Abstract
Submillimolar levels of calcium, similar to the physiological total (bound + free) intranuclear concentration (0.01–1 mM), induced a conformational change within d(TG/AC)n, one of the frequent dinucleotide repeats of the mammalian genome. This change is calcium-specific, because no other tested cation induced it and it was detected as a concentration-dependent transition from B- to a non-B-DNA conformation expanding from 3′ end toward the 5′ of the repeat. Genomic footprinting of various rat brain regions revealed the existence of similar non-B-DNA conformation within a d(TG/AC)28 repeat of the endogenous enkephalin gene only in enkephalin-expressing caudate nucleus and not in the nonexpressing thalamus. Binding assays demonstrated that DNA could bind calcium and can compete with calmodulin for calcium.
A significant proportion of cellular calcium resides in the nucleus in free ionic form, shown by in vivo imaging, and also in bound form, demonstrated by x-ray element microanalysis and ion microscopy (1–3). The estimated average physiological free (ionic) calcium concentrations range from nanomolar to micromolar (4) as opposed to the total (bound + free) calcium levels, which are in the submillimolar to millimolar range (refs. 1 and 3 and R. Leapman, B. Andrews, and D.v.A., unpublished observations). The intranuclear calcium transients detected in vivo in differentiating or regenerating cells can reflect either the movement of free ionic calcium between various subnuclear compartments and/or net calcium influx from other subcellular organelles (5). Significant changes in total intranuclear calcium concentrations have also been detected and associated with various physiological and pathological conditions. In stimulated tumor mast cells, total nuclear calcium levels can be as high as 1.2 mM (3). Very high calcium levels (3–7 mM) are found in the nucleus of hyperplastic prostate epithelial cells (1). The observed changes in free and total intranuclear calcium levels coinciding with periods of physiologically or pathologically altered nuclear activities are of particular interest because calcium can mediate a wide range of cellular functions (6). Calcium is known to affect protein–DNA interactions by regulating secondary modifications such as phosphorylation of various transcription factors with consequences for gene transcription or DNA replication (7). The calcium requirement for proteolysis of nuclear matrix proteins by m-calpain can be dramatically decreased in the presence of DNA (8). However, the role of DNA as a potential intranuclear calcium donor or direct calcium–DNA interactions and their possible effects on nuclear architecture and functions have not been discussed.
Various genomic functions, including gene transcription, require the assembly of the DNA into nuclear domains through dynamically regulated changes in DNA structure (9). The DNA structure can be altered by DNA binding proteins, by the extent of negative supercoiling, and by various ions (10–12). The DNA motif d(TG/AC)n is the most frequent tandem dinucleotide repeat in the mammalian genome (13–15). The inherent structural bias of the d(TG/AC)n repeat (16) and its involvement in superhelical stress and ion-induced structural changes (17) suggest structural role for the repeat in genomic architecture. The repeat can respond to various factors with a transition from the conventional B-DNA to a non-B-DNA forms. The transition can be induced by increased superhelical density or by 10–50 mM divalent cations, primarily magnesium (12). By using a synthetic dinucleotide repeat, Behe and Felsenfeld (18) concluded that the B–Z transition required at least 10 mM calcium. The functional consequences of such a transition could include altered DNA replication and transcriptional activity (19–22). However, intranuclear magnesium levels in vivo are much lower than required to induce conformational change of the d(TG/AC)n repeat and magnesium remains relatively unchanged during the life cycles of various cells (3) (R. Leapman, B. Andrews, and D.v.A. unpublished observations).
We have investigated the effect of calcium on the structure of d(TG/AC)28 repeat, mimicking in vivo conditions in an in vitro model system, and also tested whether similar conformational change can be found in vivo in the rat brain. Herein we provide evidence that calcium within its physiological intranuclear concentration specifically regulates DNA conformation at the d(TG/AC)n repeat and that DNA binds calcium and can compete with calmodulin (CaM) for calcium.
METHODS
OsO4 Footprinting.
Cesium chloride-purified supercoiled form of rENKpCRII756 plasmid (σ̄ = −0.05) was used as a circular plasmid or was linearized by ApaI (New England Biolabs). Both forms were dialyzed against a 1,000-fold excess volume of HPLC-grade water through a 1,000-Da cut-off membrane. One picomole of plasmid DNA (3,000 bp) was added to a solution containing 12 mM Hepes, 9 mM NaOH (pH 7.9), 100 mM KCl, various concentrations of CaCl2, 2 mM OsO4, and 2% pyridine in a total volume of 100 μl. Reactions, radioactive labeling, chemical cleavage, and separation were performed as described (23).
OsO4 Footprinting of Rat Brain Tissues.
Thalamus and caudate nuclei from the brains of 40 rats at age 14 postnatal days were dissected and pooled; 0.4 g of thoroughly minced tissues was treated with 1 mM OsO4 and 1 mM 2,2′-bipyridine (24) in Ringer’s buffer containing 10% glycerol and 10 mM Tris⋅HCl (pH 7.3) in a total volume of 10 ml for 30 min at 8°C with constant gentle mixing. After washing and centrifugation, tissue pellets were resuspended in 1 ml of PBS on ice. Genomic DNA was immediately prepared from the OsO4-treated brain tissues by using the QUICK-Geno DNA isolation kit (CLONTECH). The fragment containing positions −513 to −1,081 of the rat enkephalin (ENK) gene was released by BglII digestion and was subjected to chemical cleavage (25). Fifty micrograms (approximately 25 amol) of genomic DNA (calculated from 3 × 109 bp per haploid rat genome) were analyzed per lane on a polyacrylamide/urea gel. A sequencing reaction ladder was generated as the standard by using an identical BglII fragment of the ENK gene as the template (positions −513 to −1,081) and a primer (positions −505 to −542) that overhangs the BglII site by 8 bases. Sequencing reactions were carried out with the Sequenase Version 2.0 kit (United States Biochemical) and were diluted to an approximate concentration of 10 amol per band. After separation, the gel with the separated genomic DNA fragments was transferred onto a Zeta-Probe membrane and cross-linked. The radioactive probe for hybridization was generated in a primer–template system incorporating both [α-32P]dATP (3,000 Ci/mmol; 1 Ci = 37 GBq) and [α-32P]dCTP (3,000 Ci/mmol). The membrane was prehybridized in Hybrizol I solution (Oncor) at 40°C for 15 min and was hybridized at 40°C overnight with constant rotation, washed in 2× SSC for 15 min, followed by 0.5× SSC for 10 min and 0.1× SSC for 10 min at room temperature. The air-dried membrane was exposed to x-ray film for 2 weeks and subsequently quantified in a PhosphorImager (Molecular Dynamics). Relative OsO4 reactivities of the caudate nucleus and thalamus were obtained by dividing the intensities of the chemically cleaved fragment at position −618 (a thymidine) in the caudate nucleus and in the thalamus by the total activity loaded per lane.
Reverse Transcription-Coupled PCR.
Total cellular RNA from the dissected brain regions were isolated by using the Qiagen RNA isolation system (RNeasy, Qiagen, Hilden, Germany). Reverse transcription and PCR were performed as described (26). After separation of the PCR products on 4–20% TBE gels, the intensities of ENK-specific bands were quantified by using a PhosphorImager. The relative abundance (RA) of ENK mRNA were calculated as: RA in caudate nucleus = (cpm of ENK mRNA from caudate nucleus)/(cpm of ENK mRNA from caudate nucleus + cpm of ENK mRNA from thalamus); and RA thalamus = cpm of ENK mRNA from thalamus/(cpm of ENK mRNA from caudate nucleus + cpm of ENK mRNA from thalamus).
Calcium Binding Assay.
Thirty-nine picomoles (0.66 μg) of CaM, 0.66 μg of BSA, and 1 pmol of circular or ApaI-linearized rEnkpCRII505 plasmid DNA containing the (TG)28 repeat or 1 pmol of rENKpCRII382 circular plasmid DNA lacking the (TG)28 repeat were loaded on five identical (20 × 30 mm) Zeta-Probe membranes. The membranes were incubated with 12 mM Hepes, pH 7.9/100 mM KCl/3 mM MgCl2/50 μM 45CaCl2 (Amersham) at 24°C for 1 min and washed three times without 45CaCl2. The first membrane with no additional treatment served to assess basal 45CaCl2 binding. The last washing solutions of the remaining membranes included 50 μM EDTA, 1 mM 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA), or 1 mM unlabeled CaCl2, and one membrane was reincubated with 50 μM 45CaCl2, followed by three washes in calcium-free buffer. Filters were dried and the amount of bound 45CaCl2 was quantified with a PhosphorImager. Filters were counterstained with ethidium bromide and Coomassie blue to assess sample loss.
Ca2+/CaM Protein Kinase II (CaM-PK II) Autophosphorylation Assay.
Enzyme activities were measured in the presence of calcium (300 μM), DNA (13 μg of rEnkpCRII505 plasmid with or without BAPTA pretreatment), CaM antagonist (1 μg), BSA (13 μg), or genomic DNA (13 μg with or without BAPTA pretreatment) were added to the basic reaction mixture. Bound calcium from plasmid and genomic DNA and from BSA were removed by passing them through equilibrated BAPTA columns (Calcium Sponge S, Molecular Probes). Six nanograms of Ca2+/CaM-PK II (Sigma Ultra, Sigma) was added in a total volume of 25 μl and autophosphorylation was initiated by addition of 3 pmol of [γ-32P]ATP (3,000 Ci/mmol), and the mixtures were incubated at 24°C for 3 min. Reactions were terminated by the addition of 20 μl of CaM stop solution (10% SDS/6% 2-mercaptoethanol/0.1 M Tris⋅HCl, pH 6.0/15% glycerol) followed by heating the reaction mixtures to 95°C for 3 min. Samples were separated on a 4–20% Tris-glycine SDS gels and subjected to autoradiography.
RESULTS
The Effect of Calcium on DNA Structure in Vitro.
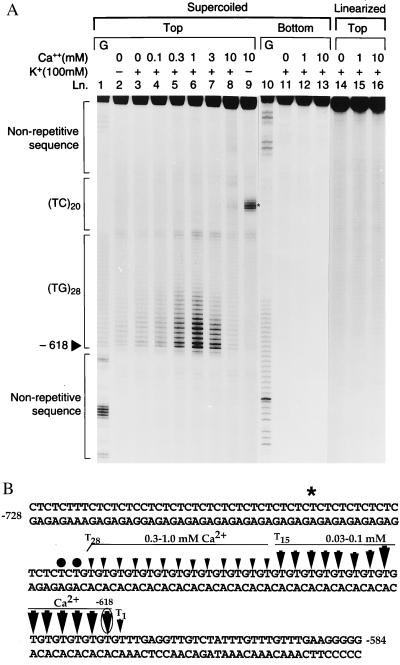
Testing the effect of calcium on various DNA regions of the rat ENK gene (data not shown) suggested that only the d(TG/AC)28 repeat, but no other DNA sequences, responded to physiological level of calcium with a concentration-dependent conformational change. The conformational change was detected by OsO4 footprinting as increased reactivity of thymidines within the repeat (Fig. 1A, lanes 3–8). OsO4 reacts with accessible thymidines on single-stranded and/or distorted double-stranded regions of the double helix (27). OsO4 footprinting therefore is a method of choice to probe conformational changes in DNA both in vitro and in vivo (for review, see ref. 24). The calcium-induced conformational change was localized within the repeat. Remarkably, reactive residues were concentrated on the 3′ end of the repeat (Fig. 1A, lanes 3–8, and B). At increasing calcium concentrations, a greater number of thymidines at more and more of the 5′ position became reactive. An adjacent dinucleotide repeat of the genome d(TC/AG)20 (known as the GAGA repeat) was not responsive to calcium, just as the flanking arbitrary sequences (Fig. 1A, lanes 3–8). Calcium was most effective at 1 mM, which is within its estimated intranuclear concentration range (bound calcium/volume of nuclei). With further elevation of calcium concentration (greater than 1 mM), the extent of strand separation vanished. In the presence of 10 mM calcium, there was no detectable strand separation (Fig. 1A, lanes 7–8). The calcium-induced response cannot be observed on linearized DNA, suggesting that the supercoiled form of DNA required for the structural transition (Fig. 1A, lanes 14–16). The reactive center in the middle of the adjacent d(TC/AG)20 repeat observed at only high calcium (3–10 mM) concentrations and in the absence of K+ (Fig. 1A, lane 9, as indicated with a star) is consistent with a triplex structure that is seen in pyrimidine-rich regions in the presence of high divalent cation concentrations (28). Two thymidines at the junction of the d(TG/AC)28 and d(TC/AG)20 sequences showed modest calcium-dependent reactivity, perhaps as the result of increased torsional stress. The bottom strand showed no ion-dependent reactivity (Fig. 1A, lanes 11–13).
Figure 1.
OsO4 footprint of the plasmid rENKpCRII756 containing (TG)28, (TC)20 repeats and nonrepetitive sequences showing calcium-induced structural changes indicated by reactive thymidines. (A) Supercoiled top (lanes 2–9) and bottom (lanes 11–13) strands and linearized top strands (lanes 14–16) were probed with OsO4 without calcium or with increasing calcium concentrations in the absence or presence of 100 mM KCl. G reaction ladders from Maxam–Gilbert sequencing of the corresponding sequences were loaded as standards (lanes 1 and 10). Solid arrowhead, 5′ reactive site at position −618; star on lane 9, Hoogsteen structure formed in the middle of (TC)20 sequences at high divalent cation concentrations. (B) The positions of OsO4 reactive thymidines in rENKpCRII756 (arrows) indicating the calcium-induced structural change, with increasing size indicating increased reactivity. The arrow pointing to the most reactive thymidine at position −618 is circled. Two reactive thymidines at the (TG)28/(TC)20 junction are indicated with solid circles. The star marks the central reactive thymidine of the Hoogsteen structure in the middle of (TC)20; T1, T15, and T28 mark the positions of the various reactive thymidines within the (TG)28 repeat. Horizontal bars mark the boundaries of the two regions within the repeat with distinct OsO4 reactivity.
The Structure of d(TG/AC)28 Repeat in Various Brain Regions.
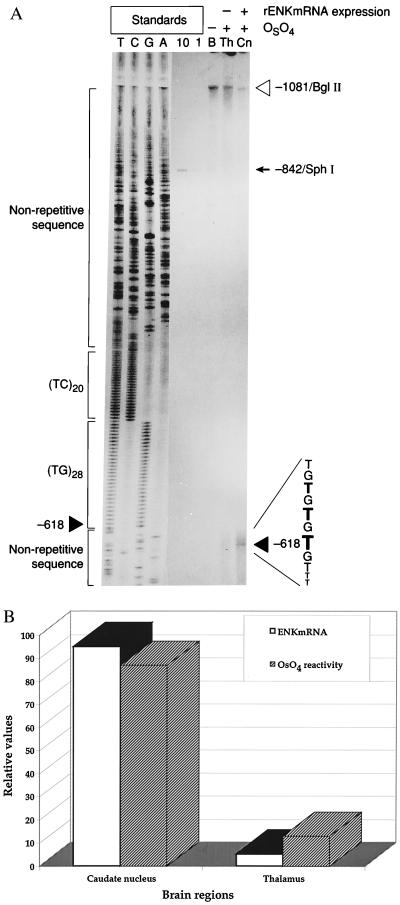
We wished to determine whether a d(TG/AC)n repeat shows similar structure within its chromatin environment and whether different OsO4 reactivity correlates with the expression of a repeat-bearing gene. We tested the d(TG/AC)28 repeat of the rat ENK gene by performing OsO4 footprinting on brain regions with high (caudate nucleus) vs. low expression (thalamus) of the ENK gene (Fig. 2A). A conformational change of the d(TG/AC)28 repeat, marked by high OsO4 reactivity at position −618 within the repeat, was found in the high ENK-expressing caudate nucleus (Fig. 2A, solid arrowhead). The most reactive thymidine at position −618 precisely corresponded to the most reactive thymidine detected on the plasmid DNA (Fig. 1). The increased OsO4 reactivity expanded to at least three additional thymidines in the 5′ direction. The reactive region of the d(TG/AC)28 in brain tissue spanned only one-half of a turn of the DNA helix in contrast to the two full helical turns in plasmid DNA.
Figure 2.
OsO4 footprint of the (TG)28 repeat of the rat ENK gene in the caudate nucleus and thalamus. (A) The solid arrowhead points to the most reactive thymidine at position −618 on the OsO4 footprint performed on caudate nucleus (see also Fig. 1). The caudate nucleus (Cn) and thalamus (Th) were dissected from 40 adult rat brains and treated with OsO4, and after the isolation of genomic DNA the 1,081-bp BglII fragment containing the endogenous (TG/AC)28 repeat of the rat ENK gene (position −1,081, indicated by open arrowhead) was probed with a radioactively labeled fragment rENK505–592 (29). An identical segment of the ENK gene isolated from rat genomic DNA without OsO4 treatment. (Lane B). Ten- and one-attomole standards (10, 1) derived from the 842-bp BglII–SphI fragment of the rENK gene (position −842 bp of the fragment is marked by arrowhead) illustrate detection sensitivity. Lanes T, C, G, and A show the sequencing ladders; the positions of the (TG)28 and (TC)20 repeat are marked. The sequencing primer overhangs the template segment by 8 bases; therefore, the sequencing ladder is shifted 8 bases above the genomic position. (B) Correlation between ENK mRNA levels and OsO4 reactivity at the d(TG/AC)28 repeat of the rENK gene in Cn and Th regions of the adult rat brain. Relative values were obtained from quantitive reverse transcription-coupled PCR and quantification of OsO4 reactivity on a PhosphorImager.
Interestingly, the OsO4 reactivity at the ENK d(TG/AC)28 repeat was obvious only in the ENK-expressing caudate nucleus. By contrast, OsO4 did not detect altered conformation of d(TG/AC)28 repeat in the low ENK-expressing thalamus (Fig. 2A). The relative abundance of ENK mRNA measured in the two brain regions was plotted with the level of relative level of OsO4 reactivities of the d(TG/AC)28 repeat on the ENK gene in the caudate nucleus vs. the thalamus of the adult rat brain. The plot indicated a high level of correlation between OsO4 reactivity at the d(TG/AC)28 repeat and the level of ENK expression (Fig. 2B).
Specificity of the Calcium Effect.
A slight but consistently increased OsO4 reactivity throughout the 3′ end of the d(TG/AC)28 region of the supercoiled plasmid was observed in the absence of exogenous calcium (Fig. 1A, lanes 2 and 3). Extensive dialysis of the DNA did not change this property. However, titration with the selective calcium chelator BAPTA resulted in the elimination of the reactivity (Fig. 3A, lanes 2–11). Titration with EGTA or EDTA had similar effect only at much higher concentrations (Fig. 3A, lanes 12–17). BAPTA has high affinity and selectivity for calcium (Kd ∼ 10−7 M) as opposed to other divalent cations such as magnesium (Kd ∼ 10−2 M) (30). Therefore, to test the specificity of calcium in inducing the observed conformational change, we first selected the lowest effective BAPTA concentration value (700 μM) that completely eliminates the OsO4-reactive conformation of the d(TG/AC)28 repeat and then tested the specificity by the addition of various exogenous divalent cations. We found that only calcium can induce the conformational change on the d(TG/AC)28 repeat and no other tested divalent cations (e.g., magnesium, manganese, and ferrous ions) had an effect (Fig. 3B). To test whether the conformation change can be attributed to the d(TG/AC)28 sequence, we replaced the repeat with a nonrepetitive sequence (rENK positions −382 to −542) in the plasmid and probed this plasmid similarily. Plasmid in which the d(TG/AC)28 element was replaced with the nonrepetitive sequence did not show the calcium-induced response (data not shown).
Figure 3.
Specificity of calcium in inducing DNA conformation change. (A) OsO4 footprint of the supercoiled plasmid rENKpCRII (positions −505 to −756) in the presence chelators. Plasmid DNA was treated with an increasing concentration of BAPTA (lanes 3–11) that resulted in an increasingly “closed” configuration of the DNA strands at the (TG)28 sequence. Chelating with EGTA (lanes 12–14) or EDTA (lanes 15–17) was effective only at higher concentrations. G indicates the Maxam–Gilbert G reaction ladder. Experimental conditions were identical to those in Fig. 1 except that chelators were added to the DNA in the indicated concentrations prior to the OsO4 treatment. (B) The effect of other divalent cations on the DNA structure at (TG)28 repeat. Calcium (lane 5), but no other divalent cation (lanes 6–8), induced DNA conformational change at the (TG)28 sequence. Experimental conditions were identical to those in Fig. 1 except that the negatively supercoiled plasmid was preincubated with 700 μM of BAPTA to remove all DNA-bound calcium where indicated. Calcium, magnesium, manganese, and ferrous ion were added to the reaction mixture.
DNA Binds Calcium.
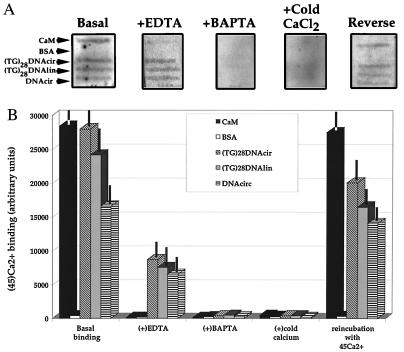
Ca2+/CaM is the most ubiquitous calcium-binding protein and its calcium-binding activity can be inhibited by micromolar concentrations of EGTA or EDTA (31). The observation that pretreatment of the DNA with BAPTA can eliminate the OsO4-reactive conformation of the d(TG/AC)28 repeat more efficiently than EDTA or EGTA suggested that DNA may bind calcium with significant affinity (Fig. 3). In a filter-binding assay, we directly compared the calcium binding of CaM to circular (plasmid) or linearized DNA containing the d(TG/AC)28 repeat with circular DNA containing no repetitive element (Fig. 4A). By assuming that CaM/Ca2+ = 1/2, we estimated that 2 μg (1 pmol) of plasmid DNA (3,000 bp) could bind the same amount of radioactive calcium 45Ca2+ as 40 pmol of CaM. Furthermore, all the 45Ca2+ bound to CaM was chelated by thoroughly washing the filter with 50 μM EDTA, whereas the same treatment only reduced the DNA bound 45Ca2+ by approximately 60% (Fig. 4B). Only 1 mM BAPTA or 1 mM (20-fold excess) nonradioactive calcium could chelate or replace all the 45Ca2+ from the DNA. The effects of chelators or unlabeled calcium could be reversed by extensive wash of the filter followed by the reincubation of the filter with 45Ca2+. The somewhat lower 45Ca2+ binding to DNA observed after the reincubation of the filter is likely due to some loss of the immobilized samples on the membrane because it was confirmed by ethidium bromide counterstaining (data not shown). There was no significant difference in 45Ca2+ binding between the linear and circular forms of DNA containing the d(TG/AC)28 repeat. However, plasmid that contained d(TG/AC)28 repeat bound a somewhat higher amount of 45Ca2+ than a plasmid of similar length but but lacking the repeat. BSA used as control bound only trace amounts of 45Ca2+ (Fig. 4B).
Figure 4.
Calcium-binding assay. (A) Representative PhosphorImager pictures of Zeta-Probe membranes with immobilized identical slots of CaM, BSA, circular plasmid DNA [(TG)28DNAcir] and linearized [(TG)28DNAlin] plasmid DNA containing the (TG)28 repeat, and circular plasmid DNA from which the (TG)28 repeat was deleted (DNAcirc). All membranes were incubated with 45CaCl2 followed by the following treatments: basal binding (Basal), remaining binding after wash with 50 μM EDTA (+EDTA), remaining binding after wash with 1 mM BAPTA (+BAPTA), remaining binding after wash with 1 mM unlabeled CaCl2 (+cold CaCl2), binding after washing out unlabeled CaCl2 followed by a reincubation with 50 μM 45CaCl2 (Reverse). (B) Graphic expression of 45CaCl2 binding. Data points are the mean ± SEM of relative activities of the different treatment groups normalized to nonspecific background binding (n = 3).
DNA Competes with CaM for Calcium and Affects Ca2+/CaM-Dependent Function.
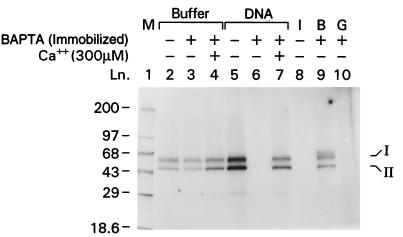
Because the filter-binding assay indicated high-affinity calcium binding to DNA, we wished to investigate how a major intranuclear calcium-binding protein, CaM, affects this binding by using Ca2+/CaM-PK II. Because this enzyme is autophosphorylated if CaM is activated through its bound calcium (32), it is a specific and sensitive way to measure calcium in the presence of DNA and CaM (Fig. 5, lanes 2–4). To obtain calcium-free DNA for this assay, we used an immobilized version of BAPTA (Calcium Sponge S). The presence of DNA had a dual effect on the activity of Ca2+/CaM-PK II depending on its calcium charge. Calcium-depleted plasmid and genomic DNA acted as chelators and completely inhibited autophosphorylation by competing for calcium with CaM (Fig. 5, lanes 6 and 10). The extent of the inhibition was comparable to the effect of a specific Ca2+/CaM-PK II inhibitor (Fig. 5, lane 8). Addition of 300 μM calcium to the reaction mixture that contained calcium-depleted DNA fully restored autophosphorylation (Fig. 5, lane 7). In contrast, calcium-charged DNA (without Calcium Sponge treatment) were capable of releasing calcium for CaM, as indicated by the increased autophosphorylation (Fig. 5, lane 5). Whether DNA acts as a calcium donor or acceptor was dependent on the molar ratios between DNA and CaM (data not shown).
Figure 5.
Ca2+/CaM-PK II autophosphorylation assay. The activity of the Ca2+/CaM-PK II is indicated by its phosphorylated 50- and 60-kDa subunits (I and II) in the assay. Basal activities are shown in the absence of DNA (lanes 2 and 3). Addition of 300 μM exogenous calcium increased phosphorylation (lane 4). Plasmid DNA containing residual calcium (dialyzed but no BAPTA treatment) donated calcium as indicated by increased autophosphorylation (lane 5), whereas calcium-free DNA completely inhibited autophosphorylation (lane 6). Exogenous calcium prevented the inhibitory effect of calcium-free plasmid DNA (lane 7). The specific inhibitor of CaM [CaM kinase II(290–309) CaM antagonist] completely blocked autophosphorylation (band I) (lane 8); addition of BSA (lane B) did not alter basal activity (lane 9); addition of calcium-free rat genomic DNA (lane G) also inhibited autophosphorylation (lane 10). The first band from the top in lane 9 is the result of BSA phosphorylation. Bands I and II indicate the two subunits of the enzyme; numbers indicate the molecular mass of the protein standards in kDa.
DISCUSSION
Herein we show that calcium within its physiological intranuclear concentration can induce a specific concentration-dependent conformational change of the DNA specific to the d(TG/AC)n repeat. This conformational change was demonstrated by OsO4 footprinting on superhelical plasmid and by genomic footprinting of different brain regions (Figs. 1 and 2). Furthermore, we have demonstrated that DNA binds calcium and can compete with CaM for available calcium, suggesting that DNA in general can also play an important role in intranuclear calcium homeostasis (Figs. 4 and 5).
Although several dinucleotide repeats can form unusual DNA structures (10, 33), the described conformational change at the d(TG/AC)n repeat is unique in its concentration-dependent response to physiological concentrations of calcium (Fig. 1). Although we think that the role of calcium in the d(TG/AC)n-mediated conformational change of the DNA is primary, the final structure of the DNA is likely affected by additional forces, primarily superhelical density, which is required for the observed calcium-induced conformational change (34–36). In the nucleus, transcription-induced local DNA supercoiling of upstream genes can affect superhelical density (37, 38). The d(TG/AC)n repeats, estimated to occur about every 30,000 bp in the mammalian genome, were found on 5′ regulatory or intronic regions (13–15), and thus calcium-sensitive loci can be very frequent in the genome. We think that the observed conformational change in vivo results in a DNA region that provides the genome with regulated flexibility permitting far-distant DNA interactions (39). Furthermore, this calcium-induced structural change could also translate into rotation or unwinding along the DNA axis. As a consequence of it, altered positioning of flanking DNA motifs could affect protein–DNA interactions by turning binding sites away (40). These calcium-induced structural changes at the d(TG/AC)n repeats could also contribute to the polymorphisms inherent in the repeat, because the altered DNA structure may affect the fidelity of replication (20).
The calcium-induced increased OsO4 reactivity at the d(TG/AC)n repeat suggests a concentration-dependent transition from B-DNA to a non-B-DNA conformation. This conformation either another form of calcium-specific Z-DNA, another non-B-DNA structure, or DNA strand separation. Several lines of evidence argues against Z-DNA formation, the calcium-specificity, the low effective concentration range (submillimolar) of calcium, the concentration dependence, and the oriented nature (3′ to 5′) of the conformational change. It should be noted that there is no consensus on the DNA structure of the d(TG/AC)n repeat (Z-DNA vs. non-Z-DNA) (41, 42). The B–Z transition of synthetic dinucleotide repeat required at least 10 mM calcium (18). However, because conditions to stabilize Z-DNA in supercoiled plasmids are different from that in polynucleotides, the latter study is not directly comparable with the present one.
Both increased calcium transients and total intranuclear calcium have been implicated in mediating various nuclear functions, especially during cellular differentiation or regeneration (43–45). The increased intranuclear calcium transients observed in various differentiating or regenerating cells in vivo likely reflect the increased translocation of free calcium between proteins and DNA (46). Previous NMR studies and our filter-binding assay have demonstrated that DNA can bind oppositely charged ions such as calcium (47). From our assay, we estimated that on average every 40 nucleotides bind one calcium ion, which is strikingly similar to the ratio of 50 to 1 found in previous NMR studies. These NMR studies have also shown that certain DNA sequences (e.g., G+C-rich) can bind elevated amount of calcium (47). Preliminary NMR experiments suggest that the d(TG/AC)n repeat can also bind elevated amount of calcium (data not shown). It should be noted that the dinucleotide repeat d(TG/AC)n used in the filter-binding assay represented a very small proportion of the total DNA moiety of the plasmid (TG)28DNAcir. However, the determination of the exact binding capacity of the repeat needs further investigation.
DNA can bind calcium and able to compete for calcium with CaM (Fig. 5). DNA can act as a calcium donor or acceptor depending on (i) its own calcium charge, (ii) the molar ratios between DNA and calcium binding proteins (e.g., CaM), and (iii) calcium binding affinity of calcium binding proteins. Our observations and previous studies showing that the calcium-binding affinity of intranuclear calcium binding proteins is influenced by secondary modifications [e.g., primarily phosphorylation (5)] raise the possibility of the existence of a previously undescribed level of intranuclear regulation.
Acknowledgments
We thank for D. Schoenberg for her editorial help, Drs. L. Hudson-Agoston and T. Balla for critical discussions, and Dr. M. Palkovits for helping with brain microdissections. We also honor the memory of Mrs. M. Schaeffer, who was assisting with this manuscript at the time of her death.
ABBREVIATIONS
- ENK
enkephalin
- BAPTA = 1
2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid
- CaM
calmodulin
- CaM-PK II
CaM protein kinase II
References
- 1.Tvedt K E, Kopstad G, Haugen O A, Halgunset J. Cancer Res. 1987;47:323–328. [PubMed] [Google Scholar]
- 2.Ornberg R L, Kuijpers G A, Leapman R D. J Biol Chem. 1988;263:1488–1493. [PubMed] [Google Scholar]
- 3.Chandra S, Fewtrell C, Millard P J, Sandison D R, Webb W W, Morrison G H. J Biol Chem. 1994;269:15186–15194. [PubMed] [Google Scholar]
- 4.Bock G R, Ackrill K. Calcium Waves, Gradients and Oscillations, CIBA Found. Symp. Vol. 188. Chichester, U.K.: Wiley; 1995. pp. 1–281. [Google Scholar]
- 5.Bootman M D, Berridge M J. Cell. 1995;83:675–678. doi: 10.1016/0092-8674(95)90179-5. [DOI] [PubMed] [Google Scholar]
- 6.Tsien R W, Tsien R Y. Annu Rev Cell Biol. 1990;6:715–760. doi: 10.1146/annurev.cb.06.110190.003435. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh A, Ginty D D, Bading H, Greenberg M E. J Neurobiol. 1994;25:294–303. doi: 10.1002/neu.480250309. [DOI] [PubMed] [Google Scholar]
- 8.Mellgren R L, Song K, Mericle M T. J Biol Chem. 1993;268:653–657. [PubMed] [Google Scholar]
- 9.Laemmli U K, Tjian R. Curr Opin Cell Biol. 1996;8:299–303. [PubMed] [Google Scholar]
- 10.Rich A, Nordheim A, Wang A H-J. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 11.Harrington R E, Winicov I. Prog Nucleic Acids Res Mol Biol. 1994;47:195–270. doi: 10.1016/s0079-6603(08)60253-6. [DOI] [PubMed] [Google Scholar]
- 12.Herbert A, Rich A. J Biol Chem. 1996;271:11595–11598. doi: 10.1074/jbc.271.20.11595. [DOI] [PubMed] [Google Scholar]
- 13.Hamada H, Kakunaga T. Nature (London) 1982;298:396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- 14.Tripathi J, Brahmachari S K. J Biomol Struct Dyn. 1991;9:387–397. doi: 10.1080/07391102.1991.10507919. [DOI] [PubMed] [Google Scholar]
- 15.Schroth G P, Chou P J, Ho P S. J Biol Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 16.Dobi A L, Mahan M A, Agoston D v. Electrophoresis. 1997;18:12–16. doi: 10.1002/elps.1150180104. [DOI] [PubMed] [Google Scholar]
- 17.Haniford D B, Pulleyblank D E. Nature (London) 1983;302:632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- 18.Behe M, Felsenfeld G. Proc Natl Acad Sci USA. 1981;78:1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruskin E A, Rich A. Biochemistry. 1993;32:2167–2176. doi: 10.1021/bi00060a007. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T A. Nature (London) 1993;365:207–208. doi: 10.1038/365207a0. [DOI] [PubMed] [Google Scholar]
- 21.Hamada H, Petrino M G, Kakunaga T, Seidman M, Stollar B D. Mol Cell Biol. 1984;4:2622–2630. doi: 10.1128/mcb.4.12.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittig B, Dorbic T, Rich A. Proc Natl Acad Sci USA. 1991;88:2259–2263. doi: 10.1073/pnas.88.6.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobi A L, Matsumoto K, Santha E, Agoston D v. Nucleic Acids Res. 1994;22:4846–4847. doi: 10.1093/nar/22.22.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palecek E. Crit Rev Biochem Mol Biol. 1991;26:151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- 25.Maxam A M, Gilbert W. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobi A L, Palkovits M, Palkovits C G, Santha E, Agoston D v. Mol Neurobiol. 1995;10:185–203. doi: 10.1007/BF02740675. [DOI] [PubMed] [Google Scholar]
- 27.Jelen F, Karlovsky P, Makaturova E, Pecinka P, Palecek E. Gen Physiol Biophys. 1991;10:461–473. [PubMed] [Google Scholar]
- 28.Shimizu M, Kubo K, Matsumoto U, Shindo H. J Mol Biol. 1994;235:185–197. doi: 10.1016/s0022-2836(05)80025-7. [DOI] [PubMed] [Google Scholar]
- 29.Durkin R C, Weisinger G, Holloway M P, LaGamma E. Biochim Biophys Acta. 1992;1131:349–351. doi: 10.1016/0167-4781(92)90040-7. [DOI] [PubMed] [Google Scholar]
- 30.Tsien R Y. Biochemistry. 1980;19:2396–2404. doi: 10.1021/bi00552a018. [DOI] [PubMed] [Google Scholar]
- 31.Klee C B, Crouch T H, Richman P G. Annu Rev Biochem. 1980;49:489–515. doi: 10.1146/annurev.bi.49.070180.002421. [DOI] [PubMed] [Google Scholar]
- 32.Nairn A, Hemmings C, Greengard P. Annu Rev Biochem. 1985;54:931–976. doi: 10.1146/annurev.bi.54.070185.004435. [DOI] [PubMed] [Google Scholar]
- 33.Gaillard C, Strauss F. Science. 1994;264:433–436. doi: 10.1126/science.8153633. [DOI] [PubMed] [Google Scholar]
- 34.Rohner K J, Hobi R, Kuenzle C C. J Biol Chem. 1990;265:19112–19115. [PubMed] [Google Scholar]
- 35.Zhang S, Lockshin C, Herbert A, Winter E, Rich A. EMBO J. 1992;11:3787–3796. doi: 10.1002/j.1460-2075.1992.tb05464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbert A G, Spitzner J R, Lowenhaupt K, Rich A. Proc Natl Acad Sci USA. 1993;90:3339–3342. doi: 10.1073/pnas.90.8.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albert A C, Spirito F, Figueroa, Bossi N, Bossi L, Rahmouni A R. Nucleic Acids Res. 1996;24:3093–3099. doi: 10.1093/nar/24.15.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfl S, Wittig B, Dorbic T, Rich A. Biochim Biophys Acta. 1997;1352:213–221. doi: 10.1016/s0167-4781(97)00015-8. [DOI] [PubMed] [Google Scholar]
- 39.Maurer R A. DNA. 1985;4:1–9. doi: 10.1089/dna.1985.4.1. [DOI] [PubMed] [Google Scholar]
- 40.Werner M H, Gronenborn A M, Clore G M. Science. 1996;271:778–784. doi: 10.1126/science.271.5250.778. [DOI] [PubMed] [Google Scholar]
- 41.Kladde M P, Kohwi Y, Kohwi-Shigematsu T, Gorski J. Proc Natl Acad Sci USA. 1994;91:1898–1902. doi: 10.1073/pnas.91.5.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shing H P. Proc Natl Acad Sci USA. 1994;91:9549–9553. doi: 10.1073/pnas.91.20.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ausserer W A, Ling Y C, Chandra S, Morrison G H. Anal Chem. 1989;61:2690–2695. doi: 10.1021/ac00199a002. [DOI] [PubMed] [Google Scholar]
- 44.Holliday J, Adams R J, Sejnowski T J, Spitzer N C. Neuron. 1991;7:787–796. doi: 10.1016/0896-6273(91)90281-4. [DOI] [PubMed] [Google Scholar]
- 45.Birch B D, Eng D L, Kocsis J D. Proc Natl Acad Sci USA. 1992;89:7978–7982. doi: 10.1073/pnas.89.17.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer T, Allbritton N L, Oancea E. CIBA Found Symp. 1995;188:252–262. doi: 10.1002/9780470514696.ch14. [DOI] [PubMed] [Google Scholar]
- 47.Braunlin W H, Drakenberg T, Nordenskiold L. J Biomol Struct Dyn. 1992;10:333–343. doi: 10.1080/07391102.1992.10508651. [DOI] [PubMed] [Google Scholar]