Abstract
Protein synthesis of an RNA template can initiate by two different known mechanisms: cap-dependent translation initiation and cap-independent translation initiation. The latter is driven by RNA sequences called internal ribosome entry sites (IRESs) that are found in both viral RNAs and cellular mRNAs. The diverse mechanisms used by IRESs are reflected in their structural diversity, and this structural diversity challenges us to develop a cohesive model linking IRES function to structure. With more direct structural information available for the viral IRESs, data suggest an inverse correlation between the degree to which an IRES RNA can form a stable structure on its own, and the number of factors that it requires to function. Lessons learned from the viral IRESs may help understand the cellular IRESs, although more structural data is needed before any strong links can be made.
Introduction
In translation, a messenger RNA (mRNA) serves first as the platform for assembly of the ribosome (initiation), then as the template for protein synthesis (elongation), and finally signals the end of the protein-coding sequence and ribosome disassembly (termination). In eukaryotes, the majority of translation initiation occurs by a complex, multi-step process that requires a modified nucleotide cap on the mRNA’s 5' end. Recognition of this cap leads to binding of eukaryotic initiation factor (eIF) proteins and the small (40S) subunit of the ribosome, which then scans the message until it finds the appropriate start codon, at which the large (60S) ribosomal subunit is recruited to form a translationally competent ribosome (review: [1]). This canonical cap- and scanning-dependent initiation does not account for all of the protein synthesis in a eukaryotic cell: translation initiation can also occur by a cap-independent process in which sequences within the mRNA recruit the translation machinery, often elimination the need for many eIF proteins (Figure 1a). This type of translational initiation occurs independent of the 5' end of the RNA, it is called internal initiation of translation and the RNA sequences responsible are called “internal ribosome entry sites (IRESs)” (reviews: [2–5]).
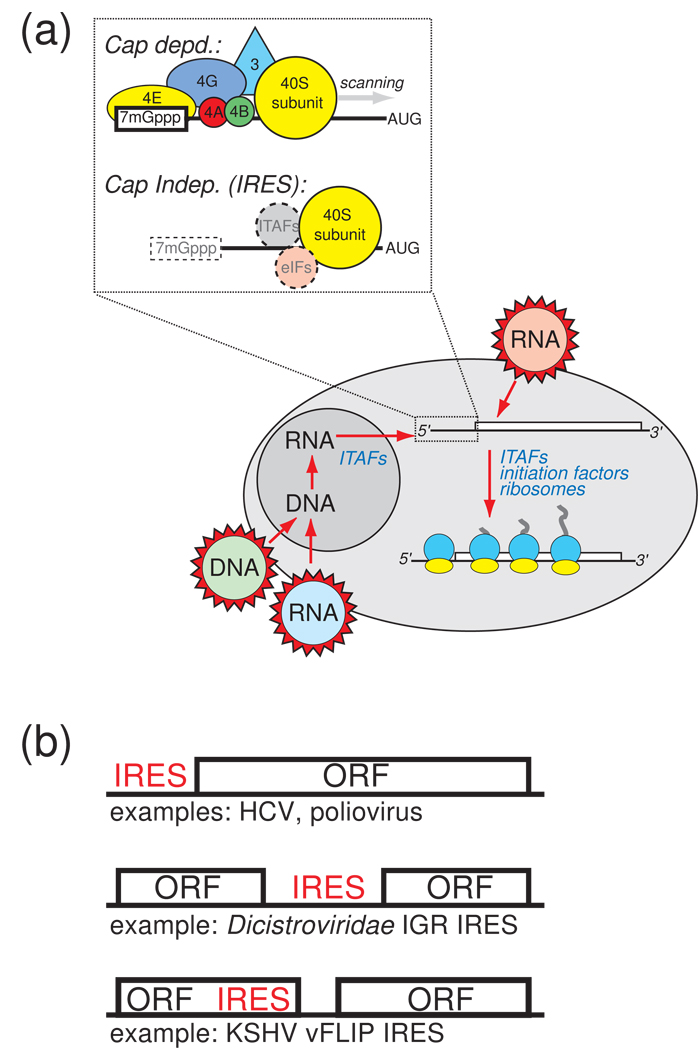
Figure 1. Cap- and IRES- dependent translation initiation.
(a) At upper left is a cartoon illustrating a simplified mechanism for IRES-driven initiation compared to cap-dependent initiation. In cap-dependent translation, the 7-methyl guanosine cap is bound by eIF4E, and this leads to the binding of many other factors, recruitment of the ribosome, and scanning to the start codon. IRESs do not use the cap structure, although IRES-containing messages can be capped (dashed box), and they may or may not use canonical eIFs and ITAFs to recruit the ribosome to the message (dashed circles). The diagram below illustrates the sources of IRES RNAs in the cell. They can come from RNA viruses that introduce RNA directly into the cytoplasm (red), and thus never experience the nuclear environment. IRESs that do have a “nuclear history” include those from RNA viruses whose genetic information is reverse transcribed and integrated into the host’s genome (blue), DNA viruses (green), and cellular IRESs. The degree to which nuclear history plays a role in the binding of specific factors to certain IRESs, and the effect it may have on the structure of the IRES, is a very important question under exploration. (c) Some mRNA contexts in which IRESs are found. Most are found in the 5’ untranslated region of the mRNA or viral RNA (top), but some are found between open reading frames in intergenic regions (middle), and they can also reside within (or partially within) coding regions (bottom). Viral IRES examples are provided for each.
IRESs were first discovered in the 5' untranslated regions (5' UTRs) of two picornavirus RNAs where they comprise highly conserved RNA sequences with a large amount of secondary structure, suggesting that these IRESs work by functionally replacing the cap and some eIFs with structured RNA. Efforts to understand the structural basis for the function of these viral IRESs began almost immediately after their discovery. Subsequent discovery of IRESs in many other viral RNAs and in a variety of cellular mRNAs uncovered substantial diversity in IRES-driven translation and IRES structure. What have we learned about the structural basis of IRES function? Are general trends emerging as more IRES-containing RNAs are identified and more structural information is gathered? Can we apply what has been learned about the structures of viral IRESs to cellular IRESs? Where should ongoing IRES structure-function studies be focused? In this short review we offer our perspective on these questions using some illustrative examples and recent discoveries, without attempting a comprehensive review of the field.
The structural diversity of viral IRES RNAs reflects functional diversity
Viral IRESs are defined by their ability to drive cap-independent translation, but the origin of the IRES RNA and the context in which it functions can vary dramatically (Figure 1a & b), correlating to a rich diversity of structures. In many cases, the viral IRES can respond to different cellular conditions. For example, the positive sense (+), single stranded RNA (ssRNA) Dicistorviridae viruses are activated by cellular conditions associated with the cell’s own antiviral response [6] (review: [7]). Retroviruses such as lentiviruses use a mix of cap- and IRES- driven translation to regulate protein synthesis during the cell cycle, indicating an ability to respond to cellular conditions [8,9]. Much information regarding the structural basis of IRES function comes from a mix of structural, biochemical, biophysical, and functional studies of IRESs from (+) ssRNA viruses. These IRESs can be divided into fairly distinct mechanistic classes based on their protein factor requirements and secondary structures (Figure 2 & Figure 3), as well as their three-dimensional structures and folding architectures (Figure 4 & Figure 5). This existing functional data in combination with structural information provides an emerging correlation between the folding characteristic of different IRES RNAs and their factor requirements, which we describe below.
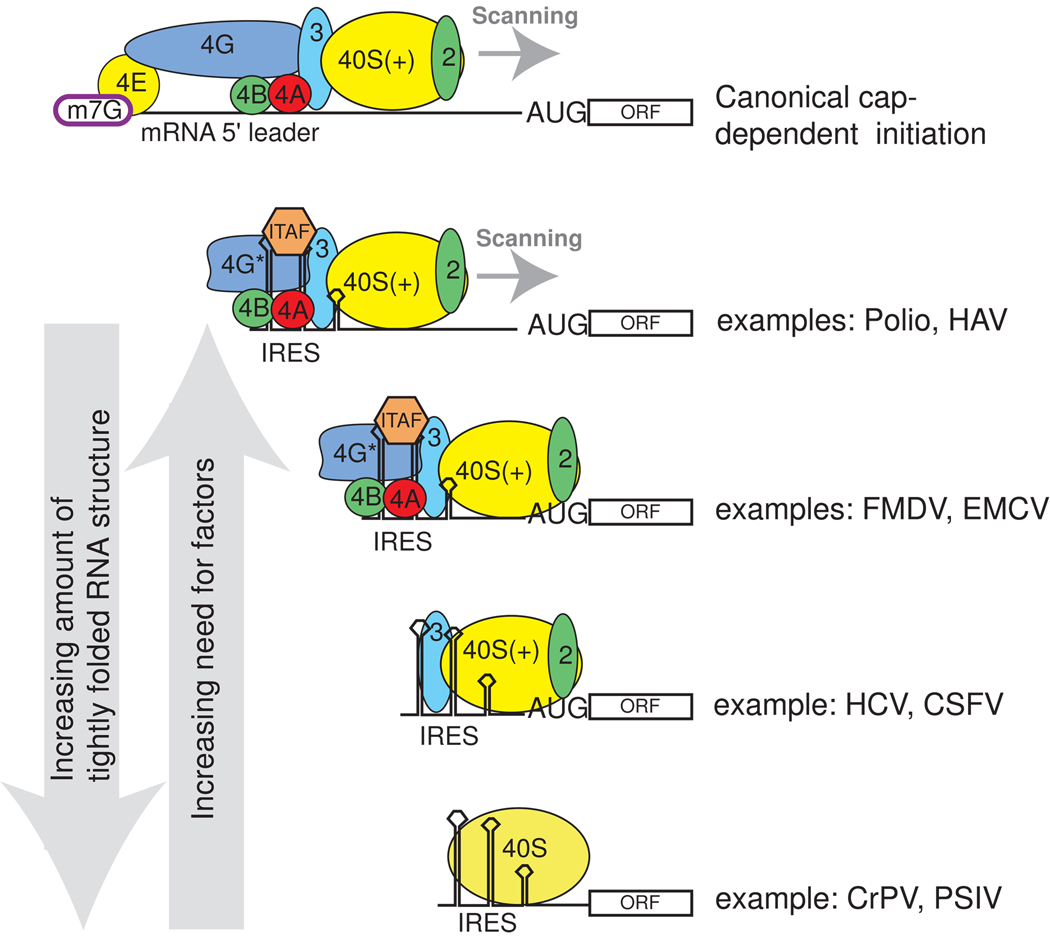
Figure 2. Examples of the diversity of viral IRES factor requirements.
Canonical initiation requires the full complement of translation eIFs (top), while IRES initiation can use subsets of these factors as well as ITAFs (below). Shown are examples of some viral IRESs with the factors each requires. For simplicity, all the factors associated with the 40S subunit are not shown. As described in the text, we note a trend in which IRES RNAs with the most inherent stably folded structure (left arrow) are those that require the fewest factors, and as the IRES become less inherently structured, more ITAFs and eIFS are needed (right arrow). The degree to which this trend will prove predictive, or can be extended to cellular IRESs, is unknown.
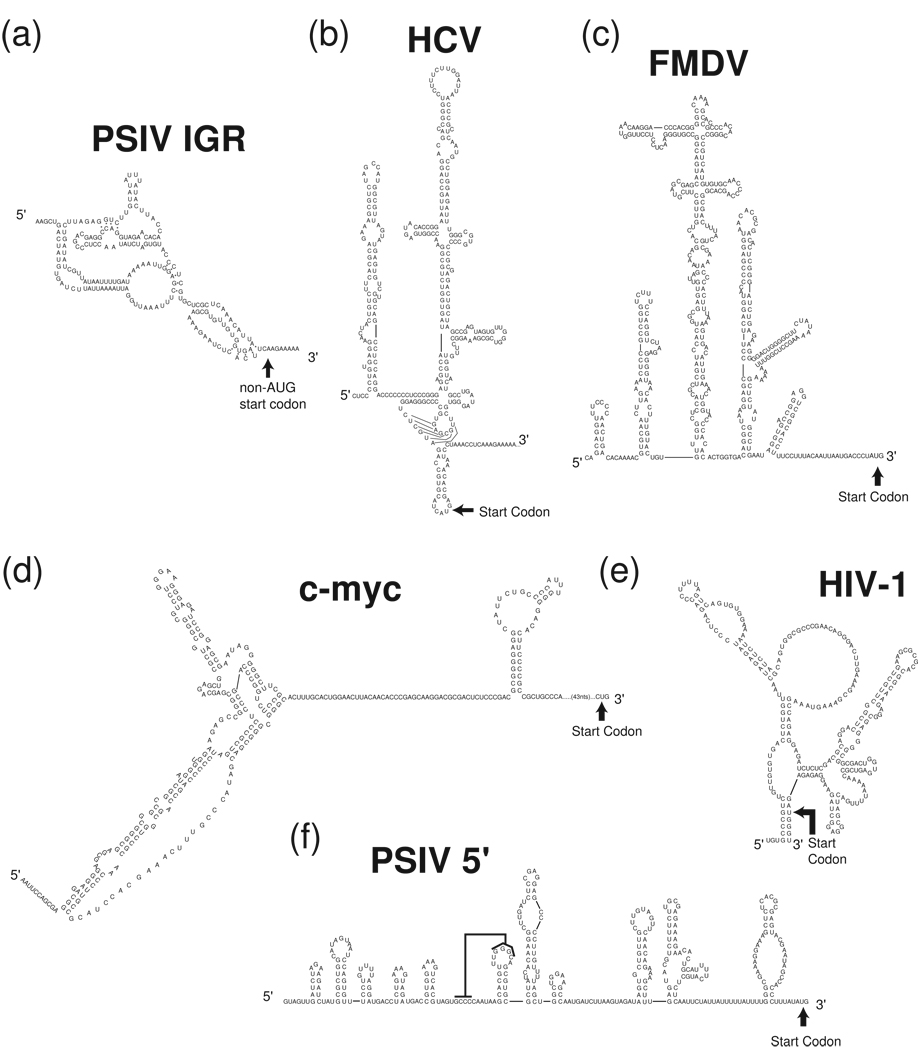
Figure 3. Examples of viral and cellular IRES secondary structures.
Experimentally tested secondary structures of several diverse viral and cellular IRES RNAs are shown. (a) Plautia stali intestine virus (PSIV) IGR IRES. (b) HCV IRES (c) FMDV IRES (d) c-myc IRES (e) Human immunodeficiency virus-1 (HIV-1) gag- IRES (f) PSIV 5' IRES, the black line indicates a proposed pseudoknot interaction. Note that these secondary structures may be revised as more information becomes available regarding differences between RNA made and folded in vitro versus that made in vivo, which folds co-transcriptionally.
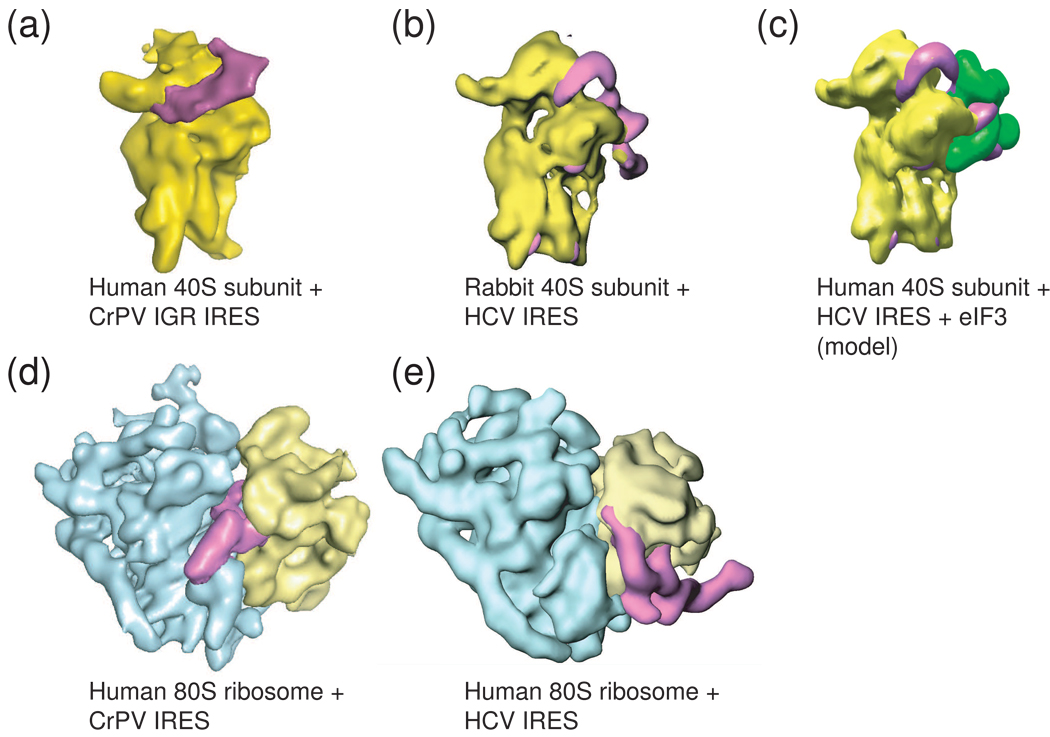
Figure 4. Cryo-EM reconstructions of two different viral IRESs bound to the ribosome.
To date, only the HCV IRES (and related) and the Dicistroviridae IRES RNAs have been shown to bind directly to ribosomal subunits, and these complexes have been studies by cryo-EM. (a) The Cricket paralysis virus (CrPV) IGR IRES (magenta) bound to the 40S subunit (yellow). (b) The HCV IRES (magenta) bound to a 40S subunit (yellow). These two IRES types occupy different sites on the subunit, but both induce a similar conformational change in the 40S subunit when they bind, suggesting some mechanistic convergence. (c) Model of the HCV IRES bound to the 40S subunit and eIF3 (green), built from several cryo-EM reconstructions. (d) The CrPV IGR IRES bound to an 80S ribosome, the view is rotated 90 degrees from the view in panel (a). Note that higher resolution reconstructions of this complex have been published, but to allow more direct comparison with the HCV IRES-bound ribosome, the middle resolution structure is shown. (e) The HCV IRES bound to an 80S ribosome. Again, the different binding modes of these two IRESs to the ribosome are obvious, as are differences in the overall folded architectures of the IRESs; the CrPV IGR IRES is more compact, while the HCV IRES is extended. Cryo-EM reconstructions of preiniation complexes containing other IRESes have not been reported.
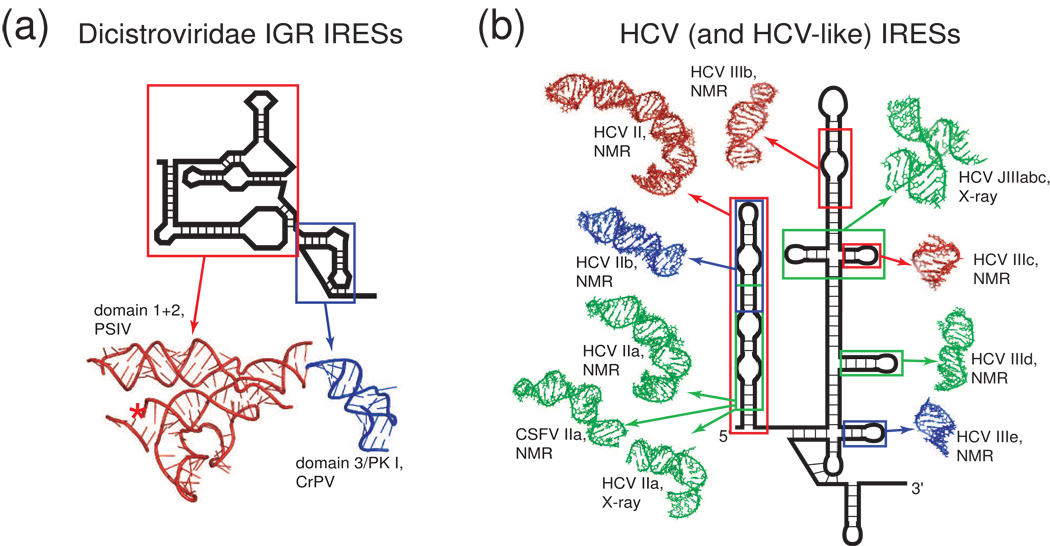
Figure 5. IRES RNA domains whose structures have been solved by X-ray crystallography or NMR.
The structures of only a handful of IRES RNA domains have been determined. (a) Secondary structure cartoon of a Dicistroviridae IGR IRES with ribbons diagrams of the two structural domains of this IRES that have been solved by X-ray crystallography. This is the only IRES for which a complete structural picture exists. (b) Secondary structure cartoon of the HCV IRES surrounded by structures of various domains (shown in various colors) from both it and the CSFV IRES that have been solved by NMR or crystallography [25–31]. For both the HCV and the Dicistroviridae IGR IRESs, these structures have been combined with cryo-EM reconstructions to develop models for how the IRES RNAs interact with the ribosome (not shown).
Compactly folded IRES RNAs
The most highly structured IRES RNAs yet identified are the (+) ssRNA Dicistroviridae intergenic region (IGR) IRESs (hereafter referred to as the IGR IRESs) that fold into a specific and compact three-dimensional structure [10]. In fact, these IRES RNAs have biophysical characteristics that are similar to catalytic RNAs, emphasizing their tightly folded character. These highly structured IRESs do not require any initiation factors, and operate essentially as an all RNA-based ribosome recruitment apparatus (for review: [6]). The folding characteristics of the IGR IRESs make them amenable to high-resolution structural studies; detailed structural information has come from a combination of X-ray crystallographic studies of the unbound IRES RNA (Figure 5a) [11,12] and cryo-EM studies of IRES RNA bound to the ribosome (Figure 4a,d) [13,14]. Even before encountering the ribosome, the IGR IRESs “prefold” into a conserved conformation that is required for binding both ribosomal subunits in a process that, remarkably, does not require GTP hydrolysis [10,15–18]. The IRES occupies part of the space within the ribosome used by tRNAs, changes the conformation of both ribosomal subunits, and directs translation initiation from a non-AUG codon in the ribosome’s A-site, suggesting that the IRES actively manipulates the ribosome (for review: [6,19,20]) possibly by mimicking a hybrid state tRNA [12,21].
Extended IRES RNAs with compact regions
The hepatitis C virus (HCV) IRES (and structurally related IRESs such as classical swine fever virus, CSFV) are part of a second structural class of IRESs that are mostly extended but maintain some structured and tightly packed regions [22] (review: [23,24]). The extended nature of the this class makes high-resolution structural studies of large portions of the RNA difficult, mandating a “divide and conquer” approach in which the structures of individual secondary structural elements are solved by NMR and crystallography (Figure 5b) [25–32], and then docked into cryo-EM reconstructions [33]. Like the IGR IRESs, the HCV IRES binds directly to the 40S ribosomal subunit and alters its conformation [34], but unlike the IGR IRESs, it also binds directly to eIF3 and it requires initiator tRNA, eIF2, and GTP hydrolysis to generate 80S ribosomes [35] (although recent reports show that under certain in vitro conditions, the HCV and CSFV IRESs can bypass some factor requirements [36,37]). Some reports implicate a role for IRES trans-acting factors (ITAFs; proteins not part of the canonical translation initiation machinery but important for the function of a specific IRES) in enhancing of inhibiting translation from the HCV IRES [38–44], but their role is unclear and there is no evidence they alter the global fold of the RNA or are necessary for 40S subunit binding [45,46]. Detailed mechanistic studies using a combination of genetic and biochemical analysis show that different parts of the HCV IRES structure are involved in different steps of the preinitiation complex assembly process. Specifically, one part is important for binding the 40S subunit [47–50], another promotes binding of eIF2 and tRNA [51,52], and yet another promotes phosphate release from eIF2 [30] (for review: [23,24]). Thus, by remaining extended even when bound to the ribosome (Figure 4b, c, e) [33,34,53], the HCV IRES correctly positions various structural elements to interact with, direct, and coordinate the action of different components of the translation initiation machinery in a process that is more complex than that used by the IGR IRESs.
Extended and largely flexible IRES RNAs
The final structural class of (+) ssRNA viral IRESs considered here do not fold into globally compact structures, but retain some conformational flexibility before binding to the ribosome and other factors. They comprise mostly picornavirus IRESs such as the foot-and-mouth disease virus (FMDV) (review: [54,55]), but some picornaviruses have HCV-like IRES [56,57] (review: [58]). Using the FMDV IRES as an example, there is evidence for long-range RNA-RNA interactions [59–61] but no direct evidence of tightly packed regions. Specifically, solvent-accessibility probing experiments with the IGR IRESs and HCV IRESs reveal protected areas of tight RNA backbone packing [10,22]; however, no such protections have been found in the FMDV IRES (unpublished data). This does not mean that the FMDV IRES is unstructured, but indicates it has a very different biophysical character than do the IGR and HCV-like IRESs. The FMDV IRES cannot bind directly to the 40S subunit, but requires the C-terminal half of eIF4G, eIF4A, eIF5B, eIF3, and two ITAFs: polypyrimidine binding protein (PTB, also called hnRNP-I), and ITAF45 [62–65]. Poliovirus, encephalomyocarditis virus (EMDV), and hepatitis A virus (HAV) IRESs also require several eIFs and often ITAFs in order to recruit the ribosome (review: [4]). The requirements of these less-structured IRES RNAs for more factors suggests that the IRES is part of a multi-component ribonucleoprotein (RNP) complex with eIFs and ITAFs organized on an RNA scaffold that then recruits the ribosome. Because there are no high-resolution structures or cryo-EM reconstructions of these IRESs, the three-dimensional structures of IRES RNPs and any conformational changes in the ribosome that occur within the preiniation complex are unknown.
When the biophysical characteristics of the IGR, HCV, and FMDV IRES RNAs are compared, we see a trend: the amount of inherent folded structure in the unbound IRES RNA is inversely correlated with the need for ITAFs and eIFs (Figure 2). We readily admit that this observation is based on a limited number of examples, and that once the three-dimensional structures, biophysical characteristics, and factor requirements of many other viral RNAs are known, this view could change.
What are the structural roles of ITAFs?
The need for ITAFs on the less-structured viral IRES RNAs raises the question of ITAF function. One hypothesis is that they stabilize a specific IRES RNA conformation that enables binding of other factors or the ribosome. Support for this “chaperone model” is found in the fact that the more structured IRESs such as the IGR IRES and the HCV-like IRES do not need ITAFs to bind the ribosome or eIFs, but that binding of PTB and ITAF45 to the FMDV IRES induces structural changes in the IRES, and synergistically enhances eIF4G and eIF4A binding [62,66]. The chaperone model is appealing and clearly explains the role of ITAFs on some IRESs, but it does not preclude the possibility that in some cases ITAFs could make direct contact to the ribosome or other factors (Figure 6). It also elicits the question of what components of the IRES RNA make up the ribosome-binding surface.
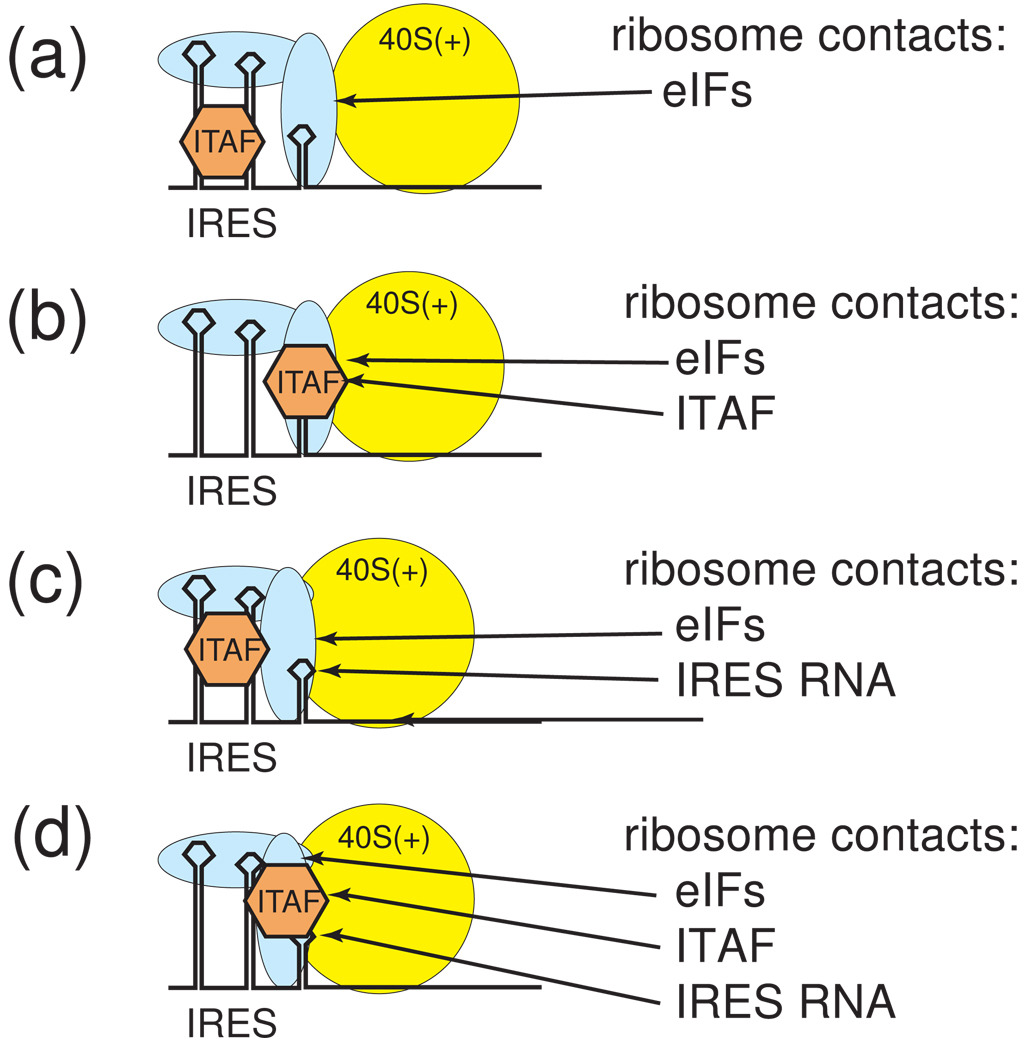
Figure 6. Models for ribosome recruitment by IRES•eIF•ITAF complexes.
Within complex “IRES RNPs,” does the ribosome contact only the bound eIFs, a combination of eIFs and the IRES RNA, a combination of eIFs and ITAFs, etc.? The answer to this question may vary depending on the IRES, and awaits more structural information. A few possibilities are diagrammed here. (a) The ITAF could stabilize the active conformation of the IRES RNA, which is bound by eIFs that interact with the ribosome, but without direct interaction between the IRES RNA and the ribosome. (b) The ITAF and eIFs could both interact directly with the ribosome, again with no direct IRES RNA-ribosome interactions. (c) The ITAF could stabilize the active conformation of the IRES RNAs, and both the IRES and eIFs could contact the ribosome. (d) The ITAF, the eIFs and the RNA could all directly contact the ribosome. Note that other combinations could occur and that these are not mutually exclusive possibilities.
Are structures of viral IRESs teaching us anything about cellular IRESs?
Unlike viral IRESs, which do not originate in the cell, cellular IRESs perform many roles associated with differentiation, mitosis, stress, proliferation, and other conditions where cap-dependent translation initiation is diminished [3]. For example, hypoxia inhibits cap-dependent protein synthesis but leads to translation of growth factors that promote blood vessel formation, including IRES-driven proteins such as the alpha subunit of hypoxia-inducible factor-1 (HIF-1) [67,68] and fibroblast growth factor 2 [69]. HIF-1 transcriptionally upregulates vascular endothelial growth factor-A, which itself uses two IRESs [70]. These examples illustrate the idea that cellular IRESs, much like viral IRESs, have evolved to respond to the state of the cell and to tightly regulate translation from individual messages, possibly in a variety of ways.
Given advances in understanding viral IRES structure, it is worth asking if we can apply what we have learned about the structural basis of viral IRES function to cellular IRESs. One area in which viral and cellular IRESs are similar is the factors they employ. The factor requirements for only a few cellular IRES have been determined, but for those it is clear they also use many eIFs and ITAFs. For example, the c-myc and N-myc IRESs use the C-terminal part of eIF4G, eIF4A, eIF3, and eIF2; while the L-myc IRES uses eIF4F complex, poly-(A) binding protein (PABP), and eIF3 [71]. Human immunoglobulin heavy-chain binding protein IRES uses the C-terminal part of eIF4G that binds eIF4A [72], and the S. cerevisiae YMR181c IRES binds PABP and recruits eIF4G [73]. Also, many of the same proteins identified as viral ITAFs are used by cellular IRESs, inlcuding PTB, hnRNP-A1 (and other hnRNP proteins), La, and unr.
The rate at which cellular IRESs have been discovered has far outpaced the rate at which they have been structurally characterized. In many cases it is not clear that a defined secondary structure exists or plays any role in the activity of the IRES, hence we hesitate to draw strong conclusions. However, the structures of the c-myc and apoptotic peptidase activating factor 1 (Apaf-1_ IRESs are illuminating. In the c-myc IRES, the tertiary structure contains several pseudoknots; interestingly, destabilization of certain structures within the IRES increases its activity, suggesting these structures are inhibitory [74]. Likewise, ITAF binding to the Apaf-1 IRES RNA causes part of the RNA to become less structured and increases IRES activity, probably by providing access for ribosomal subunits or other factors to bind [75]. Recently, it has been shown that the activity of certain yeast and Drosophila IRESs is inversely correlated to the stability of any secondary structure [76]. The pattern of less structure = higher IRES activity seen in these cellular IRESs seems very different from the functional requirements of viral IRESs to form very specific and stable structures.
The recognition that ITAFs can induce structural changes in cellular IRESs (and viral IRESs as well) leads to an appealing model of how these IRESs precisely response to changing cellular conditions. Specifically, as a cell’s state changes, the localization and cellular concentration of proteins are altered, and this is sensed by structural changes and activation of a subset of cellular IRESs, leading to translation of specific messages (review: [77]). Evidence for this model is found in studies showing that known ITAFs are regulated by changes in subcellular localization [78], but there is little direct evidence linking these localization changes to IRES RNA structure.
Conclusions
It is clear that “diverse” is the word that best describes IRES structure. No universally conserved IRES sequences or structural motifs have yet been identified (although structures such as pseudoknots appear often). In some cases, specific and stable RNA structure is necessary for IRES activity; in other cases stable structure is inhibitory. In viral IRESs, there appears to be a correlation between the biophysical characteristics of the folded architectures of IRES RNAs and their eIF/ITAF requirements. In most instances it is difficult to extend what we have learned from viral IRES structure to cellular IRES structure. However, we suspect common features will emerge in how they use some ITAFs and eIFs as more IRES structures and IRES RNP structures are studied.
The gaps in knowledge help chart a path for future structural studies of IRESs: to date there is direct structural data for only two viral IRESs; exploring the global architecture and high-resolution structures of more IRES RNAs and also eIF- and ITAF-containing IRES RNPs will be challenging but rewarding. In addition, studies to understand the dynamics of IRES RNA structures and IRES RNP composition will be critical to develop models describing how IRESs manipulate the translation machinery and how they operate within the changing conditions of the cell. Finally, as structural information is amassed on more cellular IRESs, we should gain a better understanding of the similarities and differences when compared to viral IRES.
Acknowledgements
We would like to thank Mark Johnston, Tom Blumenthal, Richard Davis, Leslie Krushel, Sten Wie, Monique Beaudoin, Terra-Dawn Plank, Kelli Kline, David Costantino, John Hammond, and Erica Nolte for critical reading of this manuscript. IRES work in the Kieft Lab is support by NIH grants R01GM072560 and R01GM081346 (JSK) and American Heart Association Protectoral Fellowship #0815655G (MEF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pestova T, Lorsch JR, Hellen CU. The Mechanism of Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, editors. Translational Control in Biology and Medicine. Hershey JWB: Cold Sring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 2.Doudna JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, editors. Translational Control in Biology and Medicine. Hershey JWB: Cold Spring Harbor Laboratory Press; 2007. pp. 129–153. [Google Scholar]
- 3.Elroy-Stein O, Merrick WC. Translation initiation via cellular internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey J, editors. Translational Control in Biology and Medicine. Hershey J: Cold Spring Harbor Laboratroy Press; 2007. pp. 155–172. [Google Scholar]
- *4.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- **5.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Sarnow P. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J Virol. 2003;77:2801–2806. doi: 10.1128/JVI.77.5.2801-2806.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricci EP, Herbreteau CH, Decimo D, Schaupp A, Datta SA, Rein A, Darlix JL, Ohlmann T. In vitro expression of the HIV-2 genomic RNA is controlled by three distinct internal ribosome entry segments that are regulated by the HIV protease and the Gag polyprotein. RNA. 2008;14:1443–1455. doi: 10.1261/rna.813608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ricci EP, Soto Rifo R, Herbreteau CH, Decimo D, Ohlmann T. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem Soc Trans. 2008;36:690–693. doi: 10.1042/BST0360690. [DOI] [PubMed] [Google Scholar]
- 10.Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **11.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **12.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **13.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM Visualization of a Viral Internal Ribosome Entry Site Bound to Human Ribosomes; The IRES Functions as an RNA-Based Translation Factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- *15.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 16.Pfingsten JS, Costantino DA, Kieft JS. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J Mol Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *17.Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 19.Pfingsten JS, Kieft JS. RNA structure-based ribosome recruitment: lessons from the Dicistroviridae intergenic region IRESes. RNA. 2008;14:1255–1263. doi: 10.1261/rna.987808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *20.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus IRES. J Biol Chem. 2007 doi: 10.1074/jbc.M610887200. [DOI] [PubMed] [Google Scholar]
- 22.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- *23.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 25.Klinck R, Westhof E, Walker S, Afshar M, Collier A, Aboul-Ela F. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA. 2000;6:1423–1431. doi: 10.1017/s1355838200000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 27.Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 28.Collier AJ, Gallego J, Klinck R, Cole PT, Harris SJ, Harrison GP, Aboul-Ela F, Varani G, Walker S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat Struct Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- **29.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 30.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. Embo J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dibrov SM, Johnston-Cox H, Weng YH, Hermann T. Functional architecture of HCV IRES domain II stabilized by divalent metal ions in the crystal and in solution. Angew Chem Int Ed Engl. 2007;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- 32.Rijnbrand R, Thiviyanathan V, Kaluarachchi K, Lemon SM, Gorenstein DG. Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites. J Mol Biol. 2004;343:805–817. doi: 10.1016/j.jmb.2004.08.095. [DOI] [PubMed] [Google Scholar]
- *33.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure (Camb) 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- **34.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- **35.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. Embo J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anwar A, Ali N, Tanveer R, Siddiqui A. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J Biol Chem. 2000;275:34231–34235. doi: 10.1074/jbc.M006343200. [DOI] [PubMed] [Google Scholar]
- 39.Tischendorf JJ, Beger C, Korf M, Manns MP, Kruger M. Polypyrimidine tract-binding protein (PTB) inhibits Hepatitis C virus internal ribosome entry site (HCV IRES)-mediated translation, but does not affect HCV replication. Arch Virol. 2004;149:1955–1970. doi: 10.1007/s00705-004-0341-8. [DOI] [PubMed] [Google Scholar]
- 40.Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5' noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initation of translation. J. Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J Biol Chem. 2000;275:27531–27540. doi: 10.1074/jbc.M001487200. [DOI] [PubMed] [Google Scholar]
- 42.Ali N, Siddiqui A. The La antigen binds 5' noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pudi R, Abhiman S, Srinivasan N, Das S. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J Biol Chem. 2003;278:12231–12240. doi: 10.1074/jbc.M210287200. [DOI] [PubMed] [Google Scholar]
- 44.Pudi R, Srinivasan P, Das S. La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J Biol Chem. 2004;279:29879–29888. doi: 10.1074/jbc.M403417200. [DOI] [PubMed] [Google Scholar]
- 45.Brocard M, Paulous S, Komarova AV, Deveaux V, Kean KM. Evidence that PTB does not stimulate HCV IRES-driven translation. Virus Genes. 2007;35:5–15. doi: 10.1007/s11262-006-0038-z. [DOI] [PubMed] [Google Scholar]
- 46.Nishimura T, Saito M, Takano T, Nomoto A, Kohara M, Tsukiyama-Kohara K. Comparative aspects on the role of polypyrimidine tract-binding protein in internal initiation of hepatitis C virus and picornavirus RNAs. Comp Immunol Microbiol Infect Dis. 2008;31:435–448. doi: 10.1016/j.cimid.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Lytle JR, Wu L, Robertson HD. The ribosome binding site of hepatitis C virus mRNA. J Virol. 2001;75:7629–7636. doi: 10.1128/JVI.75.16.7629-7636.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8:1045–1055. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–923. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **51.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 52.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci U S A. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 54.Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J Gen Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- *55.Fernandez-Miragall O, Quinto SL, Martinez-Salas E. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 2009;139:172–182. doi: 10.1016/j.virusres.2008.07.009. [DOI] [PubMed] [Google Scholar]
- *56.Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chard LS, Kaku Y, Jones B, Nayak A, Belsham GJ. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J Virol. 2006;80:1271–1279. doi: 10.1128/JVI.80.3.1271-1279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *58.Belsham GJ. Divergent picornavirus IRES elements. Virus Res. 2009;139:183–192. doi: 10.1016/j.virusres.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 59.Fernandez-Miragall O, Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez-Miragall O, Ramos R, Ramajo J, Martinez-Salas E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *61.Fernandez-Miragall O, Martinez-Salas E. In vivo footprint of a picornavirus internal ribosome entry site reveals differences in accessibility to specific RNA structural elements. J Gen Virol. 2007;88:3053–3062. doi: 10.1099/vir.0.83218-0. [DOI] [PubMed] [Google Scholar]
- **62.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 63.Saleh L, Rust RC, Fullkrug R, Beck E, Bassili G, Ochs K, Niepmann M. Functional interaction of translation initiation factor eIF4G with the foot-and-mouth disease virus internal ribosome entry site. J Gen Virol. 2001;82:757–763. doi: 10.1099/0022-1317-82-4-757. [DOI] [PubMed] [Google Scholar]
- 64.Stassinopoulos IA, Belsham GJ. A novel protein-RNA binding assay: functional interactions of the foot-and-mouth disease virus internal ribosome entry site with cellular proteins. RNA. 2001;7:114–122. doi: 10.1017/s1355838201001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopez de Quinto S, Lafuente E, Martinez-Salas E. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7:1213–1226. doi: 10.1017/s1355838201010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song Y, Tzima E, Ochs K, Bassili G, Trusheim H, Linder M, Preissner KT, Niepmann M. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bert AG, Grepin R, Vadas MA, Goodall GJ. Assessing IRES activity in the HIF-1alpha and other cellular 5' UTRs. RNA. 2006;12:1074–1083. doi: 10.1261/rna.2320506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conte C, Riant E, Toutain C, Pujol F, Arnal JF, Lenfant F, Prats AC. FGF2 translationally induced by hypoxia is involved in negative and positive feedback loops with HIF-1alpha. PLoS ONE. 2008;3:e3078. doi: 10.1371/journal.pone.0003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **70.Bornes S, Prado-Lourenco L, Bastide A, Zanibellato C, Iacovoni JS, Lacazette E, Prats AC, Touriol C, Prats H. Translational induction of VEGF internal ribosome entry site elements during the early response to ischemic stress. Circ Res. 2007;100:305–308. doi: 10.1161/01.RES.0000258873.08041.c9. [DOI] [PubMed] [Google Scholar]
- 71.Spriggs KA, Cobbold LC, Jopling CL, Cooper R, Wilson LA, Stoneley M, Coldwell MJ, Poncet D, Shen YC, Morley SJ, et al. Canonical initiation factor requirements of the Myc family of internal ribosome entry segments. Mol Cell Biol. 2009 doi: 10.1128/MCB.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thoma C, Bergamini G, Galy B, Hundsdoerfer P, Hentze MW. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol Cell. 2004;15:925–935. doi: 10.1016/j.molcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- **73.Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 74.Le Quesne JP, Stoneley M, Fraser GA, Willis AE. Derivation of a structural model for the c-myc IRES. J Mol Biol. 2001;310:111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- 75.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 76.Xia X, Holcik M. Strong eukaryotic IRESs have weak secondary structure. PLoS ONE. 2009;4:e4136. doi: 10.1371/journal.pone.0004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 78.Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033–1035. doi: 10.1038/sj.onc.1210777. [DOI] [PubMed] [Google Scholar]