Abstract
The SAM-dependent enzyme MoaA, in concert with MoaC, catalyzes the first step of molybdenum cofactor biosynthesis, the conversion of 5′-GTP into precursor Z. A published X-ray crystal structure of MoaA with the substrate 5′-GTP revealed that the substrate might be binding to the unique iron of one of two 4Fe-4S clusters, through either or both the amino and N1 nitrogen nuclei. 35 GHz CW ENDOR spectroscopy of MoaA with unlabeled and 15N-labeled substrate and a reduced [4Fe-4S]+ cluster now demonstrates that only one nitrogen nucleus is bound to the cluster. Experiments with the substrate analog 5′-ITP further demonstrate that it is the N1 nitrogen that binds. Two of the more distant nitrogen nuclei were also detected by 35 GHz pulsed ENDOR, allowing a rough approximation of their distances from the cluster to be calculated Combining this information with the crystal structure, we have proposed that the guanine base adopts the enol tautomer as N1 binds to Fe4 and the O6-H hydroxyl hydrogen-bonds to S4 of the 4Fe-4S cluster, and that this binding-induced tautomerization may have important mechanistic ramifications.
The enzymes MoaA and MoaC catalyze the first step in the biosynthesis of the biologically important molybdenum cofactor (Moco), the conversion of 5′-GTP (1) to precursor Z, an oxygen-sensitive tetrahydropyranopterin with a cyclic phosphate.1 MoaA has been identified as an S-adenosylmethionine (SAM)-dependent radical enzyme (Radical SAM) and features two catalytically important, oxygen sensitive [4Fe-4S]2+,+ clusters, each ligated by only three cysteine residues.2 The N-terminal [4Fe-4S] cluster (cluster I), present in all Radical SAM proteins, binds SAM at the unique Fe site (Fe4) and carries out the reductive cleavage of SAM to generate the 5′-deoxyadenosyl radical that subsequently initiates transformation of substrate 1 in a radical reaction. X-ray crystallography3 revealed that the triphosphate moiety of 1 is well anchored to MoaA by multiple interactions that include 12 hydrogen bonds. In contrast, the electron density for the ribose and base were not well defined. The purine ring N1 and amino N2 nitrogens appear to lie at distances of 2.8 and 2.4 Å from Fe4 of cluster II, too long for bonding, so the role, if any, of interaction with the cluster remained unresolved.3
We here report that comparison of CW4 and pulsed5 14,15N ENDOR data from 1(14,15N) and 2(14N) (Chart 1) bound to MoaA surprisingly shows that [4Fe-4S] cluster II positions its substrate by binding N1 of the purine ring to the unique Fe4. We further propose that this binding induces tautomerization of the base.
We initiated these studies by examining the C24S/C28S/C31S MoaA triple mutant, which does not contain the catalytic SAM-binding cluster I3 to eliminate possible interference by this cluster in the EPR/ENDOR spectra. We subsequently confirmed that cluster II of the wild-type enzyme shows identical ENDOR spectra to the mutant, eliminating the possibility that the mutation affects the cluster II-substrate interaction. In the absence of substrate, 35 GHz CW EPR spectra (2 K) of dithionite-reduced [Fe4S4]+ cluster II exhibit an axial signal from S = ½, g|| = 2.062 and g⊥ = 1.911 (see supplementary Fig S1); in addition, there is a barely observable signal from the cluster in a high-spin state, with g1 = 8.80, g2 = 4.20. Binding of the substrate 1 induces a slight change in the g values of the S = ½ cluster (2.063, 1.897) and an increase in the relative concentration of the S=3/2 state (Fig S1).
Fig 1 shows 2 K, 35 GHz CW ENDOR spectra collected at g2 for samples with 14N and 15N labeled 1. The spectrum for bound 1(14N) shows the ν+ branch of a 14N (I = 1) signal; it arises from overlapping signals from multiple orientations, each offset from the 14N Larmor frequency (ν(14N) = 4.04 MHz) by ½ the orientation-dependent hyperfine coupling and further split into a doublet by the orientation-dependent quadrupole interaction.
Figure 1.
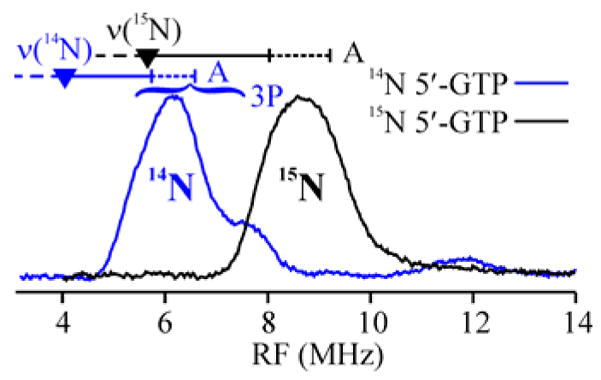
35 GHz CW ENDOR spectrum of 1(14,15N)-MoaA triple mutant at 2 K. Conditions: 4.0 G, 100 kHz modulation, 0.4 MHz/s RF sweep low-high with 100 kHz noise (15N), 0.5 MHz/s (14N). Frequency 34.752 GHz (15N), 34.109 GHz (14N).
The 14N feature is absent in the spectrum of enzyme with bound 1(15N) and is replaced by the ν+ branch for 15N; the shift in frequency matches the ratio of nuclear g values for 14/15N and the shape is simpler because 15N (I = ½) has no quadruple splitting. The 15N hyperfine coupling, A(15N) = 6.1 MHz, is equivalent to A(15N) = 5.8 MHz observed at g⊥ for the amino-nitrogen of S-adenosyl methionine (SAM) coordinated to an Fe ion of the [4Fe-4S]+ cluster of PFL-AE.6 Together, these observations prove that at least one nitrogen of 1 indeed binds to Fe4 of MoaA cluster II.
To determine the 14/15N hyperfine coupling tensors and the number of distinct nitrogen nuclei contributing to the signal, a complete 2D field-frequency pattern comprised of ENDOR spectra collected at magnetic field values across the MoaA EPR envelope was collected for both samples. Both patterns were well simulated7,8 by that for a single interacting nitrogen nucleus (Figs S2–4) whose substantial isotropic coupling, aiso(14N) = 3.6 MHz, requires the presence of a typical coordinate-covalent bond between a single N of 1 and the Fe4 ion of cluster II.
But which nitrogen of 1 is bound, N1 of the purine ring or the exocyclic N2 amino moiety? In lieu of time consuming selective 15N labeling, we addressed this question through use of the substrate analogue, 5′-ITP (2), which is identical to the natural substrate except that it lacks the exocyclic amino-group. Earlier in vitro binding studies with purine nucleotides other than 1 revealed 40% relative affinity with 5′-ATP, 60% with 2 and 100% with 5′-XTP.3 Not only does 2 bind, but it behaves similarly to 1 in increasing the rate of the reductive cleavage of the co-substrate SAM, the first step in MoaA catalysis, by a factor of about 10 and also significantly inhibits precursor Z biosynthesis (Fig S5). Together, these observations indicate that 2 binds in the active site similarly to 1, while 5′-ATP and 5′-XTP do not. This conclusion implies that if the amino group of 1(14N) binds to cluster II, then this signal should be lost in the complex with 2(14N); if purine ring nitrogen N1 binds, the signal should persist, although possibly somewhat altered by small differences in the precise binding orientation of 1 and 2.
The CW EPR spectrum of 2-MoaA (Fig S1; g|| = 2.057 and g⊥ = 1.883) again differs from that of substrate-free MoaA, and its g-values are similar to those of 1-MoaA, as expected if 2 binds in a similar fashion to 1. The mode of binding 2 to cluster II was probed by 14N ENDOR measurements. As illustrated by the g2 spectra in Fig 2 (see also Figs S2,6), 14N ENDOR signals of 2(14N)-MoaA are very similar to those of 1(14N)-MoaA. As for 1(14N), the 2D pattern for 2(14N) can be simulated with a single 14N; the coupling parameters are similar for the two complexes, with essentially the same isotropic hyperfine interaction, aiso(14N) = 3.5 MHz for 2 (Fig S6). The two substrates thus have an equivalent hyperfine-coupled 14N bound to Fe4 of cluster II.
Figure 2.
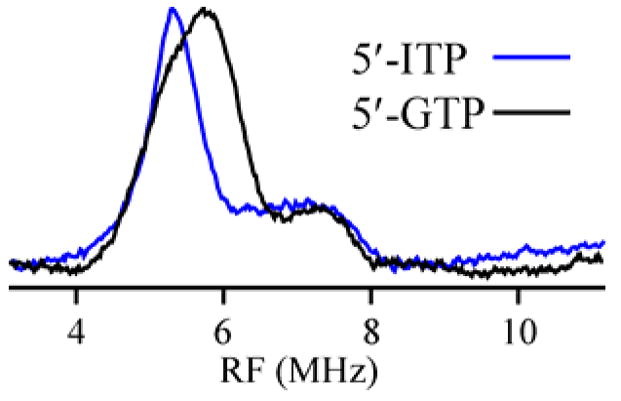
35 GHz ENDOR spectrum of 1(14N)- and 2(14N)-MoaA triple mutant at 2 K. Conditions: 4.0 G, 100 kHz modulation, 0.4 MHz/s RF sweep high-low with 100 kHz noise (GTP), 0.5 MHz/s (ITP). Frequency 35.026 GHz (GTP), 35.005 GHz (ITP).
The persistence of the substrate-derived 14N ENDOR signal upon replacement of 1 with 2 establishes that this signal does not come from the amino group of 1. In conjunction with the X-ray results, it proves that the signals of 1 and 2 both come from the purine ring nitrogen N1; the binding of any other ring nitrogen of 2 to cluster II would require a massive reorientation of 2 relative to that of 1. Given that the triphosphate of 1 is so tightly anchored,3 and that 2 clearly binds in the same general fashion as 1, we conclude that 1 (and 2) is positioned for catalysis through binding of the purine ring N1 to the unique Fe4 of cluster II.
Examination of the X-ray crystal structure of 1-MoaA suggests that when N1 binds, at least two other nitrogen atoms - N3 and the amino nitrogen - should be close enough to Fe4 that they would exhibit observable 15N dipolar hyperfine coupling to the cluster spin. Pulsed Mims ENDOR spectra of 1(15N)-MoaA reveal two 15N doublets centered at the 15N Larmor frequency, with respective maximum splittings of A(15N) = 0.5 MHz and 0.2 MHz (Fig S7). Overlap of the peaks, combined with the distortions from the Mims suppression ‘hole’ at the Larmor frequency,9 precludes a precise determination of the hyperfine tensors for the two nitrogens. However, simulations of the spectra establish the maximum and minimum dipolar components for each, from which we can set rough limits on the possible distances between the nitrogens and cluster Fe4 (Supplementary Material). This analysis places one nitrogen between 2.6 Å and 5.1 Å from the Fe, and the other at a distance greater than 3.6 Å.10
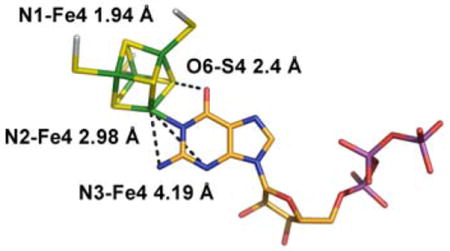
The bond between Fe4 of cluster II and N1 of 1, along with the anchoring of the triphosphate of 1 by the enzyme, strongly constrain the possible orientations of both the ribose and purine of bound 1. If we fix the triphosphate moiety as in the X-ray structure, only minor movements and rotations of the base and ribose are required to generate a structure of bound 1 consistent with the ENDOR data (Fig 3). In this model the Fe4-N1 distance is 1.94 Å, with the other two weakly-coupled nitrogens at ENDOR-compatible distances of 3.0 Å (N2) and 4.2 Å (N3). These differences from the X-ray structure may reflect the fact that the latter was obtained after soaking crystals with 1, SAM and Na-dithionite, conditions that had initiated catalysis and might have altered the binding mode. Alternatively, the ENDOR experiments are performed on an unconstrained enzyme/substrate complex formed in solution and subsequently frozen, whereas packing effects in the crystal might limit conformational changes needed to accommodate the substrate.
Figure 3.
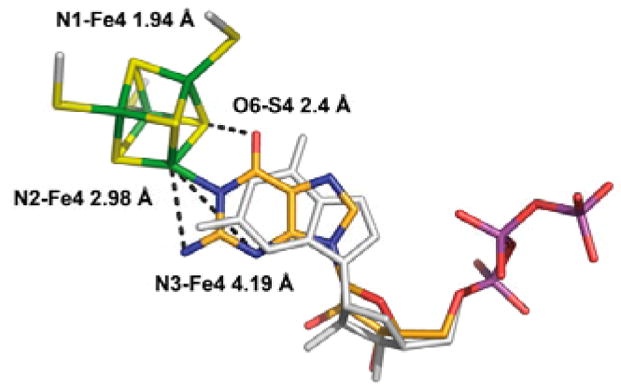
Proposed model for 5′-GTP binding (C orange, N blue, O red, P purple) to the Fe4 ion of cluster 2 (S yellow, Fe green). The 5′-GTP model derived by X-ray crystallography is shown in white (PDB code 2FB3).
The proposed structure of Fig 3 incorporates an Fe4-N1 bond that lies in the plane of the purine ring, and it thus implies that the purine binds as the enol tautomer, not the normally favored keto form. In support of this idea, S4 of cluster II and O6 of the putative guanine hydroxyl are in H-bonding distance (2.4 Å), although this was not imposed as a constraint while generating the structure. We surmise that such binding-induced tautomerization may play a mechanistic role by modulating the purine reactivity and/or stabilizing reaction intermediates.
Supplementary Material
Acknowledgments
Support by the NIH (DK54835, PH and HS; GM62524, MKJ; HL13531, BMH), Deutsche Forschungsgemeinschaft (FZ 82, PH and HS), and NSF (MCB0723330, BMH).
Footnotes
Supporting Information Available: Procedures for simulation of ENDOR spectra. In vitro activities.
References
- 1.Wuebbens MM, Rajagopalan KV. J Biol Chem. 1995;270:1082–1087. doi: 10.1074/jbc.270.3.1082. [DOI] [PubMed] [Google Scholar]
- 2.Hänzelmann P, Schindelin H. Proc Natl Acad Sci U S A. 2004;101:12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hänzelmann P, Schindelin H. Proc Natl Acad Sci U S A. 2006;103:6829–6834. doi: 10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werst MM, Davoust CE, Hoffman BM. J Am Chem Soc. 1991;113:1533–1538. [Google Scholar]
- 5.Davoust CE, Doan PE, Hoffman BM. J Magn Reson. 1996;119:38–44. [Google Scholar]
- 6.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. J Am Chem Soc. 2002;124:11270–11271. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman BM, DeRose VJ, Doan PE, Gurbiel RJ, Houseman ALP, Telser J. Biol Magn Reson. 1993;13(EMR of Paramagnetic Molecules):151–218. [Google Scholar]
- 8.Doan PE, Telser J. Paramagnetic Resonance of Metallobiomolecules. American Chemical Society. 2003:55–81. [Google Scholar]
- 9.Schweiger A, Jeschke G. Principles of Pulse Electron Paramagnetic Resonance. Oxford University Press; Oxford, UK: 2001. [Google Scholar]
- 10.If one of these nitrogen signals is the amino nitrogen, then it should not be visible in the 2(14N) sample. However, the 14N signals that correspond to the weakly coupled 15N peaks of 1(15N) proved too difficult to detect, so this could not be checked.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


