF in oligodendrocytes by RNA interference prevents expression of most CNS myelin genes; conversely, overexpression of MRF within cultured oligodendrocyte progenitors or the chick spinal cord promotes expression of myelin genes. In mice lacking MRF within the oligodendrocyte lineage, pre-myelinating oligodendrocytes are generated but display severe deficits in myelin gene expression and fail to myelinate. These mice display severe neurological abnormalities, and die due to seizures during the third postnatal week. These findings establish MRF as a critical transcriptional regulator essential for oligodendrocyte maturation and CNS myelination.
Introduction
Myelination in vertebrates has evolved to insulate axons and promote rapid, propagation of action potentials. Within the CNS, this function is carried out by oligodendrocytes (OLs). Developmentally, committed OL progenitor cells (OPCs) arise from stereotyped germinal regions of the CNS, proliferate and migrate throughout the CNS before differentiating into postmitotic pre-myelinating OLs. These pre-myelinating OLs subsequently either up-regulate myelin genes and ensheath adjacent axons, or undergo apoptosis. Several transcription factors are required for the specification of OPCs or their subsequent differentiation into postmitotic OLs, such as Nkx2.2, Sox10, and the bHLH proteins Olig1/2. (Fu et al., 2002; Kuhlbrodt et al., 1998; Lu et al., 2000; Qi et al., 2001; Stolt et al., 2002; Zhou et al., 2001). Among these, Olig2 is the only gene necessary for the OL lineage specification, with Olig2 -/- mice showing a loss of PDGFRα positive OPCs within most of the CNS (Lu et al., 2002; Zhou and Anderson, 2002). In contrast, mutants for Olig1, Sox10, and Nkx2.2 have only mild defects in OPC generation, but show impaired oligodendrocyte differentiation and myelin gene expression (Qi et al., 2001; Stolt et al., 2002; Xin et al., 2005). More recently Yin Yang1 (YY1) has also been demonstrated to play an important role in oligodendrocyte differentiation (He et al., 2007).
Few transcription factors function specifically at the postmitotic stage of the OL lineage, and those that do (e.g. Nkx6.2) (Southwood et al., 2004), do not appear to be necessary for most aspects of myelination. It is thus unclear if the known oligodendrocyte lineage transcription factors play a dual role in oligodendrocyte differentiation and myelination, or instead are required only for oligodendrocyte differentiation with other yet to be identified transcription factors playing essential roles in inducing myelin gene expression and the transition to a myelinating OL. This situation contrasts with that of PNS myelination, where the transcription factor Krox-20 expressed in myelinating Schwann cells, is both sufficient and necessary to induce myelin gene expression and myelination in these cells (Nagarajan et al., 2001; Topilko et al., 1994). As such, the nature of transcriptional control of the final stages of OL maturation including myelin gene expression remains unclear.
In a recent study, we determined the transcriptomes of acutely isolated astrocytes, neurons, and OLs from the postnatal mouse forebrain (Cahoy et al., 2008). We identified gene model 98 as a gene expressed within the CNS only by postmitotic OLs. Here, we demonstrate that gene model 98, which we have named Myelin-gene Regulatory Factor (MRF), is necessary for the generation of mature, myelin gene expressing OLs within the CNS. RNAi-mediated knockdown of MRF in cultured OPCs does not affect their exit from the cell cycle and down-regulation of OPC markers, but markedly reduces the expression of the majority of myelin genes. Conversely, overexpression of MRF within cultured OPCs is sufficient to induce myelin gene expression and it can promote myelin gene expression in the developing chick spinal cord. Moreover, we show that mice in which MRF function is disrupted within the OL lineage show severe dysmyelination. Although pre-myelinating OLs are generated in these mice, these pre-myelinating OLs fail to up-regulate myelin gene expression and ensheath axons, and instead undergo apoptosis. These results establish MRF as a transcriptional regulator vital for the generation of myelinating OLs, with an analogous role in the CNS to that of Krox-20 in the PNS.
Results
GM98/MRF is an OL specific nuclear protein within the CNS
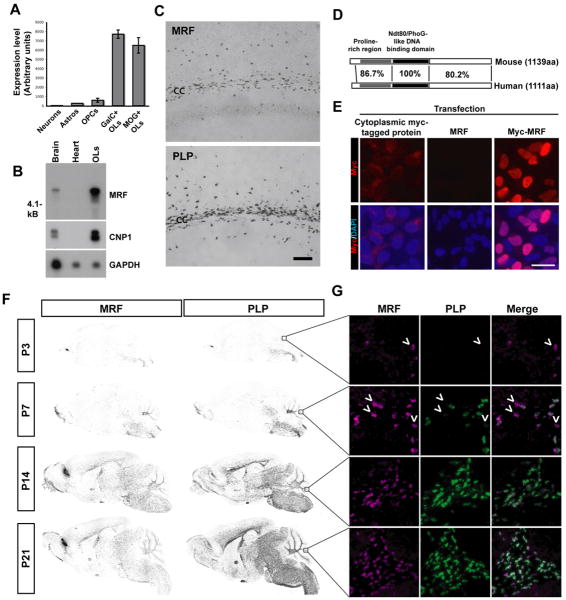
We used an immunopanning/FACS cell purification approach combined with gene profiling in order to identify cell type-specific genes within the mouse CNS (Cahoy et al., 2008). Amongst the genes displaying OL specific expression was gene model 98. The Affymetrix probe set for this gene was absent in samples derived from astrocytes and neurons, present at very low levels in OPCs, but up-regulated in newly differentiated (GC+, MOG-) and mature (MOG+) OLs (Figure 1A). Northern blotting confirmed that MRF was present in brain and OL samples as a ∼5.5 kb transcript, consistent with database predictions (Figure 1B). In situ hybridization showed that MRF displayed the same distribution as the established OL marker PLP, with expression largely confined to white matter tracts within the CNS (Figure 1C). The expression of MRF by myelinating cells was CNS-specific, with no expression detected in the PNS (Figure S1).
Figure 1. GM98/MRF is a nuclear protein specifically expressed within the CNS by postmitotic OLS.
(A) Affymetrix expression levels of GM98/MRF in acutely isolated cells from the mouse CNS (probe set 1439506_at). Expression was considered present by GCOS software for the OPC, GalC+ OL and MOG+ OL samples. (B) Northern blot confirmed expression of MRF as a single ∼5.5Kb transcript within P20 brain and cultured OLs, but not heart tissue, similar to OL marker CNP. GAPDH controls for mRNA loading. (C) In situ hybridization for PLP and MRF expression within the lateral corpus callosum (cc) and scattered cells in the overlying cortex. Scale bar=100μm (D) Schematic of the protein domains of GM98/MRF and the human orthologue C11Orf9, showing percent identity in each region. (E) Subcellular localization of myc-tagged MRF within HEK cells. Staining of Myc-MRF-transfected HEK cells with anti-myc indicates a predominantly nuclear localization of the protein, with co-localization with the nuclear counterstain DAPI in transfected cells. Non myc-tagged MRF and a myc-tagged protein displaying a cytoplasmic localization provide a negative control for the staining and a comparison, respectively. Scale bar=50um. (F) In situ hybridization for MRF and PLP in P3-21 sagittal brain sections showing their developmental expression profile. (G) Double fluorescent in situ hybridization for MRF and PLP in the white matter of the developing cerebellum (P3-21); arrowheads indicate MRF+/PLP- cells.
Sequencing of cDNA derived from cultured mouse OLs predicted a protein of 1139 amino acids, with an overall identity of 88.6% to the human gene C11Orf9 (Figure S2). The protein contained a predicted DNA binding region homologous to the yeast transcription factor Ndt80 (Montano et al., 2002); within this region MRF and C11Orf9 shared 100% sequence identity (Figure 1D). C11Orf9 has been postulated to be a transmembrane protein based on the presence of hydrophobic regions in the C-terminal region (Stohr et al., 2000). In order to assess the subcellular localization of MRF, we expressed a myc-tagged version of the protein and probed with anti-myc; the fusion protein had a nuclear localization when expressed in both HEK cells (Figure 1E) and cultured OLs (data not shown). These data provide evidence that MRF is a nuclear protein displaying OL-specific expression within the CNS.
In order to assess whether MRF displayed a developmental expression profile consistent with a role in myelination we performed in situ hybridization on brain sections of mice at postnatal days 3 to 21. At P3, the expression of MRF was restricted to cells in the hindbrain and cerebellum, subsequently spreading rostrally throughout the white matter tracks over the first two weeks postnatal, mirroring the expression of PLP (Figure 1F). Dual fluorescent in situ hybridization for MRF and PLP revealed that within the cerebellum at early ages (P3 and P7) MRF+, PLP- or weakly labeled cells could often be identified (arrowheads, Figure 1G). At later ages (P14 and P21) however, the two genes showed near 100% co-localization. This suggests that the expression of MRF may slightly precede that of myelin genes such as PLP within differentiating oligodendrocytes. Similarly, Northern blots revealed that MRF transcript was absent in the P1 brain but steadily increased from P3 to P14, peaking in expression before PLP transcript (Figure S1),. Within the embryonic spinal cord few MRF+ cells were detected by E17, in a pattern similar to PLP and MBP (Figure S1). These results confirmed that MRF has an expression pattern consistent with a potential role in myelination.
Knockdown of MRF in cultured OLs blocks myelin gene expression
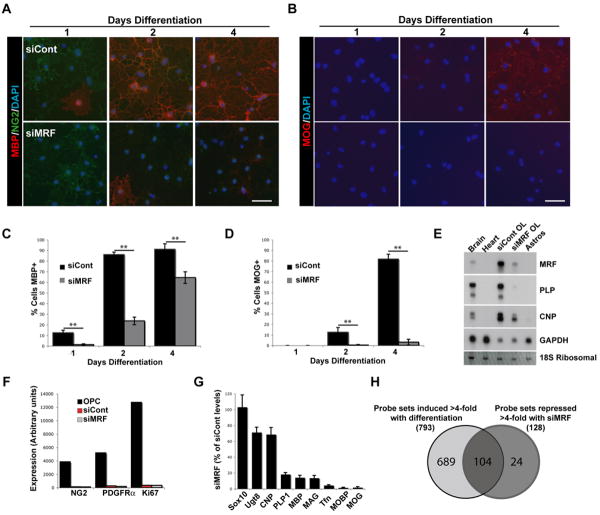
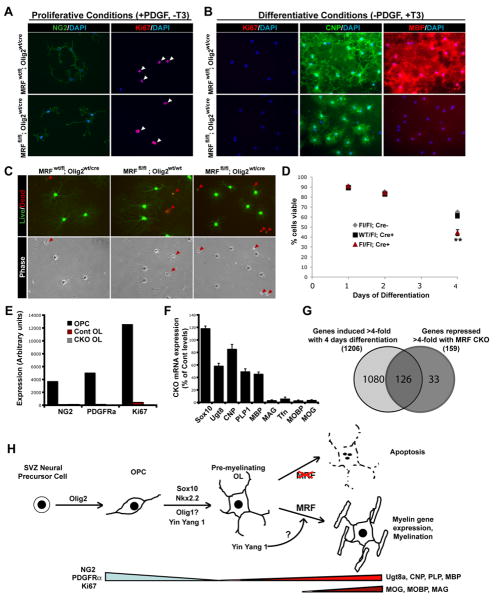
To establish whether MRF may have a role in OL differentiation or myelin gene expression, we transfected cultured mouse OPCs with control non-targeting siRNA pool (siCont) or pooled siRNAs against MRF (siMRF). Knockdown of MRF was confirmed by Northern blot and RT-PCR (Figure 2E and data not shown). siMRF-transfected OPCs displayed no obvious deficiencies when cultured in the presence of PDGF, that promotes the growth of progenitor cells (data not shown). When transfected OPCs were instead plated into media lacking PDGF and with T3 to induce their differentiation into OLs (Dugas et al., 2006), the siMRF-transfected cells showed both delayed and reduced levels of MBP expression relative to siCont-transfected cells and failed to express MOG even after 4 days of differentiation, by which time the majority of siCont-transfected cells expressed MOG (Figure 2A-D). This failure of siMRF-transfected cells to express MBP and MOG was not due to a failure of differentiation per se or a diversion to an astrocyte fate; the siMRF-transfected cells still down-regulated the OPC marker NG2, did not express the astrocyte marker GFAP, were positive for the OL marker GalC and took on the multipolar morphology characteristic of OLs, albeit with far less extensive membrane sheets than siCont-transfected cells (Figure S3C). This effect was unlikely to be mediated by off-target siRNA effects, as it was observed with 2 of 4 independent non-overlapping siRNA constructs targeting MRF (Figure S3E). Interestingly, after 2 days of differentiation the MRF knockdown resulted in an increased apoptosis relative to siCont-transfected cells (Figure S3). Although mouse OLs have a limited viability in culture after differentiation with few cells surviving more than a week, the rate of apoptosis was accelerated in cells transfected with siMRF compared to siCont. Furthermore, siMRF-transfected cells lost expression of other myelin genes CNP and PLP (Figure 2D).
Figure 2. Knockdown of MRF in OLs blocks myelin gene expression.
(A-B) Representative images of OL cultures transfected with siCont or siMRF and differentiated for 1, 2 and 4 days stained with NG2 and MBP (A) or MOG (B). Scale bars=50um. (C-D) Quantification of the proportion of siCont and siMRF transfected OLs expressing MBP (C) and MOG (D) at 1, 2 and 4 days differentiation. **P<0.01. (E) Northern blot analysis of gene expression in siCont and siMRF transfected OL cultures at 2 days differentiation. RNA from brain, heart and cultured astrocyte samples are provided for positive and negative controls, respectively. (F-G) Results of GeneChip analysis of gene expression in OPCs and OLs transfected with siCont or siMRF as OPCs then cultured for 2 days in differentiating conditions. (F) Down-regulation of selected OPC markers NG2, PDGFRα and Ki67 during differentiation is not affected by MRF knockdown. (G) Expression of OL pan-OL lineage marker Sox10, early-OL markers (Ugt8, CNP, PLP and MBP) and late-OL markers (MAG, transferrin/Tfn, MOBP and MOG) in cells transfected with siMRF expressed as a mean percentage of siCont transfected OL values ±SEM. Results are averages of 3 independent experiments. (H) Venn diagram showing overlap of genes induced >4-fold with differentiation and those repressed >4-fold by transfection with siMRF.
To further characterize the changes in gene expression resulting from inhibition of MRF expression, we performed GeneChip analysis between OL cultures differentiated for 2 days following transfection with siCont or siMRF and OPC cultures that served as a measure of baseline gene expression prior to differentiation. The GeneChip analysis confirmed that upon differentiation both siCont- and siMRF-transfected cultures completely down-regulated OPC markers (e.g NG2, PDGFRα and Ki67) (Figure 2F). In addition, siMRF-transfected cultures failed to induce most myelin genes, with the expression of late phase genes such as MAG and MOBP being entirely blocked (the 50 most repressed genes by siMRF are listed in TableS1) relative to the siCont cultures. Most early OL genes such as MBP and PLP were also inhibited, though to a lesser extent, and pan-OL lineage genes such as Sox10, Olig1, and Olig2 were unaffected by the MRF knockdown (Figure 2G). Interestingly, when the genes repressed 4-fold or more by the MRF knockdown were compared to the genes induced during OL differentiation (between OPC and siCont samples), the majority of genes dependent on MRF expression were genes usually induced specifically in OLs during their differentiation (104 of 128 probe sets). MRF-regulated genes represented only a subset of genes that are normally induced during OL differentiation (104 of 793 probe sets), while the expression of other OL-specific genes such as Cldn11/OSP or cell cycle regulators was largely unaffected (Figure 2H). These findings indicate that MRF is required for the induction of most CNS myelin genes and many, albeit not all, OL-specific genes involved in oligodendrocyte maturation.
Forced MRF expression induces myelin gene expression in vitro and in vivo
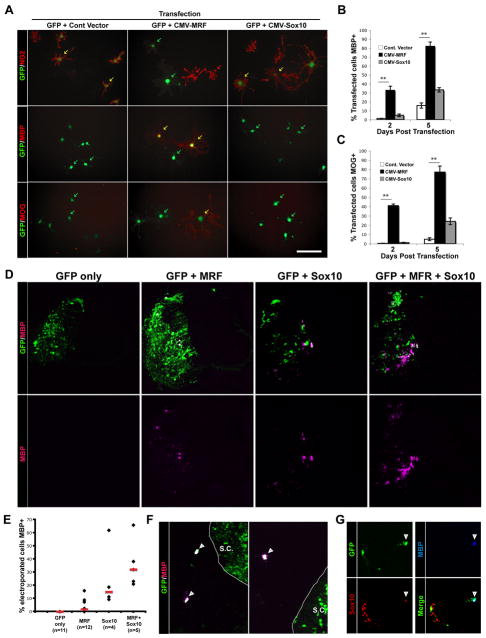
We next asked whether the expression of MRF was sufficient to induce the expression of myelin genes. We transfected mouse OPCs with a GFP expression plasmid together with either a plasmid encoding MRF or an empty control vector, and plated them in media containing PDGF to maintain their growth as OPCs. Cells transfected with the control vector (identified by GFP expression) remained NG2+, MBP/MOG- OPCs (Figure 3A). In contrast, MRF-transfected cells down-regulated NG2, up-regulated MBP and MOG, and acquired a flattened morphology (Figure S4). This effect was evident by 2 days post transfection and more pronounced at 5 days post transfection, by which time approximately 80% of the MRF-transfected cells expressed MBP and MOG (Figure 3B-C). In this assay, the induction of MBP and MOG expression by MRF was considerably more pronounced than that seen with Sox10, a gene that promotes MBP expression and OL differentiation (Li et al., 2007; Stolt et al., 2002).
Figure 3. Forced MRF expression induces myelin gene expression in vitro and in vivo.
(A) Representative images of mouse OPCs co-transfected with eGFP and control (empty) vector, or vectors encoding MRF or Sox10 and then cultured for 2 days post transfection in proliferative conditions. Cells are stained for the OPC marker NG2 or OL markers MBP and MOG. Scale bar=50um. (B-C) Quantification of the proportion of transfected cells in each condition expressing MBP (B) and MOG (C) at 2 and 5 days post transfection. **P<0.01, unpaired t-test. Values are means± SEM. (D) Representative spinal cord sections of chicken embryos electroporated with expression constructs for MRF, Sox10 or both genes at E3 (stage 12) along with a GFP construct and allowed to develop until E7. Sections are stained with anti-MBP, with GFP shown to identify electroporated cells. (E) Quantification of the proportion of electroporated cells expressing MBP for each condition; shown are values for individual animals and median values for each condition (red bars). (F-G) Examples of MRF-electroporated neural crest cells ectopically expressing MBP (F); (G) shows triple labeling for GFP, MBP and Sox10. The arrowhead indicates a triple-labeled cell, a GFP/Sox10+, MBP- cell can be seen to the left of the image.
To assess whether forced expression of MRF could also induce myelin gene expression in vivo, we electroporated HH stage 12-14 chick spinal cords with either a GFP plasmid or GFP and MRF plasmids, and analyzed the embryos 4 days later at HH stage 27-28, prior to the normal differentiation of OLs in the chick spinal cord (Zhou et al., 2001). Immunostaining revealed scattered MBP+ cells on the electroporated side of the spinal cord; such cells were never observed on the non-electroporated side or in GFP-only electroprated controls (Figure 3D-E). Misexpression of MRF in the chick spinal cord induced MBP expression only in a small subset of GFP+ cells (ranging from 0 to 15.9% of GFP+ cells in various animals, median=1.6%), a phenotype that was different to that seen in cultured mouse OPC, where forced MRF expression induced myelin gene expression in a high percentage of cells. Interestingly, a high percentage (31.7%) of neural crest-derived cells that were electroporated with MRF (GFP+ and Sox10+ cells outside the spinal cord), ceased to migrate shortly after leaving the dorsal spinal cord and up-regulated MBP expression (Figure 3F-G). MBP expression in GFP+ neural crest cells was never observed in GFP-electroporated embryos. These results suggested that these cells expressed transcription factors that in combination with MRF induced MBP expression.
To investigate this possibility, we electroporated Sox10, a neural crest marker, or Sox10 and MRF in chick spinal cord and assayed MBP expression after 4 days. Sox10 electroporation induced a larger number of MBP+ cells compared to MRF electroporation (ranging from 9.3 to 61.9% of GFP+ cells in different animals, median=14.9%). However, co-electroporation of MRF and Sox10 increased the number of MBP+ cells compared to Sox10 alone (ranging from 20.9 to 65.8% of GFP+ cells in different animals, medium=32.0%), but this difference was not statistically significant due to a high variability in MBP induction between animals. These results indicate that MRF is able to induce myelin gene expression in vivo as well as in cultured OPC cultures, but may cooperate with other transcription factors (e.g. Sox10) to promote myelination in vivo.
MRF expression is necessary for CNS myelination
To assess the role of MRF in mammalian CNS myelination we generated an MRF allele where exon 8 was flanked by loxP sites (see Figure S5A for a description of the targeting vector). Exon 8 codes part of the predicted DNA binding region of MRF; as such Cre-mediated excision of exon 8 was predicted to result in a truncated protein lacking the DNA binding region and subsequent C-terminal region of the protein due to a frame-shift (Figure S5B). Initial experiments overexpressing this predicted post-excision protein in OPCs confirmed that it had no ability to induce myelin gene expression, unlike the full-length protein (data not shown). We crossed the MRF loxP-flanked exon 8 (MRFfl/fl) mice with an Olig2-Cre knock-in mouse line (Schuller et al., 2008) to effect recombination in glia including the OL lineage from an early developmental stage. We confirmed successful deletion of MRF exon 8 by RT-PCR of cDNA samples from spinal cords and cultured OLs from MRF CKOs (Figure S9).
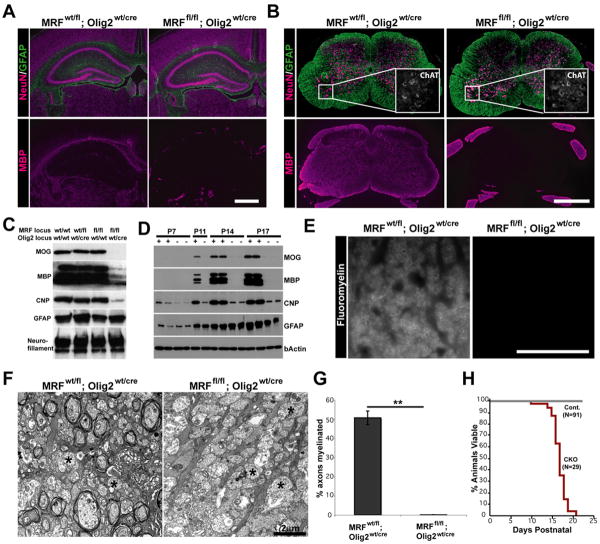
MRF conditional knockout (CKO) mice (MRFfl/fl; Olig2wt/cre) were born at Mendelian frequencies and were not overtly distinguishable from control littermates (MRFwt/fl; Olig2wt/cre and MRFfl/fl; Olig2wt/wt) for the first 10 days of life. From P11 however, CKOs developed severe tremors and ataxia (Movie S1), subsequently also developing seizures and dying during the third postnatal week (Figure 4). Immunohistochemical analysis of the brains of CKO mice at P13 (Figure 4A) indicated that NeuN and GFAP staining appeared normal and the gross CNS cytoarchitecture was not affected. In contrast, there was a severe loss of staining for MBP within the brain, with only occasional MBP+ oligodendrocytes seen within white matter tracts, compared to near ubiquitous MBP staining in the white matter tracts of control mice. CKO spinal cords also showed a severe loss of MBP immunostaining (Figure 4B), although the spinal roots (myelinated by Schwann cells) still stained intensely for MBP. As with the brains, NeuN and GFAP staining were unaffected by the conditional deletion of MRF. Similarly, ChAT immunostaining indicated that the motor neuron population was intact in CKOs, indicating that disruption of the MRF gene does not directly affect motor neurons (MNs are derived from Olig2+ progenitors and should undergo Olig2-Cre mediated recombination). At higher magnification some faintly MBP+ cells with the morphology of pre-myelinating OLs could be seen in the brains and spinal cords (Figure S6 and S7), suggesting that the early stages of OL differentiation did occur in the absence of MRF expression.
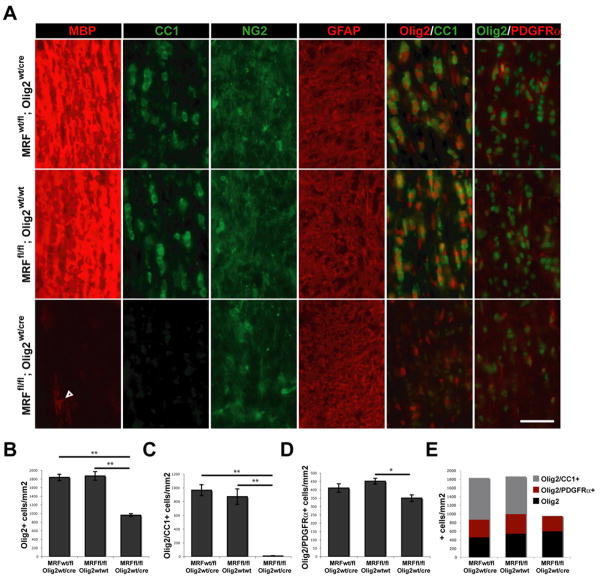
Figure 4. MRF CKOs display CNS dysmyelination.
(A-B) Representative images of the hippocampus, corpus callosum and overlying cortex (A) and spinal cord (B) of control (MRFwt/fl; Olig2wt/Cre) and MRF CKO (MRFfl/fl; Olig2wt/Cre) mice stained with MBP, NeuN and GFAP at P13. ChAT staining of spinal cord motor neurons is shown in the inserts of (B). (C) Western blot analysis of CNP, MBP, MOG, GFAP and Neurofillament expression in the spinal cords of MRF control and CKO mice at P13. (D) Developmental time-course of myelin gene expression in the brains of MRF control (+) and CKO (-) mice with CKOs showing severe deficit in MBP and MOG expression. (E) Representative images of Fluoromyelin staining of the spinal cord lateral column white matter in a control (MRFwt/fl; Olig2wt/Cre) and CKO (MRFfl/fl; Olig2wt/cre) mouse at P13. (F) Representative electron micrograph images of control and CKO optic nerves at P13. Control nerves show a significant amount of myelination in progress. In contrast, essentially all axons in the CKOs lack myelin ensheathment (examples denoted by *). Proportions of axons myelinated for control and CKOs are quantified in (G). (H) The lifespan of MRF CKOs is essentially limited to the third postnatal week. Scale bars: A and B=1mm. E=50um. E=2um. **P<0.01
We confirmed loss of myelin protein expression in the CKO by Western blot of P13 spinal cord lysates, which demonstrated substantially less MBP and CNP expression and a complete loss of MOG expression in CKOs relative to control genotypes (Figure 4C). Astrocyte and neuron proteins (GFAP and neurofilament, respectively) were not affected in the CKO. To confirm a loss of myelin proteins at other stages of development we harvested brains from CKOs and controls from P7 (at which stage myelination in the brain is just beginning) to P17 (the latest the CKOs could be reliably studied due to their death). The brains of control mice showed a substantial up-regulation of the proteins MBP, CNP and MOG during this time; at all time points MBP and MOG were near un-detectable in the CKOs (Figure 4D), although some MBP expression was seen at very long exposures (data not shown).
Fluoromyelin staining within CNS white matter tracts of CKOs revealed a complete loss of myelination in CKO animals (Figure 4E). In contrast, peripheral nerves were equivalently myelinated in both CKOs and controls as expected, since Schwann cells express neither MRF nor Olig2. We confirmed the loss of myelination by electron microscopy in the optic nerves of CKOs; whereas control optic nerves were actively myelinating by P13 with 50.7±3.5% of axons having two or more myelin wraps, virtually no myelination was observed in the optic nerves of CKO littermates, and only 0.07±0.07% of axons were ensheathed (P<0.001, Figure 4F and G). Importantly, we saw an equivalent phenotype (tremors, seizures, loss of myelin gene expression and myelination and death in the third postnatal week) when we crossed MRFfl mice with mice expressing Cre recombinase behind the CNP promoter (Lappe-Siefke et al., 2003), confirming that the CKO phenotype was not dependent on the Olig2 heterozygous background caused by insertion of the Cre gene (Figure S6).
OLs differentiate in MRF CKO mice but then undergo apoptosis
To determine which populations of the OL lineage were affected by MRF, we assessed the densities of OPCs and OLs in P13 CKO and control littermate optic nerve sections (Figure 5A). We found a ∼45% reduction in the density of Olig2+ nuclei within the optic nerves of CKOs relative to control littermates; this reduction was accounted for by the complete loss of CC1+ cells (a marker of postmitotic OLs) in CKO mice (Figure 5B, C and E). In contrast, the density of Olig2+ and PDGFRα+ cells (OPCs) was only modestly reduced (18.7%) in CKOs relative to MRFfl/fl; Olig2wt/wt mice (t-test P<0.05, but not significantly different from MRFwt/fl; Olig2wt/cre controls, Figure 5D and E). Therefore, the OPC stage was minimally affected in CKOs, an observation supported also by normal NG2 staining. Similarly, the density of Olig2+ cells negative for both CC1 and PDGFRα (presumably Olig2 expressing astrocytes) was similar between genotypes (Figure 4E). Although MBP staining was nearly absent in the CKO optic nerves a small number of weakly MBP+ cells were present, suggesting that at least some postmitotic OLs were generated in the CKO mice. The density of these faintly MBP+ cells (35±4 cells/mm2) was only ∼4% of the density of mature OLs in the control mice, and they had the morphology of pre-myelinating OLs, with thin, non-myelinating processes aligned with the axons.
Figure 5. MRF CKOs display a loss of mature OLs.
(A) Immunostaining for MBP, CC1, NG2, GFAP and Olig2 co-stained with CC1 and PDGFRα within the optic nerves of control (MRFwt/fl; Olig2wt/cre and MRFfl/fl; Olig2wt/wt) and MRF CKO (MRFfl/fl; Olig2wt/cre) mice at P13. Scale bar=50um. (B) Quantification of the density of Olig2 immunopositive nuclei within the optic nerves. (C) Quantification of the density of Olig2+/CC1+ double-immunopositive OLs within the optic nerves. (D) Quantification of the density of Olig2+/PDGFRα+ double-immunopositive OPCs within the optic nerves. All results are expressed as means ±SEM, n=4-5 per genotype. *P<0.05, **P<0.01. (E) Densities of Olig2 immunopositive cells within the optic nerves of each genotype broken down into Olig2+ cells also positive for either CC1 (OLs), PDGFRα (OPCs) or neither marker.
To assess whether myelin or mature OLs were present in the CKOs at earlier ages we examined sagittal sections of the P7 CKO and control brains for MBP, CC1 and PDGFRα expression (Figure S7). The density of OPCs was equivalent in control and CKO mice in both areas studied at P7. Control littermates displayed a caudal to rostral gradient of myelination, with substantial MBP expression in the hindbrain and cerebellum and scattered MBP+ cells in the more rostral white matter tracts. In contrast, only scattered faintly MBP+ cells were seen in the CKOs and there was an almost complete absence of MBP or CC1+ cells in the hindbrain (P<0.01). Control mice displayed a mix of pre-myelinating MBP+ OLs and myelinating OLs within the corpus callosum at P7 when myelination is proceeding (Fig S7E-F). In contrast, only pre-myelinating MBP+ but no myelinating OLs were observed in the corpus callosum of CKO mice, indicating that the absence of MRF expression caused a block in the transition from a pre-myelinating OL to a mature, myelinating OL.
During development, approximately 50% of the postmitotic OLs generated in the CNS undergo apoptosis (Barres et al., 1992; Trapp et al., 1997). The extreme paucity of myelinating OLs seen in CKO mice at P13 combined with the presence of significant numbers of pre-myelinating OLs at P7 suggested that in the absence of MRF OLs may undergo apoptosis. We observed that many weak MBP+ pre-myelinating OLs in the CKOs displayed the characteristic morphology of apoptotic OLs, with fragmented nuclei and blebbed processes. Staining of optic nerves from P10 CKO and control mice with an antibody against activated caspase-3, a marker of apoptosis, confirmed a significant ∼2-fold increase in the density of apoptotic cells in the CKO mice (P<0.05, Figure S8). These data indicate that postmitotic OLs are generated in the MRF CKO mice but then undergo apoptosis.
Dysmyelination in MRF CKOs is cell autonomous and not solely due to cell death
It has been proposed that some pre-myelinating OLs undergo apoptosis because they fail to receive axonal support. If MRF-deficit OLs are unable to upregulate proteins required for interactions with axons, this may explain their apoptosis in vivo. In order to determine if these is the case for MRF-deficient cells, we cultured highly purified OPCs isolated from P7 control and CKO mice. In the presence of PDGF both control and CKO OPCs proliferated as NG2 and Ki67+ cells, confirming MRF is not required for the OPC phase of the OL lineage (Figure 6A). Upon withdrawal of PDGF, both control and CKO cells down-regulated Ki67, took on the morphology of OLs, and expressed CNP (Figure 6B). However, CKO cells extended considerably less membrane sheets on the substrate compared to control cells, and expressed low, barely detectable levels of MBP. As in vivo, the MRF-deficient cells showed a significant acceleration of cell death relative to the control genotypes, with 44.2±2.9% of MRFfl/fl; Olig2wt/cre cells viable at 4 days differentiation, compared to 64.9±0.5% of MRFfl/fl; Olig2wt/wt cells and 61.3±1.4% of MRFwt/fl; Olig2wt/cre cells (Figure 6C-D, P<0.01). This suggests that the increased apoptosis of CKO OLs in vivo may not solely be due to a loss of axonal support when these cells fail to myelinate, but pre-myelinating OLs undergo an intrinsic apoptotic program if they cannot induce myelin gene expression and myelinate.
Figure 6. MRF deficient OPC/OL cultures display deficiencies in differentiative, but not proliferative, conditions.
(A-B) Immunostaining of control (MRFwt/fl; Olig2wt/cre) and CKO (MRFfl/fl; Olig2wt/cre) cultures for NG2 and Ki67 in proliferative (A) or differentiative (B) conditions. Arrowheads indicate Ki67+ nuclei. The same cells stained for CNP and MBP are shown in (B). Scale bars=100um. (C) Representative images of control and CKO cells stained with calcein AM (green; live cells) and ethidium homodimer (red; dead cells) at 4 days differentiation showing decreased viability and altered morphology of CKO cells. (D) Quantification of viability of control and CKO cells at 1 to 4 days differentiation. (E) Affymetrix expression levels of selected OPC markers NG2, PDGFRα and Ki67 for control and CKO cells at 4 days differentiation; CKO cells display normal down-regulation of these markers. (F) Expression levels of selected oligodendrocyte lineage markers in CKO cells at 4 days differentiation relative to control cells (G) Venn diagram showing overlap of genes induced >4-fold with OL differentiation and those repressed >4-fold in CKO relative to control cells. (H) Schematic of transcriptional control of OL lineage specification and differentiation. MRF is required for the maturation of pre-myelinating OLs into mature OLs expressing the full complement of myelin genes, with its induction possibly regulated by YY1.
We analyzed RNA from control and CKO spinal cords and OL cultures by RT-PCR and GeneChip microarrays. The expression of many myelin genes such as MAG and MOBP was abolished in both CKO spinal cords and OL cultures, demonstrating a requirement of MRF for the expression of these genes (Figure 6E-G, Table S1, Figure S7). Due to the substantial loss of OLs in the CKO spinal cords, these samples displayed a loss of essentially all genes specific to postmitotic OLs, whereas cultured CKO cells showed more restricted deficits. The effects of MRF deficiency on OL gene expression in vitro were very similar to those we observed with siMRF knockdown (Figure 6E-G, Supplemental Table 1-2), with most myelin genes and many OL maturation genes being strongly downregulated but only moderate effects on several early OL genes such as Ugt8. Interestingly several genes such as p57/cdkn1c whose expression is normally limited to transient expression by newly-formed oligodendrocytes were substantially upregulated in MRF-deficient OLs in vitro, again consistent with the idea that in the absence of MRF OLs are stalled in the immature, pre-myelinating stage.
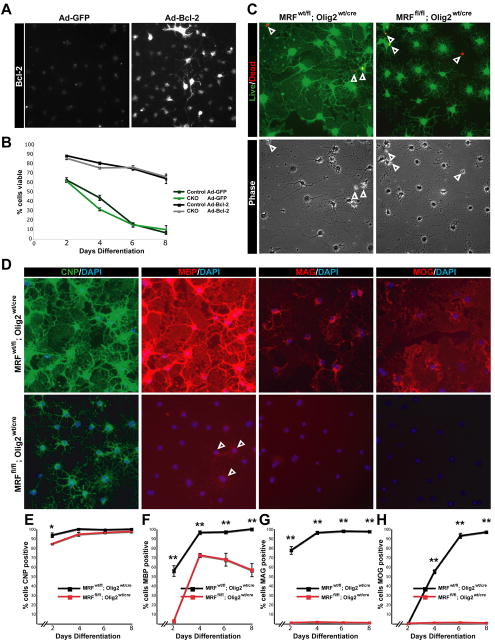
Although the increased apoptotic rate of CKO cells appeared less severe than the deficits in myelin gene expression, it was difficult to ascertain whether these cells undergo apoptosis because of a blockade to transition toward a mature phase, or whether the failure of myelin gene expression was secondary to early apoptosis. To clarify this issue, we infected control and CKO OPCs in vitro with a control adenovirus (Ad-GFP) or a Bcl-2 adenovirus (Ad-Bcl-2) to block apoptosis, induced their differentiation and assayed survival and myelin protein expression at 2-8 days of differentiation (Figure 7). Infection of cells with Ad-Bcl-2 (Figure 7A) successfully blocked the increased apoptosis of CKO cells relative to control cells, with 74.7±1.6% of control and 75.9±1.9% of CKO cells (P>0.05) remaining viable at 6 days of differentiation, a time at which nearly all of the Ad-GFP infected cells of both genotypes were dead (15.5±1.3% of control and 15.0±2.4% of CKO cells being viable, Figure 7B-C). In spite of the excellent viability of both genotypes when infected with Ad-Cre, the CKO cells still showed a clear deficit in myelin protein expression; whereas the vast majority of control cells were positive for MBP (95.8±1.9%), MAG (96.0±1.7) and MOG (94.3±2.8%); only 71.5±2.0% of CKO cells were faintly MBP+, and virtually no CKO cells were positive for MAG (1.2±0.5%) or MOG (0.8±0.3%). The CKO cells also still displayed a less mature morphology than control cells (Figure 7C) at 6 and 8 days post differentiation, a time sufficient to induce the expression of even late phase myelin genes (Figure 7D-H) (Dugas et al., 2006). Therefore, the apoptosis observed in MRF deficient OLs is likely to be a secondary effect due to their inability to transition to a mature state.
Figure 7. Blocking apoptosis does not restore myelin gene expression in MRF CKO oligodendrocytes.
(A) Confirmation of Bcl-2 overexpression in Ad-BCL-2 infected cells. (B) Viability of control and CKO cells infected with Ad-GFP or Ad-Bcl-2 over 8 days in differentiation media. (C) Calcien-AM/ethidium homodimer and phase images of Ad-Bcl-2 infected control and CKO cells at 6 days differentiation showing viability and morphology. Arrowheads indicate dead cells. (D) Expression of CNP, MBP, MAG and MOG in Ad-Bcl-2 infected control and CKO cells at 6 days differentiation. Arrowheads indicate faintly MBP+ CKO cells. (E-H) Quantification of the proportion of Ad-Bcl-2 infected control and CKO cells positive for CNP, MBP, MAG and MOG from 2-8 days differentiation. *P<0.05 **P<0.01
Discussion
MRF is a transcriptional regulator of CNS myelin gene expression and plays a role in the CNS comparable to that of Krox-20 in the PNS
In this study we have identified gene model 98/MRF as a transcriptional regulator required for OL maturation and CNS myelination. MRF is not detectably expressed by OPCs, neurons or astrocytes and is upregulated strongly upon OL differentiation. In the absence of MRF, OPCs are able to withdraw from the cell cycle and differentiate into pre-myelinating OLs but are unable to mature, express the full complement of myelin genes or myelinate and instead undergo apoptosis.
Montano et al., (2002) suggest that the yeast transcriptional activator Ndt80 may be the defining member of a family of transcription factors sharing a conserved DNA binding domain that includes MRF/GM98 and C11Orf9. Here, we confirm a role for MRF in myelin gene regulation in OLs, and thus CNS myelination. The high degree of homology between MRF and C11Orf9, in addition to the ability of MRF to induce MBP expression in the chick, suggests that its function is likely to be conserved between mammalian and avian homologs. C11Orf9 mRNA is downregulated in fetal motor neuron/oligodendroglial dysfunction in humans (Pakkasjarvi et al., 2005), indicating it may be expressed in OLs.
MRF's role is distinct from that of other transcription factors previously implicated in the OL lineage. MRF expression is restricted to postmitotic OLs and is required for expression of the vast majority of CNS myelin genes but not for OL specification or differentiation per se. Several OL lineage transcription regulators such as Nxk2.2, Olig1/2, Sox10 and YY1 are all expressed by OPCs as well as OLs, or in other CNS cell types (e.g. YY1). Although both MRF CKO and Olig1-/- mice have similar phenotypes with severe dysmyelination and death in the third week postnatal, it is not clear whether OLs differentiate in the Olig1-/- mice (Xin et al., 2005). Similarly, analysis of the spinal cords in Sox10-/- embryos suggests a role for Sox10 in promoting OL differentiation (Stolt et al., 2002). In contrast, MRF CKOs contain substantial numbers of pre-myelinating OLs at earlier ages but few myelinating OLs. Although Olig1 and Sox10 can activate specific myelin genes (Li et al., 2007; Stolt et al., 2002), Olig1 is not localized to the nucleus of myelinating oligodendrocytes (Arnett et al., 2004), suggesting that it may not directly induce myelin genes. So far, Nkx6.2 is the only transcription factor shown to be specific to postmitotic OLs. However Nkx6.2-/- mice show surprisingly few myelin deficits, with aberrations largely confined to the paranodal region (Southwood et al., 2004), which contrasts with those seen in MRF CKOs. Whereas, the majority of transcription factors within the OL lineage are critical for the specification of OPCs or initial differentiation of OLs, MRF is unique in coordinating subsequent maturation/myelination of postmitotic OLs.
The role of MRF in the CNS appears analogous to the role of Krox-20 in the PNS. Krox-20 is expressed by myelinating Schwann cells and is both sufficient and necessary to drive the transition of a non-myelinating Schwann cell to a myelinating state (Nagarajan et al., 2001; Topilko et al., 1994), and activate myelin gene promoters (Jang et al., 2006). As such, Krox-20 has been ascribed the role of a “master regulator” of PNS myelination. However, Krox-20 does is not expressed within the CNS. The specificity of MRF expression to postmitotic OLs, both in the newly formed and more mature stages, in combination with the loss of myelin gene expression seen upon disruption of MRF function, suggests that MRF may have a functionally equivalent role to Krox-20 within the CNS. Interestingly, it is possible that MRF and Krox20 share a common molecular switch that induces expression of both genes at the onset of myelination. Krox20 expression is induced by the transcriptional regulator YY1 (He and Casaccia-Bonnefil, 2008). The MRF gene contains a YY1 consensus site within a highly conserved region in intron 1. Since YY1 has been shown to link the exit of OPCs from the cell cycle with their differentiation (He et al., 2007), the induction of MRF by YY1 or similar factors upon OPC differentiation may be a mechanism by which cell cycle exit is linked with OL differentiation and myelin gene expression.
How does MRF induce myelination?
How does MRF induce myelin gene expression and myelination? Given its nuclear localization and conservation of the DNA binding domain with the yeast transcriptional activator Ndt80, MRF may function as a transcription factor in OLs. Consistent with this role, overexpression of a truncated form of MRF protein lacking the DNA binding domain in cultured OPCs failed to induce MBP expression (data not shown). Whether MRF functions as a bona fide transcriptional activator is an important question for future studies.
Although MRF is specific to OLs within the CNS, it is also expressed in other tissues such as lung and pancreas. Embryos lacking MRF in all tissues from germline deletion of MRF using a ubiquitous Cre driver failed to survive past ∼E12.5 (data not shown). This indicates that MRF has additional roles unrelated to myelin gene expression outside the CNS. As such, MRF may cooperate with other factors to specifically induce myelin gene expression in OLs. Our observation that MRF overexpression in OPCs induced MBP and MOG in the majority of cells, whereas it induced only a small number of MBP cells in the spinal cord, strongly suggests that MRF may require other factors (e.g. Sox10) to drive myelin gene expression. Conversely, the confinement of MRF expression to the postmitotic phase of the OL lineage may explain why Sox10 (present at all stages of the OL lineage) can induce myelin gene expression only in postmitotic cells. MRF may control the expression of myelin genes directly by binding to their promoters, or alternatively it may indirectly induce the transition to a myelinating state by gating the expression of other transcription factors. Finally, in addition to myelin genes, MRF is required for the expression of a subset of other genes normally associated with OL maturation including Adamts4 and TPPP, a tubulin polymerase. It is therefore possible that MRF induces a cohort of genes during OL maturation that work together to assemble and wrap CNS myelin sheathes. Further analysis of MRF function and MRF deficient mice will provide insights into these processes of CNS myelination independent of OL differentiation.
Experimental Protocols
Isolation and culture of mouse OPCs
Detailed protocols for the purification and culture of mouse OL lineage cells are available upon request. Mouse OPCs were isolated from enzymatically dissociated P7 mouse brains as described (Cahoy et al., 2008), positively immunopanned for PDGFRα following a depletion of microglia with BSL1. Cells were grown in serum-free media as previously described (Dugas et al., 2006), but with the addition of 2% B-27 (Invitrogen). PDGF-AA (10 ng/ml, PeproTech) was added to the media to proliferate OPCs; PDGF-AA was removed and triiodothyronine (T3) (40 ng/ml; Sigma) added to the media to induce differentiation.
Transfections of OPCs
Cultures of mouse OPCs were passaged and resuspended to 50×106 cells/ml in OPC Nucleofection solution (Amaxa). 100 μl (5×106 cells) was added to expression plasmids (∼4ug/transfection) or siRNAs (10ul of 20uM of siRNAs against MRF or siControl nontargeting siRNA pool) and electroporated with the Amaxa nucleofection apparatus. See supplemental methods for more details.
Adenoviral infection of OPCs
1 million OPCs of each genotype were cultured in proliferative conditions for 4 hours in the presence of Ad-Bcl2 or Ad-GFP (Vector Biolabs) at an MOI of 100 in 10ml of media. After 4 hours the media was changed and cells were passaged into differentiative conditions. Efficiency of infection was analyzed at 2 days post infection using GFP expression; infection efficiencies of >95% were obtained.
In ovo electroporations of chick embryos
Approximately 1ul of DNA solutions (∼4ug/ul of of a 1:4 mix of pCIG and pCAGGS-MRF and pCAGGS-Sox10 constructs) in TE buffer with 0.2% fast green to permit visualization were injected into the neural tube of HH stage 12 embryos. Electroporation was performed as described (Zhou et al., 2001). Embryos were harvested 4 days post electroporation and processed for in situ hybridization or immunohistochemistry.
Immunohistochemistry
Ten μm tissue cryosections or cells grown on PDL-coated coverslips were fixed for 10 minutes in 4% paraformaldehyde in PBS for 10 minutes, washed in PBS and then incubated for 30 minutes in blocking solution (10% FCS in PBS for surface antigens, with 0.3% triton-X for intracellular antigens). See supplemental experimental procedures for list of antibodies used. Coverslips or slides were washed in PBS and incubated with the appropriate fluorophore conjugated secondary (Molecular Probes, 1:500 in blocking solution), washed in PBS and mounted in DAKO mounting medium with DAPI. Fluoromyelin (Invitrogen) staining was performed on 10um cryosections according to manufacturer's instructions.
In situ hybridization
An in situ hybridization probe corresponding to 941bp from the MRF 3′ UTR was amplified from OL cDNA using the primers GGTGGGTTTGAGTTTGGAGGTT and GGGGAAACGCTCTATGAACAGG and subcloned into the PCR-II-Topo vector (Invitrogen). The PLP probe was a kind gift of William Richardson. In situ hybridizations were performed as described (Cahoy et al., 2008).
TEM Microscopy
Anesthetized P13 mice were perfused with PBS followed by 2% gluteraldehyde/4% paraformaldehyde in sodium cacodylate buffer. Optic nerves were dissected out and postfixed overnight at 4°C. Following treatment with 1% osmium tetroxide and 1% uranyl acetate, nerves were embedded in epon. Sectioning and electron microscopy was performed at Stanford Microbiology and Immunology Electron Microscopy Facility. Blind counts of myelinated axons were performed on images of randomly selected areas of the optic nerves. At least 100 axons of 0.3um or greater diameter were counted from 5 images per animal.
Northern hybridization
Total RNA (5-15ug) was run on a denaturing gel, transferred to Hybond-N+ (Amersham) and UV crosslinked. Membranes were pre-hybridized for 2hrs in 100ml hybridization solution (7% SDS, 0.5 M Na2HPO4 pH 7.2, 100ug/ml herring sperm DNA) at 68 °C. Radioactively labeled DNA probes were generated from plasmids containing approximately 1kb of MRF, CNP, PLP or GAPDH cDNA using the Prime It II random Primer kit (Stratagene) as per manufacturer's instructions with α-32P-dCTP (GE Healthcare). Hybridization was allowed to occur at 68 °C overnight, washed 2×10min in 1×SSC, 0.1% SDS at RT then 3×10min in 0.5×SSC, 0.1% SDS at 68°C. X-ray films (Amersham) were then exposed to the membrane at -80 °C before being developed.
Affymetrix Analysis
Total RNA was isolated from cells with the RNeasy micro kit (Qiagen, Valencia, CA) using Qiagen on-column DNase treatment to remove any contaminating genomic DNA. The integrity of RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Rockland, DE). 1ug total RNA was used for Affymetrix two-cycle target labeling and 430 2.0 Array hybridization according to manufacturer's instructions, see supplemental methods for further details. Array data has been deposited at http://www.ncbi.nlm.nih.gov/geo/ (accession number GSE15303).
RT-PCR
RNA prepared as per above was subject to reverse transcription using Invitrogen Superscript III reverse transcriptase as per manufacturer's instructions. cDNA was subject to amplification for 25 or 30 cycles with gene-specific primers and run on 2% agarose gels.
Generation of MRF CKO mice
Mice in which exon 8 of MRF was flanked by loxP sites were generated my cloning exon 8 into the Sal1 site of the Pez-Frt-lox-DT targeting vector. A 5kb 5′ arm and 3kb 3′ arm were cloned into the Not1 and Xho1 sites, respectively, to enable targeting of homologous recombination into E14 ES cell line. Correctly targeted neomycin resistant clones were identified by Southern blotting of HindIII digested DNA and PCR verification of the insertion of the 5′ loxP site. Targeted cells were injected into blastocytes to generate chimeric mice. MRFfl/+ mice were crossed onto the FlpER strain (Farley et al., 2000) to delete the neomycin cassette. Heterozygous MRF floxed mice were crossed for two generations onto Olig2-Cre mice (Schuller et al., 2008) or CNP-Cre mice (Lappe-Siefke et al., 2003) to obtain MRFfl/fl; Olig2wt/cre or MRFfl/fl; CNPwt/cre mice. See supplemental experimental procedures for genotyping primers.
Quantification and Statistics
Blind counts of cultured cells were performed on at least 3 coverslips per condition, with 10 fields of vision (20× objective) counted per coverslip. For quantification of tissue sections, 4-6 mice were used per genotype. Three sections were analyzed per mouse, with sections 60-100um apart photographed (×20 objective) for analysis (areas of counted regions were quantified in ImageJ). For quantification of activated Caspase 3+ cells the number of immunopositive cells was quantified for 3 longitudinal optic nerve sections 60um apart per mouse at ×20 objective, the nerves were then photographed at low power (4× objective) and areas of the nerves determined in ImageJ. Averages were calculated for each coverslip or mouse, and these results used to calculate means and SEMs for each experimental condition or genotype. Groups were compared using unpaired 2-way t-tests and Bonferroni's correction for multiple comparisons.
Supplementary Material
Acknowledgments
We thank Professor Klaus-Armin Nave for kindly providing the CNP-Cre mouse line. We are indebted to Lydia-Marie Joubert for processing samples for TEM, and Elizabeth Zou for processing samples for Affymetrix analysis. This work was funded by the National Eye Institute (RO1 EY10257, BAB) and the Myelin Repair Foundation. B.E. is supported by an Australian NHMRC CJ Martin award.
Footnotes
Supplemental Data: Supplemental data include supplemental experimental procedures, 9 figures, 2 tables and a movie.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Barres BA, Hart IK, Coles HS, Burne JF, Voyvodic JT, Richardson WD, Raff MC. Cell death and control of cell survival in the oligodendrocyte lineage. Cell. 1992;70:31–46. doi: 10.1016/0092-8674(92)90531-g. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Fu H, Qi Y, Tan M, Cai J, Takebayashi H, Nakafuku M, Richardson W, Qiu M. Dual origin of spinal oligodendrocyte progenitors and evidence for the cooperative role of Olig2 and Nkx2.2 in the control of oligodendrocyte differentiation. Development. 2002;129:681–693. doi: 10.1242/dev.129.3.681. [DOI] [PubMed] [Google Scholar]
- He Y, Casaccia-Bonnefil P. The Yin and Yang of YY1 in the nervous system. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi YN, K A, Casaccia-Bonnefil P. The Transcription Factor Yin Yang 1 Is Essential for Oligodendrocyte Progenitor Differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SW, LeBlanc SE, Roopra A, Wrabetz L, Svaren J. In vivo detection of Egr2 binding to target genes during peripheral nerve myelination. J Neurochem. 2006;98:1678–1687. doi: 10.1111/j.1471-4159.2006.04069.x. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog--regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Montano SP, Cote ML, Fingerman I, Pierce M, Vershon AK, Georgiadis MM. Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast. Proc Natl Acad Sci U S A. 2002;99:14041–14046. doi: 10.1073/pnas.222312199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Svaren J, Le N, Araki T, Watson M, Milbrandt J. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 2001;30:355–368. doi: 10.1016/s0896-6273(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Pakkasjarvi N, Gentile M, Saharinen J, Honkanen J, Herva R, Peltonen L, Kestila M. Indicative oligodendrocyte dysfunction in spinal cords of human fetuses suffering from a lethal motoneuron disease. J Neurobiol. 2005;65:269–281. doi: 10.1002/neu.20185. [DOI] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood C, He C, Garbern J, Kamholz J, Arroyo E, Gow A. CNS myelin paranodes require Nkx6-2 homeoprotein transcriptional activity for normal structure. J Neurosci. 2004;24:11215–11225. doi: 10.1523/JNEUROSCI.3479-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr H, Marquardt A, White K, Weber BH. cDNA cloning and genomic structure of a novel gene (C11orf9) localized to chromosome 11q12-->q13.1 which encodes a highly conserved, potential membrane-associated protein. Cytogenet Cell Genet. 2000;88:211–216. doi: 10.1159/000015552. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, Babinet C, Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- Trapp BD, Nishiyama A, Cheng D, Macklin W. Differentiation and death of premyelinating oligodendrocytes in developing rodent brain. J Cell Biol. 1997;137:459–468. doi: 10.1083/jcb.137.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.