Abstract
Background and Purpose
Ischemic stroke is a multifactorial disease with a strong genetic component. Pathways including lipid metabolism, systemic chronic inflammation, coagulation, blood pressure regulation, and cellular adhesion have been implicated in stroke pathophysiology, and candidate gene polymorphisms in these pathways have been proposed as genetic risk factors.
Methods
We genotyped 105 simple deletions and single nucleotide polymorphisms from 64 candidate genes in 3550 patients and 6560 controls from six case-control association studies conducted in the United States, Europe and China. Genotyping was performed using the same immobilized probe typing system and meta-analyses were based on summary logistic regressions for each study. The primary analyses were fixed-effects meta-analyses adjusting for age and sex with additive, dominant and recessive models of inheritance.
Results
Although seven polymorphisms showed a nominal additive association, none remained statistically significant after adjustment for multiple comparisons. In contrast, after stratification for hypertension, two lymphotoxin-alpha polymorphisms which are in strong linkage disequilibrium were significantly associated among non-hypertensive individuals: for LTA 252A>G (additive model), OR=1.41 with 95% CI, 1.20 to 1.65, p=0.00002; for LTA 26Thr>Asn, OR 1.19 with 95% CI, 1.06 to 1.34, p=0.003. LTA 252A>G remained significant after adjustment for multiple testing using either the false discover rate or by permutation testing. The two SNPs showed no association in hypertensive subjects (eg, LTA 252A>G, OR=0.93; 95%CI, 0.84 to 1.03, p=0.17).
Conclusions
These observations may indicate an important role of LTA-mediated inflammatory processes in the pathogenesis of ischemic stroke.
Indexing terms: ischemic stroke, hypertension, inflammation, genetics
Ischemic stroke is a complex multi-factorial and polygenic disorder thought to reflect interactions between an individual’s genetic background and various environmental components. Previous studies have established hypertension, smoking, diabetes mellitus, body mass index and age as reliable stroke-risk predictors1,2. However, these conventional stroke risk factors do not fully account for the overall risk of stroke. Several physiological pathways, including lipid metabolism, blood pressure regulation, coagulation, and cellular adhesion are thought to play critical roles in stroke pathophysiology.
Among the known risk factors for ischemic stroke, hypertension contributes significantly to the onset of disease. Increased risk of stroke is not, however, limited to those with hypertension and the conventional stroke risk factors do not fully explain the risk among normotensives. Strategies to identify additional risk factors include stratification by hypertension3,4 and using blood pressure as a matching criterion for cases and controls5. A key role for inflammation is suggested by observations that hypertensive patients have elevated circulating levels of markers of inflammation and that some anti-hypertensive therapies reduce both levels of pro-inflammatory markers and the risk of ischemic stroke, in addition to lowering blood pressure6.
Both systemic and local inflammatory processes are implicated in the etiology of ischemic cerebrovascular disease and in the pathophysiology of cerebral ischemia7. Viral and bacterial infections are independent risk factors for ischemic stroke8 and increased levels of systemic inflammatory markers such as C-reactive protein (CRP), leukocyte count, and fibrinogen are associated with increased risk of ischemic stroke9. Moreover, many stroke-related diseases such as Alzheimer’s disease and atherosclerosis are initiated or worsened by systemic inflammation10,11. Polymorphisms in the CRP gene have been recently associated with both circulating protein levels and cardiovascular events12, demonstrating the potential impact of genetic variation. Pro-inflammatory cytokines are believed to play a pathogenic role in these diseases, and variations in cytokine genes have also been shown to influence both predisposition and penetrance by altering the transcription profile and pattern of pro-inflammatory cytokine production13. For example, polymorphism in the lymphotoxin-alpha gene can enhance transcription and susceptibility to myocardial infarction14. At the local level, migration of inflammatory cells to the vascular wall is associated with vascular changes leading to atherosclerosis, and early atherosclerotic lesions are preceded by inflammatory cell deposition in the sub-endothelial layer of major cerebral arteries and in small brain vessels15. Genetic variants influencing inflammatory processes could potentially contribute to the etiology of stroke.
The complex etiology of stroke suggests that individual genetic polymorphisms have modest effects that are difficult to detect, as has been observed to date16. Large studies are needed to assess these polymorphisms as risk factors. Here we report a six-study meta-analysis to investigate the associations of 105 simple deletions and single nucleotide polymorphisms (SNPs) in inflammatory and cardiovascular system-related genes with susceptibility to ischemic stroke. To search for genetic risk factors contributing to ischemic stroke beyond hypertension, we stratified the study cohort on hypertension status.
Materials and Methods
Study Sample Description
As part of the Roche Stroke SNP Consortium, results of six independent studies (Table 1) were pooled for this analysis. All six study samples were comprised of individuals with proven ischemic stroke status and healthy controls. All studies were approved by the local ethics committees and all participants gave informed consent. Briefly, the study subjects were recruited as follows:
Table 1.
Characteristics of the Study Subjects. Summary of population characteristics for each of the six studies used in the meta-analysis. All cases were diagnosed by CT/MRI.
| PHS17,18 | SOF19 | Vienna23 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Subjects (n) | 319 cases, 2092 controls | 247 cases, 559 controls | 844 cases, 979 controls | ||||||
| Study Type | prospective | prospective | cross-sectional | ||||||
| Recruitment Period | 1982; follow-up through 1995 | 1986 – 1988; follow-up through 1998 | 1998 – 2001 | ||||||
| Population | Caucasians from USA | Caucasians from USA | Caucasians from Vienna, Austria | ||||||
| Cases | Controls | p‡ | Cases | Controls | p‡ | Cases | Controls | p‡ | |
| Age* (yrs) | 61.1±8.3 | 58.8±8.5 | <.0001 | 73.9±5.9 | 70.4±4.5 | <.0001 | 66.1±14.4 | 48.8±13.0 | <.0001 |
| Sex (% male) | 100 | 100 | 0 | 0 | 50.6 | 53.4 | 0.23 | ||
|
Hypertension (% yes) |
48.6 | 26.8 | <.0001 | 79.8 | 62.1 | <.0001 | 72.2 | 34.2 | <.0001 |
| Diabetes (%yes) | 10.7 | 2.7 | <.0001 | 19.3 | 4.3 | <.0001 | 32.8 | 2.5 | <.0001 |
|
Smoking (% ever) |
60.7 | 56.6 | 0.15 | 29.8 | 37.5 | 0.15 | 54.9 | 41.7 | <.0001 |
| BMI*,† (kg/m2) | 25.6±3.3 | 25.0±3.0 | 0.002 | 26.9±4.6 | 26.8±4.9 | 0.83 | 26.7±4.6 | 25.3±3.9 | <.0001 |
| SBP*,† (mm Hg) | 133.6±12.8 | 127.4±11.9 | <.0001 | 151.9±23.1 | 140.4±17.5 | <.0001 | NA | 132.3±20.7 | |
| DBP*,† (mm Hg) | 82.1±7.0 | 79.0±7.2 | <.0001 | 78.2±11.6 | 76.9±8.9 | 0.13 | NA | 82.7±11.7 | |
| Westphalia20,21 | Pomerania22 | SHINING5 | |||||||
| Subjects (n) | 700 cases, 757 controls | 277 cases, 702 controls | 1163 cases, 1471 controls | ||||||
| Study Type | cross-sectional | cross-sectional | cross-sectional | ||||||
| Recruitment Period | 2000 – 2003 | 1996 – 2001 | 1997–2000 | ||||||
| Population | Caucasians from northwestern Germany | Caucasians from northeastern Germany | Chinese primarily from Beijing, China | ||||||
| Cases | Controls | p‡ | Cases | Controls | p‡ | Cases§ | Controls§ | p‡ | |
| Age* (yrs) | 61.0±16.9 | 58.7±9.9 | 0.002 | 62.8±13.2 | 62.7±11.8 | 0.88 | 59.3±10.7 | 61.1 ±10.7 | <.0001 |
| Sex (% male) | 56.3 | 46.6 | <.0001 | 55.1 | 50.8 | 0.23 | 60.3 | 60.1 | 0.93 |
|
Hypertension (% yes) |
68.0 | 41.0 | <.0001 | 66.8 | 52.9 | <.0001 | 69.6 | 77.1 | <.0001 |
| Diabetes (%yes) | 21 | 8.9 | <.0001 | 30.1 | 14.5 | <.0001 | 15.3 | 8.5 | <.0001 |
|
Smoking (% ever) |
NA | 54.1 | 87.6 | 58.3 | <.0001 | 39.7 | 32.3 | <.0001 | |
| BMI*,† (kg/m2) | NA | NA | NA | NA | 24.4±3.0 | 25.0±3.3 | <.0001 | ||
| SBP*,† (mm Hg) | NA | 145.9±21.4 | NA | 148.7±21.3 | 145.4±23.2 | 143.2±23.7 | 0.018 | ||
| DBP*,† (mm Hg) | NA | 89.1±12.2 | NA | 86.1±11.1 | 87.0±12.8 | 86.1±13.0 | 0.087 | ||
Continuous variables are given as mean ±SD.
BMI = Body mass index, SBP = Systolic blood pressure prior to stroke, DBP =Diastolic blood pressure prior to stroke.
Using chi-square test for categorical variables, t-test for continuous variables.
Partially matched by age and BP group during recruitment
Physician’s Health Study (PHS)
A nested case-control sample (319 cases, 2092 controls) was derived from the PHS cohort consisting of 22,071 predominantly Caucasian U.S. male physicians initially free of prior myocardial infarction, stroke, transient ischemic attack and cancer, who were enrolled in a placebo controlled trial of aspirin and beta-carotene for the primary prevention of cardiovascular disease and cancer17. DNA was isolated from baseline blood samples provided by 14,916 (68%) of the participants. Incident cases of ischemic stroke were identified during an average 13-year follow-up, and confirmed by medical record review. Controls were selected from study participants remaining free of reported cardiovascular disease, and matched to cases of any cardiovascular disease by age, smoking, and time since study entry18.
Study of Osteoporotic Fractures (SOF)
Ambulatory women were recruited from four clinical centers in Portland, Oregon; Minneapolis, Minnesota; Baltimore, Maryland; and the Monongahela Valley, Pennsylvania19. The SOF cohort consists of 9615 white women of at least 65 years of age who had not had bilateral hip replacement or earlier hip fracture at the time of recruitment. The stroke subgroup included here consists of 247 who suffered adjudicated ischemic strokes and 559 controls who remained free of stroke through the mean follow-up of 5.4 years. Individuals who died during follow-up were included in both cases and controls, avoiding survivor bias.
Westphalia, Germany
Cases (n = 700) were recruited through the regional Westphalian Stroke Register in northwestern Germany20. Standardized patient documentation included socio-demographic characteristics, comorbidities, stroke type and severity as well as details regarding the diagnostic and therapeutic procedures and complications; 96.8% had at least one CT or MRI of the brain during hospitalization. Controls (n = 757) were recruited from the population-based Dortmund Health Study, conducted in the same region21. Participants in this study were randomly drawn from the city’s registration office within 5-year age groups and stratified by sex. Medical histories were assessed in face to face interviews.
Pomerania, Germany
Cases (n = 277) were recruited with a standardized patient assessment form; 96.5% had at least one CT or MRI during hospitalization. Controls for this region were recruited from the population-based Study of Health in Pomerania (SHIP)22. Participants in SHIP were 20- to 79-year-olds randomly sampled from registration offices in the area. Face-to-face interviews with each participant included a short stroke symptom questionnaire. A random sample of 702 SHIP participants who were free of self-reported stroke and within the same age range and sex distribution as the cases, formed the control group.
Vienna Stroke Study
In the Vienna Stroke Registry, cases (n= 844) consisted of consecutive Caucasian patients submitted to one of nine stroke units within 72 hours of symptom onset of acute ischemic stroke. Patients who died on the way to the hospital or were first admitted to an intensive care unit were not included23. All patients underwent cranial CT or MRI and were documented according to a standardized protocol including stroke severity, risk factors and medical history (with particular reference to vascular diseases). Controls (n = 979) were voluntary participants in a health care program offered by the city of Vienna, were free of clinically manifest arterial vascular disease and reported no arterial vascular diseases in first degree relatives.
Stroke Hypertension Investigation in Genetics (SHINING)
Individuals were recruited from six geographical regions within China; 70% came from in and near Beijing. Cases (n = 1163) were individuals who had suffered a stroke within the previous 5 years, as diagnosed by brain CT/MRI. The original goal was to identify SNPs that predispose to stroke independent of blood pressure, thus randomly drawn population-based controls were initially individually matched to cases by sex, birth year ±3 yrs, geographic location, and blood pressure category (<140/90, ≥140/90 and ≤180/105, >180/105). Because some cases could not be matched, additional controls were recruited for a total of 1471 controls5.
Genotyping
A total of 105 polymorphisms from 64 genes were selected based on reported associations in the literature, as well as on evidence of gene product involvement in cardiovascular disease and inflammatory processes. As previously described24,25, three separate multi-locus polymerase chain reactions (PCRs) were carried out using biotinylated primer pools (Roche Molecular Systems, Inc., CA). The resulting PCR product pools were denatured and hybridized to linear arrays of immobilized, sequence-specific oligonucleotide probes. Hybridized amplicon was detected using a streptavidin-horseradish peroxidase conjugate and a chromogenic substrate. Laboratory technicians were blinded to the case-control status of each sample. Genotype assignments were made either manually and independently by two researchers or made by capturing images with a flatbed scanner and then using proprietary software developed by Roche Molecular Systems to resolve probe signals into genotypes for all polymorphisms. Discordant or ambiguous results were resolved by repeat PCR or hybridization. Twenty polymorphisms were not available for PHS and 23 polymorphisms were genotyped in only a subset of the Vienna Stroke participants; for these, genotypes were obtained for >90.5% of the individuals typed. For each of the 62 polymorphisms genotyped for all 10,110 subjects across all six studies, the final genotype database was ≥97.5% complete. For the LTA [MIM 153440] 252A>G and LTA 26Thr>Asn polymorphisms, in particular, the database contained 6090 (not available for PHS) and 10,091 genotypes (99.8% complete), respectively.
Statistical analysis
Individual level data were provided from each study site to the Coordinating Center at the Brigham and Women’s Hospital in Boston. Pre-specified inclusion criteria for the meta-analyses were: age of at least 20 years and no prior history of myocardial infarction. Cases were restricted to those experiencing an ischemic stroke and controls had no previous history of stroke.
Allele and genotype frequencies were estimated by study site among cases and controls separately using SAS GENETICS. Tests for Hardy-Weinberg equilibrium (HWE), both large-sample and exact, were conducted among cases and controls for each site (Supplementary Table 1). Results for the effect of each SNP on ischemic stroke were estimated for each study separately using logistic regression. Each analysis controlled for age and sex, and assessed genetic effects under three modes of inheritance: additive, dominant, and recessive. In addition, analyses were conducted for each site using all three genotypes, using a two-degree of freedom test.
Meta-analyses were conducted based on the summary logistic regression results for each study site26. The primary analyses were fixed effects meta-analyses adjusting for age and sex. These meta-analyses were also conducted across Caucasians only (data not shown), since these comprised the majority of participants for five of the six studies. Effects for each of the three modes of inheritance were estimated. PROC MIXED of SAS was used for effect estimation. Tests for heterogeneity of the genetic effect across sites were conducted using the Q-statistic27. For comparison, random effects models were estimated which allowed the genetic effect to vary across sites using study-specific effect estimates and PROC MIXED of SAS. To adjust for multiple comparisons, the false discovery rate (FDR)28 was computed and stepdown permutation tests were conducted for selected comparisons29.
Other pre-specified analyses adjusted for hypertension as well as age and sex. Across all studies, hypertensives were defined as having current or past anti-hypertensive medication, systolic blood pressure ≥140 mm Hg, or diastolic blood pressure ≥90 mm Hg. Additional smoking-adjusted analyses were limited to five studies due to the limited availability of smoking data for the Westphalian study participants. Subgroup analyses were conducted according to age, sex, presence of hypertension, or smoking (ever vs. never).
Results
A total of 3550 stroke patients and 6560 controls were genotyped with inflammation and cardiovascular SNP panels in six study sites with common methodology and genotyping software. The characteristics of all participants from six study sites are listed in Table 1. Two studies were drawn from prospective cohorts; the PHS study followed only male subjects and the SOF study followed only female subjects. The SHINING study was comprised of subjects of Han ethnicity, while the five other study populations were >99% Caucasian. There was a greater proportion of hypertension among cases than in controls, except in the SHINING study subset, for which blood pressure had been matched between the majority of cases and controls.
Table 2 lists the results obtained in the primary fixed-effects meta-analysis for all polymorphic sites under dominant, additive and recessive genetic modes of inheritance; similar results were observed under a random effects meta-analysis (data not shown). Nine SNPs were nominally significant (P < 0.05) under at least one mode of inheritance: ADRB3 [MIM 109691] Trp64Arg, CETP [MIM 118470] (−629)C>A, GNB3 [MIM 139130] 825C>T, IL4 [MIM 147780] (−590)C>T, LIPC [MIM 151670] (−480)C>T, LPL [MIM 609708] Ser447Ter, NOS3 [MIM 163729](−690)C>T, PON2 [MIM 602447] Ser311Cys and TGFB1 [MIM 190180] (−509)C>T. To account for multiple hypothesis testing, the false discovery rate or permutation testing was applied and none of these SNPs remained statistically significantly associated with ischemic stroke. Among Caucasian participants only, the same GNB3, LPL, NOS3, PON2 and TGFB1 SNPs were nominally significant under at least one mode of inheritance, in addition to eight others (APOB [MIM 107730] 71Ile>Thr, APOC3 [MIM 107720] 3175C>G, CCR5 [MIM 601373] (−2459)G>A, IL6 [MIM 147620] (−174)G>C, IL10 [MIM 124092] (−571)C>A , ITGA3 [MIM 192974] 873G>A, NOS2A [MIM 163730] 231C>T, TNF [MIM 191160] (−376)G>A), but none of the SNPs remained statistically significant after the false discovery rate was applied (data not shown).
Table 2.
Unstratified meta-analysis for risk of ischemic stroke: all SNPs under three modes of inheritance.
| Additive | Dominant | Recessive | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene [MIM number] | SNP rs number | SNP Name | n* | Sites† | OR (LCL,UCL)‡ | p‡ | Het p§ | OR (LCL,UCL)‡ | p‡ | Het p§ | Sites∥ | OR (LCL,UCL)‡ | p‡ | Het p§ |
| Adducin 1 (alpha) [102680] | rs4961 | ADD1 460Gly>Trp | 9936 | 6 | 1.02 (0.95,1.10) | 0.638 | 0.960 | 1.04 (0.95,1.15) | 0.403 | 0.876 | 6 | 0.98 (0.84,1.14) | 0.762 | 0.692 |
| Adrenergic, beta-2-, receptor, surface [109690] | rs1042713 | ADRB2 16Gly>Arg | 10082 | 6 | 0.94 (0.89,1.01) | 0.080 | 0.271 | 0.95 (0.86,1.05) | 0.303 | 0.437 | 6 | 0.90 (0.80,1.01) | 0.064 | 0.502 |
| Adrenergic, beta-2-, receptor, surface [109690] | rs1042714 | ADRB2 27Gln>Glu | 10087 | 6 | 1.04 (0.97,1.11) | 0.323 | 0.121 | 1.04 (0.94,1.15) | 0.461 | 0.177 | 6 | 1.06 (0.93,1.22) | 0.381 | 0.222 |
| Adrenergic, beta-2-, receptor, surface [109690] | rs1800888 | ADRB2 164Thr>Ile | 8535 | 6 | 1.33 (0.88,2.02) | 0.175 | 0.308 | 1.29 (0.85,1.98) | 0.236 | 0.324 | Too few variant homozygotes observed | |||
| Adrenergic, beta-3-, receptor [109691] | rs4994 | ADRB3 64Trp>Arg | 10093 | 6 | 0.91 (0.82,1.02) | 0.090 | 0.679 | 0.89 (0.79,1.00) | 0.046 | 0.803 | 6 | 1.13 (0.75,1.69) | 0.570 | 0.265 |
| Angiotensin II receptor type 1 [106165] | rs5186 | AGTR1 1166A>C | 9942 | 6 | 1.04 (0.96,1.13) | 0.346 | 0.068 | 1.03 (0.93,1.14) | 0.546 | 0.015 | 6 | 1.11 (0.92,1.35) | 0.284 | 0.734 |
| Angiotensinogen [106150] | rs699 | AGT 235Met>Thr | 9939 | 6 | 1.00 (0.94,1.07) | 0.973 | 0.371 | 0.97 (0.87,1.08) | 0.586 | 0.279 | 6 | 1.03 (0.93,1.15) | 0.581 | 0.344 |
| Angiotensin-converting enzyme [106180] | rs1799752 | ACE IVS16 Del>Ins | 9862 | 6 | 0.99 (0.93,1.06) | 0.841 | 0.069 | 1.02 (0.91,1.13) | 0.758 | 0.473 | 6 | 0.97 (0.88,1.07) | 0.556 | 0.029 |
| Apolipoprotein A-IV [107690] | rs675 | APOA4 347Thr>Ser | 10091 | 6 | 0.94 (0.85,1.03) | 0.193 | 0.672 | 0.96 (0.86,1.07) | 0.446 | 0.551 | 5 | 0.76 (0.57,1.03) | 0.074 | 0.512 |
| Apolipoprotein A-IV [107690] | rs5110 | APOA4 360Gln>His | 10088 | 6 | 0.95 (0.82,1.11) | 0.545 | 0.297 | 0.97 (0.83,1.14) | 0.730 | 0.288 | 5 | 0.81 (0.32,2.03) | 0.653 | 0.999 |
| Apolipoprotein B [107730] | rs1367117 | APOB 71Thr>Ile | 10091 | 6 | 1.04 (0.97,1.12) | 0.259 | 0.807 | 1.02 (0.93,1.12) | 0.675 | 0.416 | 6 | 1.18 (1.00,1.41) | 0.056 | 0.849 |
| Apolipoprotein B [107730] | rs5742904 | APOB 3500Arg>Gln | 10093 | 3 | 6.11 (0.45,82.6) | 0.173 | 0.999 | 6.11 (0.45,82.6) | 0.173 | 0.999 | No variant homozygotes observed | |||
| Apolipoprotein C-III [107720] | rs2542052 | APOC3 (−641)C>A | 10064 | 6 | 1.03 (0.97,1.10) | 0.303 | 0.864 | 1.04 (0.95,1.15) | 0.377 | 0.595 | 6 | 1.05 (0.93,1.18) | 0.434 | 0.834 |
| Apolipoprotein C-III [107720] | rs2854117 | APOC3 (−482)C>T | 10089 | 6 | 1.06 (0.99,1.13) | 0.096 | 0.266 | 1.06 (0.97,1.16) | 0.226 | 0.137 | 6 | 1.12 (0.97,1.28) | 0.114 | 0.728 |
| Apolipoprotein C-III [107720] | rs2854116 | APOC3 (−455)T>C | 10087 | 6 | 1.04 (0.97,1.11) | 0.244 | 0.775 | 1.04 (0.95,1.15) | 0.372 | 0.544 | 6 | 1.06 (0.94,1.20) | 0.311 | 0.871 |
| Apolipoprotein C-III [107720] | rs4520 | APOC3 1100C>T | 10089 | 6 | 1.02 (0.96,1.09) | 0.517 | 0.313 | 1.03 (0.93,1.13) | 0.609 | 0.244 | 6 | 1.04 (0.92,1.18) | 0.562 | 0.111 |
| Apolipoprotein C-III [107720] | rs5128 | APOC3 3175C>G | 10091 | 6 | 1.01 (0.93,1.10) | 0.776 | 0.124 | 1.05 (0.95,1.16) | 0.351 | 0.153 | 6 | 0.84 (0.66,1.08) | 0.173 | 0.389 |
| Apolipoprotein C-III [107720] | rs4225 | APOC3 3206T>G | 10073 | 6 | 1.04 (0.97,1.11) | 0.274 | 0.372 | 1.09 (0.97,1.21) | 0.137 | 0.606 | 6 | 1.02 (0.91,1.13) | 0.780 | 0.405 |
| Apolipoprotein E [107741] | rs429358 | APOE 112Cys>Arg | 10050 | 6 | 1.04 (0.94,1.14) | 0.447 | 0.341 | 1.03 (0.92,1.14) | 0.624 | 0.469 | 6 | 1.25 (0.88,1.78) | 0.206 | 0.223 |
| Apolipoprotein E [107741] | rs7412 | APOE 158Arg>Cys | 10055 | 6 | 0.96 (0.85,1.07) | 0.424 | 0.171 | 0.97 (0.86,1.09) | 0.570 | 0.221 | 6 | 0.71 (0.41,1.23) | 0.223 | 0.531 |
| CD14 molecule [158120] | rs2569190 | CD14 (−260)C>T | 8536 | 6 | 1.03 (0.96,1.11) | 0.397 | 0.474 | 1.02 (0.90,1.14) | 0.801 | 0.274 | 6 | 1.06 (0.95,1.19) | 0.285 | 0.952 |
| Chemokine (C-C motif) ligand 11 [601156] | rs4795895 | CCL11 (−1328)G>A | 6123 | 5 | 0.96 (0.85,1.08) | 0.476 | 0.202 | 0.97 (0.84,1.11) | 0.602 | 0.083 | 5 | 0.88 (0.58,1.33) | 0.536 | 0.707 |
| Chemokine (C-C motif) ligand 11 [601156] | rs3744508 | CCL11 23Ala>Thr | 8536 | 6 | 1.08 (0.99,1.19) | 0.099 | 0.856 | 1.09 (0.98,1.21) | 0.113 | 0.985 | 6 | 1.16 (0.86,1.57) | 0.341 | 0.224 |
| Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) [600835] | rs1801157 | CXCL12 (+800)G>A | 6032 | 5 | 0.97 (0.88,1.06) | 0.454 | 0.599 | 0.96 (0.86,1.07) | 0.446 | 0.748 | 5 | 0.99 (0.76,1.27) | 0.911 | 0.079 |
| Chemokine receptor 2 [601267] | rs1799864 | CCR2 62Val>Ile | 8532 | 6 | 1.08 (0.98,1.19) | 0.118 | 0.661 | 1.07 (0.96,1.20) | 0.241 | 0.670 | 6 | 1.28 (0.96,1.69) | 0.090 | 0.717 |
| Chemokine receptor 3 [601268] | rs5742906 | CCR3 39Pro>Leu | 8542 | 5 | 0.86 (0.37,2.02) | 0.730 | 0.565 | 0.86 (0.37,2.02) | 0.730 | 0.565 | No variant homozygotes observed | |||
| Chemokine receptor 5 [601373] | rs1799987 | CCR5 (−2459)A>G | 8541 | 6 | 1.01 (0.94,1.08) | 0.850 | 0.040 | 1.00 (0.89,1.12) | 0.960 | 0.045 | 6 | 1.02 (0.91,1.15) | 0.701 | 0.095 |
| Chemokine receptor 5 [601373] | rs333 | CCR5 580Ins>Del32 | 8523 | 5 | 0.94 (0.81,1.08) | 0.372 | 0.264 | 0.94 (0.81,1.10) | 0.461 | 0.174 | 5 | 0.82 (0.44,1.51) | 0.517 | 0.706 |
| Cholesteryl ester transfer protein, plasma [118470] | rs1800776 | CETP (−631)C>A | 10088 | 6 | 1.00 (0.87,1.15) | 0.977 | 0.616 | 1.02 (0.88,1.19) | 0.792 | 0.718 | 5 | 0.92 (0.46,1.83) | 0.800 | 0.565 |
| Cholesteryl ester transfer protein, plasma [118470] | rs1800775 | CETP (−629)C>A | 10076 | 6 | 0.95 (0.90,1.01) | 0.129 | 0.203 | 0.90 (0.81,1.00) | 0.046 | 0.595 | 6 | 0.98 (0.88,1.08) | 0.653 | 0.134 |
| Cholesteryl ester transfer protein, plasma [118470] | rs5882 | CETP 405Ile>Val | 10086 | 6 | 0.99 (0.92,1.05) | 0.661 | 0.748 | 0.99 (0.91,1.09) | 0.895 | 0.635 | 6 | 0.96 (0.84,1.09) | 0.498 | 0.791 |
| Cholesteryl ester transfer protein, plasma [118470] | rs2303790 | CETP 442Asp>Gly | 10089 | 5 | 1.28 (0.87,1.89) | 0.207 | 1.000 | 1.41 (0.93,2.13) | 0.108 | 1.000 | Too few variant homozygotes observed | |||
| Coagulation factor II (thrombin) [176930] | rs1799963 | F2 20210G>A | 9902 | 5 | 1.01 (0.74,1.39) | 0.941 | 0.108 | 1.02 (0.74,1.40) | 0.928 | 0.099 | Too few variant homozygotes observed | |||
| Coagulation factor V (proaccelerin, labile factor) [227400] | rs6025 | F5 506Arg>Gln | 9941 | 6 | 0.96 (0.77,1.20) | 0.712 | 0.867 | 0.96 (0.76,1.20) | 0.698 | 0.848 | Too few variant homozygotes observed | |||
| Coagulation factor VII (serum prothrombin conversion accelerator) [227500] | rs5742910 | F7 (−323) Del>Ins10 | 9941 | 6 | 0.99 (0.89,1.11) | 0.909 | 0.057 | 1.00 (0.89,1.12) | 0.984 | 0.072 | 6 | 1.05 (0.67,1.64) | 0.832 | 0.908 |
| Coagulation factor VII (serum prothrombin conversion accelerator) [227500] | rs6046 | F7 353Arg>Gln | 9938 | 6 | 0.96 (0.86,1.08) | 0.510 | 0.193 | 0.97 (0.86,1.10) | 0.638 | 0.163 | 6 | 0.83 (0.53,1.31) | 0.427 | 0.736 |
| Complement component 3 [120700] | rs2230199 | C3 102Arg>Gly | 6083 | 5 | 0.99 (0.87,1.13) | 0.891 | 0.488 | 0.96 (0.83,1.12) | 0.609 | 0.429 | 5 | 1.18 (0.80,1.76) | 0.408 | 0.798 |
| Complement component 5 [120900] | rs17611 | C5 802Val>Ile | 6117 | 5 | 1.02 (0.95,1.10) | 0.604 | 0.087 | 1.07 (0.94,1.21) | 0.328 | 0.162 | 5 | 0.99 (0.88,1.12) | 0.897 | 0.328 |
| Colony stimulating factor 2 (granulocyte-macrophage) [138960] | rs25882 | CSF2 117Ile>Thr | 6127 | 5 | 1.00 (0.92,1.09) | 0.961 | 0.610 | 1.03 (0.91,1.17) | 0.601 | 0.285 | 5 | 0.96 (0.83,1.12) | 0.626 | 0.535 |
| Cytotoxic T-lymphocyte-associated protein 4 [123890] | rs5742909 | CTLA4 (−318)C>T | 6119 | 5 | 0.91 (0.80,1.02) | 0.100 | 0.688 | 0.91 (0.79,1.03) | 0.135 | 0.544 | 5 | 0.81 (0.51,1.30) | 0.387 | 0.197 |
| Cytotoxic T-lymphocyte-associated protein 4 [123890] | rs231775 | CTLA4 17Thr>Ala | 6128 | 5 | 1.02 (0.94,1.11) | 0.580 | 0.139 | 0.99 (0.87,1.13) | 0.862 | 0.154 | 5 | 1.07 (0.94,1.21) | 0.301 | 0.251 |
| Fibrinogen beta [134830] | rs1800790 | FGB (−455)G>A | 9933 | 6 | 1.01 (0.93,1.09) | 0.905 | 0.877 | 1.01 (0.92,1.11) | 0.866 | 0.646 | 6 | 1.01 (0.81,1.25) | 0.963 | 0.503 |
| Group-specific component (vitamin D binding protein) [139200] | rs7041 | GC 416Glu>Asp | 6121 | 5 | 1.02 (0.94,1.10) | 0.719 | 0.224 | 0.97 (0.85,1.12) | 0.718 | 0.306 | 5 | 1.05 (0.93,1.19) | 0.395 | 0.336 |
| Group-specific component (vitamin D binding protein) [139200] | rs4588 | GC 420Thr>Lys | 6122 | 5 | 1.02 (0.94,1.11) | 0.598 | 0.436 | 1.01 (0.89,1.14) | 0.928 | 0.382 | 5 | 1.06 (0.92,1.21) | 0.437 | 0.443 |
| Guanine nucleotide binding protein (G protein), beta polypeptide 3 [139130] | rs5443 | GNB3 825C>T | 9941 | 6 | 1.08 (1.01,1.15) | 0.031 | 0.759 | 1.06 (0.96,1.16) | 0.243 | 0.864 | 6 | 1.19 (1.05,1.36) | 0.009 | 0.134 |
| Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) [192974] | rs1062535 | ITGA2 873G>A | 9941 | 6 | 1.04 (0.97,1.11) | 0.268 | 0.157 | 1.04 (0.95,1.14) | 0.367 | 0.327 | 6 | 1.06 (0.93,1.21) | 0.356 | 0.163 |
| Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) [173470] | rs5918 | ITGB3 33Leu>Pro | 9937 | 6 | 0.98 (0.88,1.08) | 0.648 | 0.349 | 0.97 (0.87,1.10) | 0.662 | 0.423 | 5 | 1.00 (0.70,1.44) | 0.993 | 0.375 |
| Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor [147840] | rs5491 | ICAM1 56Lys>Met | 8531 | 6 | 1.04 (0.84,1.29) | 0.717 | 0.928 | 1.05 (0.84,1.31) | 0.690 | 0.940 | 3 | 0.98 (0.26,3.70) | 0.980 | 0.907 |
| Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor [147840] | rs1799969 | ICAM1 241Gly>Arg | 10088 | 6 | 1.09 (0.97,1.23) | 0.139 | 0.466 | 1.09 (0.95,1.24) | 0.208 | 0.398 | 6 | 1.31 (0.86,1.99) | 0.203 | 0.916 |
| Interleukin 1, alpha [147760] | rs1800587 | IL1A (−889)C>T | 8508 | 6 | 0.93 (0.85,1.02) | 0.110 | 0.778 | 0.91 (0.82,1.01) | 0.083 | 0.834 | 6 | 0.95 (0.76,1.19) | 0.659 | 0.686 |
| Interleukin 1, beta [147720] | rs16944 | IL1B (−1418)C>T | 6094 | 5 | 1.05 (0.97,1.13) | 0.240 | 0.506 | 1.06 (0.95,1.19) | 0.288 | 0.473 | 5 | 1.06 (0.92,1.23) | 0.407 | 0.541 |
| Interleukin 1, beta [147720] | rs1143634 | IL1B 4336C>T | 8534 | 6 | 0.95 (0.86,1.05) | 0.331 | 0.415 | 0.92 (0.82,1.04) | 0.188 | 0.286 | 6 | 1.07 (0.81,1.41) | 0.642 | 0.595 |
| Interleukin 4 [147780] | rs2243250 | IL4 (−590)C>T | 8541 | 6 | 1.05 (0.97,1.15) | 0.237 | 0.015 | 1.19 (1.05,1.35) | 0.007 | 0.101 | 6 | 0.92 (0.8,1.07) | 0.275 | 0.081 |
| Interleukin 4 receptor [147781] | rs1805010 | IL4R 75Ile>Val | 8538 | 6 | 1.00 (0.93,1.07) | 0.987 | 0.910 | 1.02 (0.91,1.14) | 0.722 | 0.790 | 6 | 0.98 (0.87,1.10) | 0.720 | 0.915 |
| Interleukin 4 receptor [147781] | rs1805015 | IL4R 503Ser>Pro | 6130 | 5 | 1.00 (0.89,1.12) | 0.945 | 0.286 | 1.00 (0.88,1.14) | 0.979 | 0.536 | 5 | 1.00 (0.63,1.59) | 0.997 | 0.168 |
| Interleukin 4 receptor [147781] | rs1801275 | IL4R 576Gln>Arg | 8525 | 6 | 1.04 (0.95,1.13) | 0.437 | 0.058 | 1.06 (0.96,1.18) | 0.247 | 0.088 | 6 | 0.92 (0.70,1.21) | 0.546 | 0.273 |
| Interleukin 5 receptor alpha [147851] | rs2290608 | IL5RA (−80)G>A | 8528 | 6 | 1.02 (0.94,1.11) | 0.608 | 0.182 | 1.02 (0.92,1.12) | 0.754 | 0.275 | 6 | 1.08 (0.87,1.34) | 0.482 | 0.114 |
| Interleukin 6 [147620] | rs1800796 | IL6 (−572)G>C | 6124 | 5 | 0.92 (0.82,1.02) | 0.103 | 0.061 | 0.84 (0.70,1.00) | 0.052 | 0.113 | 4 | 0.95 (0.81,1.10) | 0.480 | 1.000 |
| Interleukin 6 [147620] | rs1800795 | IL6 (−174)G>C | 8543 | 6 | 1.08 (0.99,1.18) | 0.095 | 0.687 | 1.08 (0.95,1.23) | 0.232 | 0.248 | 5 | 1.14 (0.97,1.34) | 0.121 | 0.718 |
| Interleukin 9 [146931] | rs2069885 | IL9 113Thr>Met | 8544 | 6 | 1.04 (0.92,1.18) | 0.559 | 0.543 | 1.10 (0.95,1.26) | 0.209 | 0.772 | 5 | 0.73 (0.44,1.19) | 0.205 | 0.131 |
| Interleukin 10 [124092] | rs1800872 | IL10 (−571)C>A | 8538 | 6 | 0.96 (0.89,1.04) | 0.307 | 0.012 | 0.96 (0.86,1.07) | 0.463 | 0.028 | 6 | 0.94 (0.82,1.08) | 0.375 | 0.190 |
| Interleukin 13 [147683] | rs1295686 | IL13 4045C>T | 5366 | 4 | 1.01 (0.92,1.11) | 0.894 | 0.751 | 0.99 (0.89,1.12) | 0.917 | 0.854 | 4 | 1.07 (0.85,1.35) | 0.561 | 0.380 |
| Leukotriene C4 synthase [246530] | rs730012 | LTC4S (−444)A>C | 6044 | 5 | 0.99 (0.90,1.09) | 0.797 | 0.241 | 1.01 (0.91,1.13) | 0.874 | 0.193 | 5 | 0.88 (0.69,1.13) | 0.308 | 0.940 |
| Lipase, hepatic [151670] | rs1800588 | LIPC (−480)C>T | 10092 | 6 | 0.93 (0.87,1.00) | 0.048 | 0.468 | 0.94 (0.86,1.03) | 0.182 | 0.636 | 6 | 0.85 (0.71,1.00) | 0.051 | 0.091 |
| Lipoprotein, Lp(a) [152200] | rs1853021 | LPA 93C>T | 10082 | 6 | 0.96 (0.88,1.04) | 0.309 | 0.524 | 0.97 (0.88,1.06) | 0.475 | 0.588 | 6 | 0.89 (0.66,1.19) | 0.424 | 0.155 |
| Lipoprotein, Lp(a) [152200] | rs1800769 | LPA 121G>A | 10080 | 6 | 0.95 (0.88,1.03) | 0.206 | 0.404 | 0.94 (0.85,1.03) | 0.194 | 0.647 | 6 | 0.97 (0.81,1.15) | 0.680 | 0.127 |
| Lipoprotein lipase [609708] | rs1800590 | LPL (−93)T>G | 10091 | 6 | 1.12 (0.84,1.49) | 0.433 | 0.460 | 1.13 (0.83,1.54) | 0.444 | 0.450 | 3 | 1.64 (0.42,6.36) | 0.473 | 0.861 |
| Lipoprotein lipase [609708] | rs1801177 | LPL 9Asp>Asn | 10091 | 6 | 1.15 (0.80,1.65) | 0.464 | 0.610 | 1.15 (0.80,1.66) | 0.443 | 0.611 | Too few variant homozygotes observed | |||
| Lipoprotein lipase [609708] | rs268 | LPL 291Asn>Ser | 10090 | 6 | 1.24 (0.94,1.64) | 0.133 | 0.887 | 1.29 (0.97,1.72) | 0.080 | 0.925 | Too few variant homozygotes observed | |||
| Lipoprotein lipase [609708] | rs328 | LPL 447Ser>Term | 10088 | 6 | 0.89 (0.80,0.99) | 0.033 | 0.455 | 0.88 (0.79,0.99) | 0.033 | 0.542 | 6 | 0.89 (0.57,1.40) | 0.623 | 0.244 |
| Low density lipoprotein receptor [606945] | rs5742911 | LDLR NcoI +/− | 10086 | 6 | 1.01 (0.94,1.08) | 0.882 | 0.869 | 0.99 (0.91,1.09) | 0.860 | 0.862 | 6 | 1.05 (0.91,1.21) | 0.540 | 0.472 |
| Lymphotoxin alpha; Tumor necrosis factor beta [153440] | rs909253 | LTA 252A>G | 6090 | 5 | 1.02 (0.94,1.10) | 0.682 | 0.404 | 1.07 (0.96,1.20) | 0.226 | 0.561 | 5 | 0.93 (0.80,1.09) | 0.381 | 0.294 |
| Lymphotoxin alpha; Tumor necrosis factor beta [153440] | rs1041981 | LTA 26Thr>Asn | 10091 | 6 | 1.01 (0.95,1.08) | 0.742 | 0.387 | 1.02 (0.93,1.11) | 0.715 | 0.274 | 6 | 1.01 (0.88,1.16) | 0.872 | 0.344 |
| Membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) [147138] | rs569108 | MS4A2 237Glu>Gly | 8517 | 6 | 1.08 (0.95,1.23) | 0.256 | 0.479 | 1.10 (0.95,1.27) | 0.220 | 0.480 | 3 | 1.03 (0.65,1.64) | 0.891 | 0.999 |
| 5, 10-methylenetetrahydrofolate reductase [607093] | rs1801133 | MTHFR 677C>T | 9943 | 6 | 1.03 (0.96,1.10) | 0.383 | 0.389 | 1.05 (0.95,1.15) | 0.360 | 0.514 | 6 | 1.03 (0.91,1.16) | 0.629 | 0.165 |
| Natriuretic peptide precursor A [108780] | rs5063 | NPPA 664G>A | 9940 | 6 | 0.96 (0.83,1.11) | 0.555 | 0.016 | 0.94 (0.81,1.10) | 0.448 | 0.022 | 5 | 1.54 (0.73,3.26) | 0.257 | 0.689 |
| Natriuretic peptide precursor A [108780] | rs5065 | NPPA 2238T>C | 9939 | 6 | 1.03 (0.92,1.15) | 0.612 | 0.781 | 1.05 (0.93,1.18) | 0.452 | 0.924 | 6 | 0.89 (0.59,1.33) | 0.560 | 0.453 |
| Nitric oxide synthase 2A (inducible, hepatocytes) asp346asp [163730] | rs1137933 | NOS2A 231C>T | 6131 | 5 | 0.91 (0.82,1.01) | 0.062 | 0.659 | 0.91 (0.81,1.02) | 0.111 | 0.439 | 5 | 0.81 (0.60,1.09) | 0.166 | 0.516 |
| Nitric oxide synthase 3 (endothelial cell) [163729] | rs1800779 | NOS3 (−922)A>G | 10093 | 6 | 1.02 (0.95,1.10) | 0.575 | 0.676 | 1.03 (0.93,1.14) | 0.562 | 0.358 | 6 | 1.03 (0.88,1.20) | 0.714 | 0.129 |
| Nitric oxide synthase 3 (endothelial cell) [163729] | rs3918226 | NOS3 (−690)C>T | 9936 | 6 | 1.15 (1.00,1.32) | 0.047 | 0.479 | 1.16 (1.00,1.34) | 0.052 | 0.562 | 5 | 1.30 (0.70,2.41) | 0.408 | 0.726 |
| Nitric oxide synthase 3 [163729] | rs1799983 | NOS3 298Glu>Asp | 10094 | 6 | 1.05 (0.97,1.13) | 0.224 | 0.176 | 1.05 (0.95,1.15) | 0.327 | 0.145 | 6 | 1.10 (0.92,1.31) | 0.293 | 0.569 |
| Paraoxonase 1 [168820] | rs854560 | PON1 55Leu>Met | 10090 | 6 | 0.97 (0.91,1.05) | 0.467 | 0.510 | 0.97 (0.88,1.08) | 0.584 | 0.495 | 6 | 0.95 (0.81,1.12) | 0.554 | 0.157 |
| Paraoxonase 1 [168820] | rs662 | PON1 192Gln>Arg | 10088 | 6 | 1.05 (0.98,1.13) | 0.155 | 0.774 | 1.07 (0.97,1.18) | 0.167 | 0.463 | 6 | 1.05 (0.93,1.20) | 0.412 | 0.268 |
| Paraoxonase 2 [602447] | rs6954345 | PON2 311Ser>Cys | 10092 | 6 | 1.09 (1.01,1.18) | 0.025 | 0.793 | 1.09 (1.00,1.20) | 0.060 | 0.628 | 6 | 1.22 (0.99,1.49) | 0.063 | 0.460 |
| Peroxisome proliferator activated-receptor gamma [601487] | rs1801282 | PPARG 12Pro>Ala | 10093 | 6 | 1.05 (0.95,1.16) | 0.362 | 0.702 | 1.07 (0.96,1.20) | 0.220 | 0.425 | 6 | 0.88 (0.59,1.32) | 0.532 | 0.490 |
| Secretoglobin, family 1A, member 1 (uteroglobin) [192020] | rs3741240 | SCGB1A1 (+38)G>A | 8538 | 6 | 0.99 (0.92,1.06) | 0.797 | 0.103 | 0.98 (0.89,1.09) | 0.701 | 0.431 | 6 | 1.01 (0.88,1.16) | 0.884 | 0.040 |
| Selectin E (endothelial adhesion molecule 1) [131210] | rs5361 | SELE 128Ser>Arg | 10094 | 6 | 1.00 (0.89,1.13) | 0.965 | 0.666 | 1.01 (0.89,1.15) | 0.903 | 0.642 | 6 | 1.05 (0.61,1.81) | 0.850 | 0.673 |
| Selectin E (endothelial adhesion molecule 1) [131210] | rs5355 | SELE 554Leu>Phe | 9904 | 6 | 0.91 (0.76,1.08) | 0.266 | 0.004 | 0.92 (0.77,1.10) | 0.362 | 0.005 | 5 | 0.37 (0.07,1.88) | 0.228 | 0.998 |
| Selectin P (granule membrane protein 140kDa, antigen CD62) [173610] | rs6131 | SELP 330Ser>Asn | 8539 | 6 | 1.04 (0.95,1.13) | 0.424 | 0.296 | 1.03 (0.93,1.14) | 0.565 | 0.483 | 6 | 1.12 (0.88,1.43) | 0.365 | 0.506 |
| Selectin P (granule membrane protein 140kDa, antigen CD62) [173610] | rs6133 | SELP 640Val>Leu | 8537 | 6 | 1.09 (0.95,1.25) | 0.229 | 0.037 | 1.10 (0.94,1.28) | 0.226 | 0.028 | 5 | 1.22 (0.71,2.12) | 0.471 | 0.876 |
| Serine (or cysteine) proteinase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 [173360] | rs1799768 | SERPINE1 (−675)Del>InsG | 9931 | 6 | 0.99 (0.93,1.06) | 0.769 | 0.541 | 1.04 (0.95,1.15) | 0.387 | 0.525 | 6 | 0.92 (0.82,1.03) | 0.126 | 0.669 |
| Serine (or cysteine) proteinase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 [173360] | rs7242 | SERPINE1 11053T>G | 9940 | 6 | 0.99 (0.93,1.05) | 0.699 | 0.266 | 1.02 (0.92,1.12) | 0.766 | 0.257 | 6 | 0.95 (0.84,1.06) | 0.324 | 0.413 |
| Sodium channel, nonvoltage-gated 1 alpha [600228] | rs5742912 | SCNN1A 493Trp>Arg | 9940 | 6 | 0.99 (0.76,1.27) | 0.907 | 0.004 | 0.97 (0.75,1.26) | 0.817 | 0.002 | 4 | 1.73 (0.10,28.9) | 0.705 | 1.000 |
| Sodium channel, nonvoltage-gated 1 alpha [600228] | rs2228576 | SCNN1A 663Ala>Thr | 9932 | 6 | 1.00 (0.93,1.07) | 0.958 | 0.958 | 1.00 (0.91,1.10) | 1.000 | 0.921 | 6 | 1.00 (0.87,1.14) | 0.939 | 0.275 |
| Matrix metalloproteinase 3 (stromelysin 1, progelatinase) [185250] | rs3025058 | MMP3 (−1171) Ins>DelA | 9932 | 6 | 0.97 (0.91,1.04) | 0.384 | 0.074 | 0.95 (0.85,1.05) | 0.279 | 0.126 | 6 | 0.98 (0.87,1.11) | 0.801 | 0.273 |
| Transcription factor 7 (T-cell specific, HMG-box) [189908] | rs244656 | TCF7 (−1459)A>T | 6118 | 5 | 0.96 (0.86,1.07) | 0.438 | 0.033 | 0.97 (0.86,1.10) | 0.674 | 0.037 | 5 | 0.75 (0.50,1.14) | 0.174 | 0.610 |
| Transcription factor 7 (T-cell specific, HMG-box) [189908] | rs5742913 | TCF7 19Pro>Thr | 6117 | 5 | 1.05 (0.88,1.25) | 0.614 | 0.404 | 1.04 (0.86,1.25) | 0.724 | 0.498 | 4 | 1.40 (0.65,3.02) | 0.388 | 0.570 |
| Transforming growth factor, beta 1 [190180] | rs1800469 | TGFB1 (−509)C>T | 8486 | 6 | 0.92 (0.86,0.99) | 0.028 | 0.750 | 0.89 (0.8,0.98) | 0.023 | 0.148 | 6 | 0.92 (0.81,1.06) | 0.252 | 0.658 |
| Tumor necrosis factor (TNF superfamily, member 2) [191160] | rs1800750 | TNF (−376)G>A | 9941 | 6 | 0.72 (0.50,1.04) | 0.077 | 0.371 | 0.72 (0.5,1.04) | 0.081 | 0.363 | Too few variant homozygotes observed | |||
| Tumor necrosis factor (TNF superfamily, member 2) [191160] | rs1800629 | TNF (−308)G>A | 10096 | 6 | 1.04 (0.95,1.15) | 0.395 | 0.253 | 1.04 (0.93,1.16) | 0.479 | 0.326 | 6 | 1.14 (0.83,1.59) | 0.420 | 0.506 |
| Tumor necrosis factor (TNF superfamily, member 2) [191160] | rs673 | TNF (−244)G>A | 9938 | 5 | 1.51 (0.38,6.01) | 0.562 | 0.944 | 1.51 (0.38,6.01) | 0.562 | 0.944 | Too few variant homozygotes observed | |||
| Tumor necrosis factor (TNF superfamily, member 2) [191160] | rs361525 | TNF (−238)G>A | 10050 | 6 | 1.02 (0.88,1.18) | 0.815 | 0.231 | 1.02 (0.87,1.19) | 0.797 | 0.280 | 6 | 1.04 (0.38,2.82) | 0.946 | 0.694 |
| Vascular cell adhesion molecule 1 [192225] | rs1041163 | VCAM1 (−1594)T>C | 8525 | 6 | 0.98 (0.89,1.07) | 0.595 | 0.891 | 0.99 (0.89,1.10) | 0.805 | 0.871 | 6 | 0.86 (0.64,1.16) | 0.325 | 0.967 |
| Vitamin D (1,25- dihydroxyvitamin D3) receptor [601769] | rs2228570 | VDR 1Thr>Met | 6104 | 5 | 1.07 (0.99,1.16) | 0.078 | 0.424 | 1.12 (1.00,1.25) | 0.057 | 0.867 | 5 | 1.07 (0.93,1.23) | 0.367 | 0.237 |
| Vitamin D (1,25- dihydroxyvitamin D3) receptor [601769] | rs1544410 | VDR 45082 G>A | 6120 | 5 | 1.01 (0.92,1.12) | 0.798 | 0.504 | 1.00 (0.88,1.14) | 0.974 | 0.173 | 5 | 1.06 (0.87,1.31) | 0.558 | 0.508 |
Total number of genotypes available across all six study populations.
Number of studies observing sufficient polymorphism at SNP site for analysis under additive and dominant modes.
OR = Odds Ratio, LCL = Lower confidence limit, UCL = Upper confidence limit, p= corresponding p-value
P-value for heterogeneity of the genetic effect across all studies.
Number of studies observing sufficient polymorphism at SNP site for analysis under recessive mode.
The data were then stratified on age, sex, hypertension or smoking status. No statistically significant associations were observed in the age- (Supplementary Table 2A) or sex-stratified (Supplementary Table 2B) analyses, nor among those with current or past hypertension (Table 3) after adjusting for the FDR. In contrast, a large number of nominally significant associations with ischemic stroke among normotensives were observed (Table 4). The strongest associations under the additive and dominant models were for LTA 252A>G and LTA 26Thr>Asn, two SNPs in strong linkage disequilibrium, while NOS3 298Glu>Asp had the strongest association under the recessive model. After adjusting for FDR and permutation testing, only the LTA 252A>G SNP showed significant association among those without hypertension. In the additive mode, the estimated relative risk across the three LTA 252 genotypes was 1.41 (p=0.00002) in the fixed effects analysis and the FDR was 0.002, with p<0.01 in permutation testing. Results for the dominant model were similar (OR = 1.57, FDR = 0.005). In the random effects meta-analysis (data not shown), the LTA 252A>G association with stroke under the dominant model (OR = 1.56) had an FDR of 0.02. Among Caucasians only, LTA 252A>G was similarly associated with ischemic stroke among those without hypertension under additive and dominant models (OR = 1.28, p=0.016 and OR = 1.39, p=0.019, respectively). Minor allele frequencies among non-hypertensive controls are given in Table 4; frequencies among non-hypertensive cases were 0.37, 0.38, 0.31, 0.41, and 0.46 in SOF, Vienna, Westphalia, Pomerania, and SHINING, respectively.
Table 3.
Nominally significant (P<0.05) fixed-effects meta-analysis results among those with current or past hypertension
| SNP | Mode* | # Studies | OR† | Lower CL† | Upper CL† | p | FDR‡ |
|---|---|---|---|---|---|---|---|
| LPA 121G>A | ADD | 6 | 0.886 | 0.801 | 0.979 | 0.017 | 0.915 |
| TGFB1 (−509)C>T | ADD | 6 | 0.894 | 0.812 | 0.983 | 0.021 | 0.915 |
| CTLA4 (−318)C>T | ADD | 5 | 0.851 | 0.730 | 0.993 | 0.040 | 0.915 |
| TGFB1 (−509)C>T | DOM | 6 | 0.845 | 0.737 | 0.969 | 0.016 | 0.968 |
| LPA 121G>A | DOM | 6 | 0.872 | 0.768 | 0.992 | 0.037 | 0.968 |
| APOC3 3175C>G | REC | 6 | 0.708 | 0.514 | 0.975 | 0.034 | 0.824 |
| SERPINE1 (−675)Del>InsG | REC | 6 | 0.851 | 0.731 | 0.991 | 0.038 | 0.824 |
| LTA 252A>G | REC | 5 | 0.806 | 0.657 | 0.989 | 0.039 | 0.824 |
| ITGA2 873G>A | REC | 6 | 1.202 | 1.003 | 1.442 | 0.047 | 0.824 |
ADD = additive, DOM = dominant, REC = recessive mode of inheritance
Odds ratio (OR) and confidence limits (CL)
False discovery rate
Table 4.
SNPs achieving P<0.05 for association with ischemic stroke among normotensives (1068 cases, 3390 controls) under three modes of inheritance.
| PHS (163 cs, 1522 ctrl)* | SOF (50 cs, 212 ctrl)* | Vienna (232 cs, 632 ctrl) | Westphalia (224 cs, 444 ctrl) | Pomerania (92 cs, 330 ctrl) | SHINING (307 cs, 250 ctrl) | Meta* | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mode | SNP | OR† | P | MAF‡ | OR† | P | MAF‡ | OR† | P | MAF‡ | OR† | P | MAF‡ | OR† | P | MAF‡ | OR† | P | MAF‡ | OR† | P | FDR† | Het§ |
| LTA 252A>G | not genotyped | 0.97 | 0.913 | 0.33 | 1.30 | 0.182 | 0.32 | 1.11 | 0.604 | 0.32 | 1.77 | 0.003 | 0.29 | 1.62 | <0.001 | 0.36 | 1.41 | <0.0001 | 0.002 | * | |||
| LTA 26Thr>Asn | 1.07 | 0.603 | 0.32 | 0.99 | 0.970 | 0.33 | 1.10 | 0.475 | 0.32 | 0.96 | 0.771 | 0.33 | 1.63 | 0.008 | 0.29 | 1.60 | <0.001 | 0.35 | 1.19 | 0.003 | 0.161 | 0.03 | |
| ACE Del>Ins | 0.83 | 0.107 | 0.45 | 0.86 | 0.508 | 0.49 | 0.93 | 0.518 | 0.47 | 0.69 | 0.003 | 0.55 | 0.98 | 0.884 | 0.50 | 0.97 | 0.826 | 0.65 | 0.86 | 0.007 | 0.244 | * | |
| CETP (−631)C>A | 0.53 | 0.036 | 0.07 | 0.93 | 0.871 | 0.09 | 0.99 | 0.976 | 0.09 | 0.45 | 0.003 | 0.08 | 0.78 | 0.47 | 0.08 | not polymorphic | 0.00 | 0.71 | 0.009 | 0.244 | * | ||
| SELP 640Val>Leu | 1.73 | <0.001 | 0.11 | 1.20 | 0.618 | 0.12 | 0.91 | 0.759 | 0.11 | 1.02 | 0.947 | 0.11 | 0.92 | 0.777 | 0.11 | 1.52 | 0.637 | 0.004 | 1.31 | 0.013 | 0.255 | * | |
| Additive | TCF7 19 Pro>Thr | not genotyped | 0.90 | 0.788 | 0.11 | 1.94 | 0.024 | 0.07 | 1.03 | 0.922 | 0.10 | 1.91 | 0.023 | 0.08 | not polymorphic | 0.00 | 1.45 | 0.017 | 0.255 | * | |||
| ADD 460Gly>Trp | 1.05 | 0.758 | 0.19 | 1.70 | 0.063 | 0.17 | 0.96 | 0.797 | 0.18 | 1.22 | 0.203 | 0.19 | 1.56 | 0.022 | 0.20 | 1.16 | 0.241 | 0.51 | 1.17 | 0.017 | 0.255 | * | |
| IL10 (−571)C>A | 0.65 | 0.006 | 0.24 | 0.53 | 0.034 | 0.26 | 1.15 | 0.508 | 0.23 | 0.78 | 0.249 | 0.26 | 1.02 | 0.915 | 0.22 | 0.93 | 0.548 | 0.63 | 0.84 | 0.020 | 0.255 | * | |
| AGT 235Met>Thr | 0.69 | 0.002 | 0.43 | 1.03 | 0.917 | 0.40 | 1.13 | 0.309 | 0.46 | 0.91 | 0.436 | 0.45 | 0.64 | 0.011 | 0.43 | 0.94 | 0.670 | 0.81 | 0.87 | 0.021 | 0.255 | 0.03 | |
| PON2 311Ser>Cys | 1.07 | 0.610 | 0.23 | 0.90 | 0.706 | 0.24 | 1.27 | 0.096 | 0.23 | 1.21 | 0.195 | 0.23 | 1.11 | 0.608 | 0.24 | 1.15 | 0.366 | 0.19 | 1.15 | 0.038 | 0.389 | * | |
| IL6 (−572)G>C | not genotyped | 1.07 | 0.894 | 0.05 | 0.74 | 0.485 | 0.06 | 0.54 | 0.189 | 0.06 | 1.30 | 0.499 | 0.05 | 0.76 | 0.039 | 0.72 | 0.79 | 0.041 | 0.389 | * | |||
| APOB 71Thr>Ile | 1.23 | 0.091 | 0.29 | 0.84 | 0.493 | 0.32 | 1.25 | 0.089 | 0.28 | 1.32 | 0.036 | 0.28 | 0.93 | 0.682 | 0.33 | 0.84 | 0.333 | 0.14 | 1.13 | 0.046 | 0.389 | * | |
| LTA 252A>G | not genotyped | 1.24 | 0.542 | 0.33 | 1.53 | 0.116 | 0.32 | 1.09 | 0.746 | 0.32 | 1.77 | 0.025 | 0.29 | 1.94 | <0.001 | 0.36 | 1.57 | <0.0001 | 0.005 | * | |||
| LTA 26Thr>Asn | 1.14 | 0.437 | 0.32 | 1.29 | 0.461 | 0.33 | 1.13 | 0.462 | 0.32 | 0.88 | 0.454 | 0.33 | 1.63 | 0.045 | 0.29 | 1.87 | 0.001 | 0.35 | 1.24 | 0.007 | 0.253 | 0.06 | |
| ADD1 460Gly>Trp | 1.14 | 0.443 | 0.19 | 1.50 | 0.235 | 0.17 | 0.97 | 0.860 | 0.18 | 1.40 | 0.066 | 0.19 | 1.57 | 0.059 | 0.20 | 1.35 | 0.161 | 0.51 | 1.25 | 0.008 | 0.253 | * | |
| SELP 640Val>Leu | 1.94 | <0.001 | 0.11 | 1.10 | 0.815 | 0.12 | 0.98 | 0.936 | 0.11 | 1.01 | 0.983 | 0.11 | 0.96 | 0.896 | 0.11 | 1.52 | 0.637 | 0.004 | 1.36 | 0.011 | 0.253 | * | |
| CETP (−631)C>A | 0.54 | 0.045 | 0.07 | 0.99 | 0.982 | 0.09 | 1.01 | 0.954 | 0.09 | 0.38 | 0.001 | 0.08 | 0.79 | 0.494 | 0.08 | not polymorphic | 0.00 | 0.71 | 0.011 | 0.253 | 0.08 | ||
| Dominant | IL4 (−590)C>T | 1.45 | 0.031 | 0.17 | 1.16 | 0.682 | 0.14 | 0.86 | 0.570 | 0.18 | 1.00 | 0.993 | 0.15 | 1.35 | 0.245 | 0.15 | 4.44 | 0.002 | 0.80 | 1.30 | 0.015 | 0.275 | 0.07 |
| TCF7 19 Pro>Thr | not genotyped | 0.96 | 0.920 | 0.11 | 2.14 | 0.017 | 0.07 | 0.96 | 0.888 | 0.10 | 1.84 | 0.038 | 0.08 | not polymorphic | 0.00 | 1.46 | 0.022 | 0.353 | * | ||||
| IL10 (−571)C>A | 0.62 | 0.007 | 0.24 | 0.48 | 0.042 | 0.26 | 1.24 | 0.388 | 0.23 | 0.78 | 0.356 | 0.26 | 0.86 | 0.569 | 0.22 | 1.06 | 0.821 | 0.63 | 0.80 | 0.028 | 0.363 | * | |
| PON2 311Ser>Cys | 0.99 | 0.948 | 0.23 | 0.89 | 0.720 | 0.24 | 1.31 | 0.116 | 0.23 | 1.38 | 0.073 | 0.23 | 1.12 | 0.646 | 0.24 | 1.27 | 0.184 | 0.19 | 1.19 | 0.031 | 0.363 | * | |
| AGT 235Met>Thr | 0.61 | 0.003 | 0.43 | 1.15 | 0.704 | 0.40 | 1.17 | 0.406 | 0.46 | 0.90 | 0.580 | 0.45 | 0.57 | 0.023 | 0.43 | 1.26 | 0.609 | 0.81 | 0.82 | 0.033 | 0.363 | 0.05 | |
| ACE Del>Ins | 0.86 | 0.390 | 0.45 | 0.66 | 0.241 | 0.49 | 0.949 | 0.778 | 0.47 | 0.74 | 0.149 | 0.55 | 0.93 | 0.794 | 0.50 | 0.73 | 0.249 | 0.65 | 0.83 | 0.046 | 0.413 | * | |
| APOE 112Cys>Arg | 1.18 | 0.369 | 0.14 | 1.19 | 0.648 | 0.13 | 1.23 | 0.327 | 0.11 | 1.54 | 0.025 | 0.14 | 1.09 | 0.754 | 0.12 | 0.92 | 0.700 | 0.11 | 1.20 | 0.048 | 0.413 | * | |
| NOS3 298Glu>Asp | 1.35 | 0.244 | 0.31 | 1.80 | 0.205 | 0.31 | 1.52 | 0.125 | 0.29 | 1.63 | 0.084 | 0.31 | 2.49 | 0.013 | 0.27 | 0.58 | 0.423 | 0.11 | 1.56 | 0.001 | 0.102 | * | |
| LTA 252A>G | not genotyped | 0.53 | 0.272 | 0.33 | 1.15 | 0.744 | 0.32 | 1.29 | 0.548 | 0.32 | 2.90 | 0.005 | 0.29 | 1.72 | 0.029 | 0.36 | 1.55 | 0.007 | 0.360 | * | |||
| Recessive | ACE Del>Ins | 0.65 | 0.066 | 0.45 | 1.04 | 0.926 | 0.49 | 0.85 | 0.420 | 0.47 | 0.48 | 0.001 | 0.55 | 1.01 | 0.98 | 0.50 | 1.09 | 0.635 | 0.65 | 0.80 | 0.020 | 0.536 | 0.06 |
| GNB3 825C>T | 0.97 | 0.918 | 0.32 | 2.07 | 0.164 | 0.30 | 1.03 | 0.928 | 0.31 | 1.75 | 0.045 | 0.31 | 1.21 | 0.661 | 0.28 | 1.42 | 0.112 | 0.44 | 1.31 | 0.028 | 0.569 | * | |
| LTA 26Thr>Asn | 0.96 | 0.882 | 0.32 | 0.51 | 0.254 | 0.33 | 1.10 | 0.737 | 0.32 | 1.20 | 0.533 | 0.33 | 2.50 | 0.013 | 0.29 | 1.80 | 0.017 | 0.35 | 1.31 | 0.032 | 0.569 | * | |
| APOB 71Thr>Ile | 1.80 | 0.016 | 0.29 | 1.14 | 0.805 | 0.32 | 0.88 | 0.676 | 0.28 | 1.51 | 0.154 | 0.28 | 1.14 | 0.721 | 0.33 | 1.09 | 0.897 | 0.14 | 1.33 | 0.039 | 0.592 | * | |
Fixed effect meta-analyses adjusted for age and sex were conducted based on the summary logistic regression results for each study site.Cs = cases, ctrl = controls.
OR= Odds Ratio, FDR = False Discovery Rate
Minor allele frequency observed among controls. For ACE, the insertion allele was defined as the minor allele based upon the PHS population.
P-values <0.1 for heterogeneity of the genetic effect across six studies.
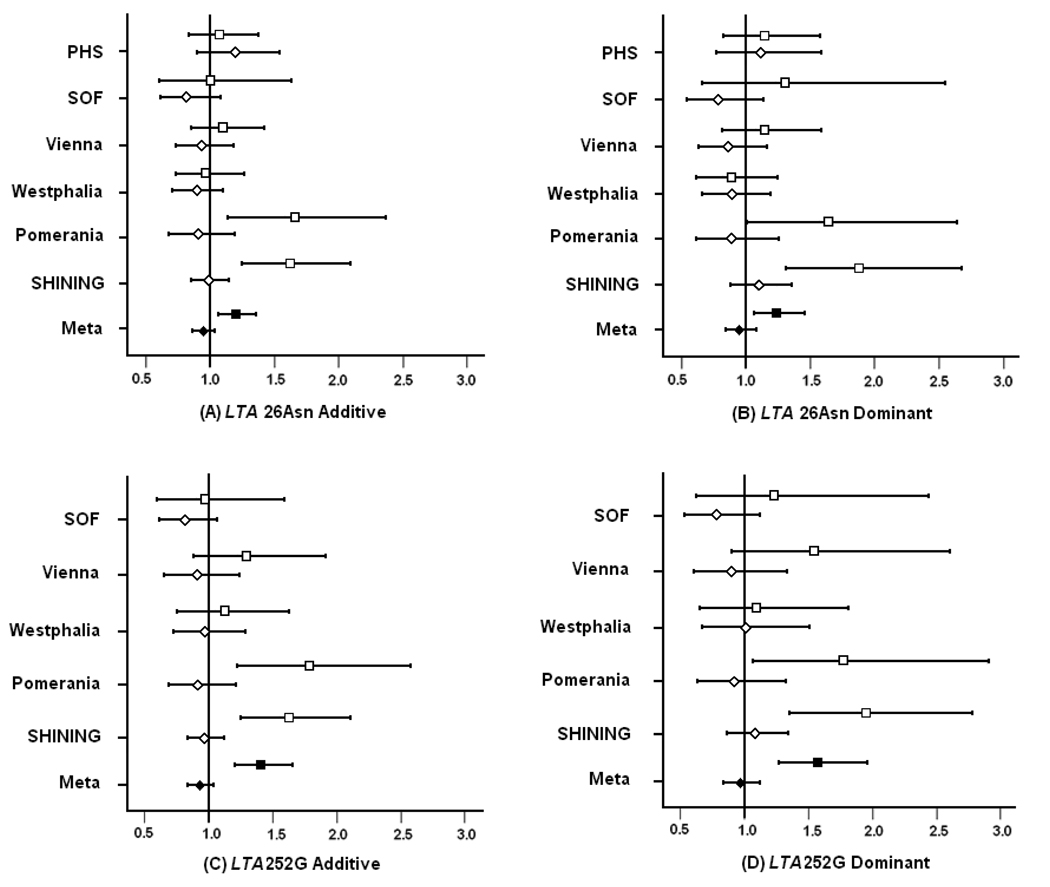
The point estimates for the OR were somewhat higher for LTA 252A>G, a polymorphism in intron 1, than for the non-synonymous polymorphism LTA 26Thr>Asn, although the confidence intervals overlapped after adjusting for age and sex (Figure1, A–D). The FDR values for the Thr>Asn polymorphism were also greater than 0.05. The associations with stroke risk for both LTA SNPs reached statistical significance among normotensives within the individual studies of SHINING and Pomerania, whereas among hypertensives, the OR point estimates were usually just below 1 and were not statistically significant (Figure 1). This trend for increased stroke risk associated with the LTA SNPs among normotensives relative to hypertensives was observed across the other studies, although none of these individual associations was statistically significant. We note that the PHS cohort was genotyped only for the LTA 26Thr>Asn polymorphism under the expectation that this coding SNP could be functional and would be an effective “tag” for LTA 252, based on the very strong LD between these two polymorphisms; furthermore, in the Vienna and Westphalia studies, some samples had missing genotypes for LTA 252A>G. When the meta-analysis was repeated with only those samples that had been genotyped for both LTA SNPs, the OR estimates were virtually identical (1.572 and 1.565 among normotensives under the dominant model for LTA 252 and LTA 26, respectively; data not shown). In addition, if the LTA 26 result for the PHS were imputed for the missing LTA 252 data, the additive result for LTA 252 would remain highly significant (OR=1.30, p=0.0001). Alternatively, if a completely null estimate for the PHS were imputed, the overall result would remain significant (OR=1.27, p=0.0004) and would continue to pass the stringent multiple comparisons testing.
Figure 1.
Risk of ischemic stroke associated with LTA polymorphism. Odds ratios under additive and dominant modes of inheritance for the LTA 252A>G and LTA 26Thr>Asn polymorphisms among normotensive (square) and hypertensive (diamond) subjects are plotted for each study (open squares/diamonds) and the fixed effects meta-analysis (filled squares/diamonds). Horizontal lines extend across the 95% confidence limits.
In the smoking-stratified analyses, no associations remained statistically significant after the false discovery rate was applied, although under the dominant model, CD14 (−260)C>T was suggestively associated (OR 1.24, P = 0.001, FDR = 0.058) with ischemic stroke among never-smokers (Supplementary Table 2C). Both LTA SNPs were associated with a greater risk for ischemic stroke in never-smokers than ever-smokers under the dominant model and although these associations were not statistically significant after accounting for multiple testing, this trend was consistent across five studies (data not shown); the Westphalian study was excluded due to limited smoking data.
Discussion
In this meta-analysis, we evaluated the association between 105 polymorphisms in 64 inflammation and cardiovascular-related genes and ischemic stroke in 3550 case and 6560 control subjects across six different studies. Although we could not further define subtypes of ischemic stroke, key strengths of our stroke consortium are that these analyses were not subject to publication bias and all studies used common genotyping reagents. In the primary meta-analysis, modest associations with stroke became non-significant after adjustment for multiple testing using the FDR or permutation testing. Stratification on sex or age also revealed no significant associations. Notably, subjects in two of our studies were limited to one sex and the consortium encompassed subjects recruited from different regions in Europe, North America, and China. We observed similar results among Caucasian participants only, but study population differences resulting in heterogeneity in stroke etiology could have obscured genetic associations.
Stratification on hypertension status did, however, reveal a statistically significant association for LTA 252A>G that remained after adjustment for multiple testing. Across four Caucasian populations and one Chinese population, the odds ratio for LTA 252G was consistently greater among normotensive than hypertensive subjects. LTA 26Thr>Asn yielded similar results among study participants with genotypes at both sites, as expected, given the strong linkage disequilibrium between these two LTA SNPs. Although a recent Japanese study observed no association of these SNPs with any subtype of ischemic stroke30, a smaller Hungarian study had previously reported LTA 252G as a risk factor for large-vessel ischemic stroke31 and an earlier Korean study had identified the LTA 252AA genotype as a risk factor for cerebral infarction32. We were unable to analyze ischemic stroke subtypes, but there is some evidence that subtypes may differ depending upon hypertensive status33. Although the number of non-hypertensives cases was limited to 1068, stratification by hypertension across our six populations may have reduced heterogeneity and thus enabled us to discern the modest risk associated with LTA polymorphism.
A role for LTA in chronic inflammation has been suggested by its ability to induce expression of ICAM-1 and VCAM-1 on endothelial cells in vitro34,35. LTA expression results in a localized infiltrate consisting of T cells, B cells, follicular and interdigitating dendritic cells and macrophages36. A recent mouse model study indicated that LTA was expressed in atherosclerotic lesions whose size correlated with concentration. Moreover, loss of the adjacent gene TNF did not affect development of lesions in mice fed an atherogenic diet37. The A252G site is intronic, but has been associated with higher transcriptional activity in a luciferase assay, while the variant protein bearing the LTA 26 threonine to asparagine substitution has been observed to induce greater expression of VCAM1 and SELE mRNA in vascular smooth-muscle cells. Since these two LTA SNPs are in almost complete LD, the variant protein level was estimated to be 1.5-fold higher than wildtype10. An increased level of the variant protein may contribute to the increased risk for ischemic stroke through inflammatory processes. Although the mechanism by which LTA polymorphisms influence inflammatory pathways is not clear, the meta-analysis presented here indicated that these LTA variants were associated with ischemic stroke in non-hypertensive patients.
It is believed that subjects with hypertension tend to develop chronic, low-grade systemic inflammation38–40. Severity of inflammation caused by genetic variation could independently modify predisposition to ischemic stroke. Recent reports on the association of PDE4D variants with ischemic stroke among normotensives3,4 are consistent with the hypothesis that hypertension may obscure or mask the effect of inflammation-related genetic variants and that such genetic effects can be most readily observed in the absence of this major risk factor.
Smoking, like hypertension, can elicit an inflammatory response41. In our study, the effect of LTA variation on stroke was more discernable among never-smokers than ever-smokers. Whether pro-inflammatory risks for ischemic stroke caused by hypertension, smoking, or carrying a risk allele are additive remains to be addressed by a carefully designed study.
Summary
Our six-study analysis surveyed inflammatory and cardiovascular gene polymorphisms in examining the risk for ischemic stroke. Our results indicate that the LTA 252A>G and LTA 26Thr>Asn polymorphisms have significant effects on the risk for ischemic stroke in non-hypertensive subjects. We cannot rule out the possible importance of these polymorphisms in hypertensive subjects, but a much larger cohort may be needed to clarify the interaction of hypertension and inflammation in the etiology of ischemic stroke.
Supplementary Material
On-line Supplementary Table 1. Frequencies of the less common or “variant” allele among the (A) control and ischemic stroke case (B) subgroups for each study. P values for large-sample and exact tests for Hardy-Weinberg equilibrium (HWE).
Online Supplementary Table 2. Nominally significant (P<0.05) fixed-effects meta-analysis results with (A) age, (B) sex, or (C) smoking-status stratification.
Acknowledgments
The RMS Stroke SNP Consortium (see Appendix for full list of investigators) thanks the staff and many participants of the Physician’s Health Study, Study of Osteoporotic Fractures, Vienna Stroke Registry, Westphalian Stroke Register, Dortmund Health Study, Pomeranian Stroke Study, Study of Health in Pomerania, and SHINING for their dedication and cooperation. We thank the RMS CVD project team for their expertise in developing the genotyping reagents used for these studies, the RMS High-Throughput Genotyping Group for their expert technical assistance in genotyping the SOF samples and Michael Janisiw for assistance in genotyping the Viennese samples.
The Physician’s Health Study (PHS) is supported by grants from the National Heart Lung and Blood Institute (HL-58755, and HL-63293), the Doris Duke Charitable Foundation, the American Heart Association, and the Donald W. Reynolds Foundation, Las Vegas, Nevada. Additional funding provided by the Fondation Leducq, Paris FR (Dr Ridker).
The Study of Osteoporotic Fractures (SOF) is supported by National Institutes of Health funding. The following institutes provide support: the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) and the National Institute on Aging (NIA) under the following grant numbers: AG05407, AR35582, AG05394, AR35584, AR35583, R01 AG005407, R01 AG027576-22, 2 R01 AG005394-22A1, and 2 R01 AG027574-22A1.
Case assessments in the Westphalian and Pomeranian studies are part of the German ‘Competence Net Stroke’ which is supported by the German Federal Ministry of Education and Research (01GI9909/3). Data collection in the Dortmund Health Study was funded by the German Migraine- and Headache Society and unrestricted grants of equal share of a consortium of 7 pharmaceutical companies. The Study of Health in Pomerania (SHIP) is funded by grants from the German Federal Ministry of Education and Research (BMBF,01ZZ96030), and from the Ministry for Education, Research and Cultural Affairs and the Ministry for Social Affairs of the Federal State of Mecklenburg-Vorpommern.
The Vienna Stroke Registry (VSR) is supported by research grants of the Medizinisch-Wissenschaftlicher Fonds des Bürgermeisters der Bundeshauptstadt Wien (project numbers 1540, 1829, 1970), of the Jubiläumsfonds der Oesterreichischen Nationalbank (project numbers 6866, 7115, 8281,9344), and the Austrian Science Foundation (P13902-MED). The VSR is also supported by the Wiener Krankenanstaltenverbund.
SHINING was funded through the Beijing Hypertension League Institute, in part through the National Infrastructure Program of Chinese Genetic Resource (2005DKA21300) and an unrestricted educational grant from F. Hoffmann-La Roche.
Design and data acquisition for initial individual genetic association studies: XW, SC, HAE, CM, KB, WL, WSB, YS, ERB, CK, JL, KL, LL, PMR, RYLZ, NRC
Design and interpretation of meta-analysis: XW, SC, VHB, HAE, CM, KB, WL, PMR, RYLZ, NRC
Statistical analysis: NRC
Manuscript writing and approval: all
Conflicts of Interest Disclosures: SC, VHB and HAE are employees of Roche Molecular Systems, Inc, which provided reagents and support for genotyping to all study sites under research collaborations and partial funding for the meta-analysis. KL is an employee of F. Hoffmann-La Roche, Ltd, which provided an unrestricted educational grant to the Beijing Hypertension League Institute.
Acknowledgments Appendix
RMS Stroke SNP Consortium Investigators:
Physicians’ Health Study (PHS)
Nancy R. Cook, Paul M Ridker and Robert Y. L. Zee from the Center for Cardiovascular Disease Prevention and Division of Preventive Medicine, Brigham and Women’s Hospital, Boston, MA, USA
Study of Osteoporotic Fractures (SOF)
Warren S. Browner, Steve R. Cummings and Li-Yung Lui from the S.F. Coordinating Center, California Pacific Medical Center, Research Institute, San Francisco, CA, USA
Vienna Stroke Study
Georg Endler and Christine Mannhalter from the Department of Medical and Chemical Laboratory Diagnostics, Medical University Vienna, Vienna, Austria; Stefan Greisenegger and
Wolfgang Lalouschek from the University Clinic of Neurology, Medical University Vienna, Vienna, Austria
Pomerania and Westphalia Studies
Klaus Berger, Institute of Epidemiology and Social Medicine, University of Muenster, Germany; Harald Funke, Department of Molecular Haemostaseology, University of Jena, Germany; E. Bernd Ringelstein, Department of Neurology, University of Muenster, Germany; Florian Stoegbauer, Department of Neurology, University of Muenster and Klinikum Osnabrück, Germany; Jan Luedemann, Institutes of Clinical Chemistry and Laboratory Medicine, University of Greifswald, Germany; Christoph Kessler, Department of Neurology, University of Greifswald, Germany
SHINING
Lisheng Liu, Yu Shi and Xingyu Wang from the Laboratory of Human Genetics, Beijing Hypertension League Institute, Beijing, China
Roche Molecular Systems (RMS)
Victoria H. Brophy, Suzanne Cheng, Henry A. Erlich, Andrea M. Johnson and Brian K. Rhees from the Department of Human Genetics, Roche Molecular Systems, Inc., Pleasanton, CA, USA
References
- 1.Rohr J, Kittner S, Feeser B, Hebel JR, Whyte MG, Weinstein A, Kanarak N, Buchholz D, Earley C, Johnson C, Macko R, Price T, Sloan M, Stern B, Wityk R, Wozniak M, Sherwin R. Traditional risk factors and ischemic stroke in young adults: the Baltimore-Washington Cooperative Young Stroke Study. Arch Neurol. 1996;53:603–607. doi: 10.1001/archneur.1996.00550070041010. [DOI] [PubMed] [Google Scholar]
- 2.Zhang LF, Yang J, Hong Z, Yuan GG, Zhou BF, Zhao LC, Huang YN, Chen J, Wu YF Collaborative Group of China Multicenter Study of Cardiovascular Epidemiology. Proportion of different subtypes of stroke in China. Stroke. 2003;34:2091–2096. doi: 10.1161/01.STR.0000087149.42294.8C. [DOI] [PubMed] [Google Scholar]
- 3.Brophy VH, Ro SK, Rhees BK, Lui LY, Lee JM, Umblas N, Bentley LG, Li J, Cheng S, Browner WS, Erlich HA. Association of phosphodiesterase 4D polymorphisms with ischemic stroke in a US population stratified by hypertension status. Stroke. 2006;37:1385–1390. doi: 10.1161/01.STR.0000221788.10723.66. [DOI] [PubMed] [Google Scholar]
- 4.Zee RY, Brophy VH, Cheng S, Hegener HH, Erlich HA, Ridker PM. Polymorphisms of the phosphodiesterase 4D, cAMP-specific (PDE4D) gene and risk of ischemic stroke: a prospective, nested case-control evaluation. Stroke. 2006;37:2012–2017. doi: 10.1161/01.STR.0000230608.56048.38. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Ma LY, Liu YX, Wang XY, Liu LS, Lindpaintner K. [Relationship between alpha-ENaC gene Thr663Ala polymorphism and ischemic stroke] Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2001;23:499–501. [PubMed] [Google Scholar]
- 6.Schmieder RE, Hilgers KF, Schlaich MP, Schmidt BM. Renin-angiotensin system and cardiovascular risk. Lancet. 2007;369:1208–1219. doi: 10.1016/S0140-6736(07)60242-6. [DOI] [PubMed] [Google Scholar]
- 7.Lindsberg PJ, Grau AJ. Inflammation and infections as risk factors for ischemic stroke. Stroke. 2003;34:2518–2532. doi: 10.1161/01.STR.0000089015.51603.CC. [DOI] [PubMed] [Google Scholar]
- 8.Bova IY, Bornstein NM, Korczyn AD. Acute infection as a risk factor for ischemic stroke. Stroke. 1996;27:2204–2206. doi: 10.1161/01.str.27.12.2204. [DOI] [PubMed] [Google Scholar]
- 9.Grau AJ, Buggle F, Becher H, Werle E, Hacke W. The association of leukocyte count, fibrinogen and C-reactive protein with vascular risk factors and ischemic vascular diseases. Thromb Res. 1996;82:245–255. doi: 10.1016/0049-3848(96)00071-0. [DOI] [PubMed] [Google Scholar]
- 10.Hansson GK, Robertson AK, Söderberg-Nauclér C. Inflammation and atherosclerosis. Annu Rev Pathol. 2006;1:297–329. doi: 10.1146/annurev.pathol.1.110304.100100. [DOI] [PubMed] [Google Scholar]
- 11.McGeer PL, Rogers J, McGeer EG. Inflammation, anti-inflammatory agents and Alzheimer disease: the last 12 years. J Alzheimers Dis. 2006;9:271–276. doi: 10.3233/jad-2006-9s330. [DOI] [PubMed] [Google Scholar]
- 12.Lange LA, Carlson CS, Hindorff LA, Lange EM, Walston J, Durda JP, Cushman M, Bis JC, Zeng D, Lin D, Kuller LH, Nickerson DA, Psaty BM, Tracy RP, Reiner AP. Association of polymorphisms in the CRP gene with circulating C-reactive protein levels and cardiovascular events. JAMA. 2006;296:2703–2711. doi: 10.1001/jama.296.22.2703. [DOI] [PubMed] [Google Scholar]
- 13.Hollegaard MV, Bidwell JL. Cytokine gene polymorphism in human disease: on-line databases, Supplement 3. Genes Immun. 2006;7:269–276. doi: 10.1038/sj.gene.6364301. [DOI] [PubMed] [Google Scholar]
- 14.Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- 15.Endres M, Laufs U, Merz H, Kaps M. Focal expression of intercellular adhesion molecule-1 in the human carotid bifurcation. Stroke. 1997;28:77–82. doi: 10.1161/01.str.28.1.77. [DOI] [PubMed] [Google Scholar]
- 16.Dichgans M, Markus HS. Genetic association studies in stroke: methodological issues and proposed standard criteria. Stroke. 2005;36:2027–2031. doi: 10.1161/01.STR.0000177498.21594.9e. [DOI] [PubMed] [Google Scholar]
- 17.Steering Committee of the Physicians' Health Study Research Group. Final report of the aspirin component of the ongoing Physicians' Health Study. N Engl J Med. 1989;321:129–135. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 18.Zee RY, Cook NR, Cheng S, Reynolds R, Erlich HA, Lindpaintner K, Ridker PM. Polymorphism in the P-selectin and interleukin-4 genes as determinants of stroke: a population-based, prospective genetic analysis. Hum Mol Genet. 2004;13:389–396. doi: 10.1093/hmg/ddh039. [DOI] [PubMed] [Google Scholar]
- 19.Cummings SR, Nevitt MC, Browner WS, Stone K, Fox KM, Ensrud KE, Cauley J, Black D, Vogt TM. Risk factors for hip fracture in white women. Study of Osteoporotic Fractures Research Group. N Engl J Med. 1995;332:767–773. doi: 10.1056/NEJM199503233321202. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt W-P, Heuschmann P, Taeger D, Henningsen H, Bücker-Nott HJ, Berger K. Determinants of IV heparin treatment in patients with ischemic stroke. Neurology. 2004;63:2407–2409. doi: 10.1212/01.wnl.0000147542.37657.2f. [DOI] [PubMed] [Google Scholar]
- 21.Evers S, Fischera M, May A, Berger K. Prevalence of cluster headache in Germany: results of the epidemiological DMKG study. J Neurol Neurosurg Psychiatry. 2007;78:1289–1290. doi: 10.1136/jnnp.2007.124206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luedemann J, Schminke U, Berger K, Piek M, Willich SN, Döring A, John U, Kessler C. Association between behavior-dependent cardiovascular risk factors and asymptomatic carotid atherosclerosis in a general population. Stroke. 2002;33:2929–2935. doi: 10.1161/01.str.0000038422.57919.7f. [DOI] [PubMed] [Google Scholar]
- 23.Lang W, Lalouschek W on behalf of the Vienna Stroke Study Group. The Vienna Stroke Registry – objectives and methodology. Wien Klin Wochenschr. 2001;113:141–147. [PubMed] [Google Scholar]
- 24.Cheng S, Grow MA, Pallaud C, Klitz W, Erlich HA, Visvikis S, Chen JJ, Pullinger CR, Malloy MJ, Siest G, Kane JP. A multilocus genotyping assay for candidate markers of cardiovascular disease risk. Genome Res. 1999;9:936–949. doi: 10.1101/gr.9.10.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barcellos LF, Begovich AB, Reynolds RL, Caillier SJ, Brassat D, Schmidt S, Grams SE, Walker K, Steiner LL, Cree BA, Stillman A, Lincoln RR, Pericak-Vance MA, Haines JL, Erlich HA, Hauser SL, Oksenberg JR. Linkage and association with the NOS2A locus on chromosome 17q11 in multiple sclerosis. Ann Neurol. 2004;55:793–800. doi: 10.1002/ana.20092. [DOI] [PubMed] [Google Scholar]
- 26.Whitehead A. Meta-Analysis of Controlled Clinical Trials. New York: Wiley; 2002. [Google Scholar]
- 27.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 29.Westfall PH, Young SS. P value adjustments for multiple tests in multivariable binomial models. J Amer Stat Assoc. 1989;84:780–786. [Google Scholar]
- 30.Hagiwara N, Kitazono T, Kamouchi M, Kuroda J, Ago T, Hata J, Ninomiya T, Ooboshi H, Kumai Y, Yoshimura S, Tamaki K, Fujii K, Nagao T, Okada Y, Toyoda K, Nakane H, Sugimori H, Yamashita Y, Wakugawa Y, Kubo M, Tanizaki Y, Kiyohara Y, Ibayashi S, Iida M. Polymorphisms in the Lymphotoxin Alpha Gene and the Risk of Ischemic Stroke in the Japanese Population. The Fukuoka Stroke Registry and the Hisayama Study. Cerebrovasc Dis. 2008;25:417–422. doi: 10.1159/000121342. [DOI] [PubMed] [Google Scholar]
- 31.Szolnoki Z, Havasi V, Talian G, Bene J, Komlosi K, Somogyvari F, Kondacs A, Szabo M, Fodor L, Bodor A, Melegh B. Lymphotoxin-alpha gene 252G allelic variant is a risk factor for large-vessel-associated ischemic stroke. J Mol Neurosci. 2005;27:205–211. doi: 10.1385/JMN:27:2:205. [DOI] [PubMed] [Google Scholar]
- 32.Um JY, An NH, Kim HM. TNF-alpha and TNF-beta gene polymorphisms in cerebral infarction. J Mol Neurosci. 2003;21:167–171. doi: 10.1385/JMN:21:2:167. [DOI] [PubMed] [Google Scholar]
- 33.Arboix A, Roig H, Rossich R, Martínez EM, García-Eroles L. Differences between hypertensive and non-hypertensive ischemic stroke. Eur J Neurol. 2004;11:687–692. doi: 10.1111/j.1468-1331.2004.00910.x. [DOI] [PubMed] [Google Scholar]
- 34.Pober JS, Lapierre LA, Stolpen AH, Brock TA, Springer TA, Fiers W, Bevilacqua MP, Mendrick DL, Gimbrone MA., Jr Activation of cultured human endothelial cells by recombinant lymphotoxin: comparison with tumor necrosis factor and interleukin 1 species. J Immunol. 1987;138:3319–3324. [PubMed] [Google Scholar]
- 35.Cavender DE, Edelbaum D, Ziff M. Endothelial cell activation induced by tumor necrosis factor and lymphotoxin. Am J Pathol. 1989;134:551–560. [PMC free article] [PubMed] [Google Scholar]
- 36.Kratz A, Campos-Neto A, Hanson MS, Ruddle NH. Chronic inflammation caused by lymphotoxin is lymphoid neogenesis. J Exp Med. 1996;183:1461–1472. doi: 10.1084/jem.183.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schreyer SA, Vick CM, LeBoeuf RC. Loss of lymphotoxin-alpha but not tumor necrosis factor-alpha reduces atherosclerosis in mice. J Biol Chem. 2002;277:12364–12368. doi: 10.1074/jbc.M111727200. [DOI] [PubMed] [Google Scholar]
- 38.Kampus P, Muda P, Kals J, Ristimae T, Fischer K, Teesalu R, Zilmer M. The relationship between inflammation and arterial stiffness in patients with essential hypertension. Int J Cardiol. 2006;112:46–51. doi: 10.1016/j.ijcard.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 39.Li JJ, Chen JL. Inflammation may be a bridge connecting hypertension and atherosclerosis. Med Hypotheses. 2005;64:925–929. doi: 10.1016/j.mehy.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Morishita R. Is vascular endothelial growth factor a missing link between hypertension and inflammation? Hypertension. 2004;44:253–254. doi: 10.1161/01.HYP.0000138689.29876.b3. [DOI] [PubMed] [Google Scholar]
- 41.Yanbaeva DG, Dentener MA, Creutzberg EC, Wesselingm G, Wouters EF. Systematic effects of smoking. Chest. 2007;131:1557–1566. doi: 10.1378/chest.06-2179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
On-line Supplementary Table 1. Frequencies of the less common or “variant” allele among the (A) control and ischemic stroke case (B) subgroups for each study. P values for large-sample and exact tests for Hardy-Weinberg equilibrium (HWE).
Online Supplementary Table 2. Nominally significant (P<0.05) fixed-effects meta-analysis results with (A) age, (B) sex, or (C) smoking-status stratification.