SUMMARY
Structural changes in specific chromatin domains are essential to the orderly progression of numerous nuclear processes, including transcription. We report that the nuclear protein NSBP1 (HMGN5), a recently discovered member of the HMGN nucleosome-binding protein family, is specifically targeted by its C-terminal domain to nucleosomes in euchromatin. We find that the interaction of NSBP1 with nucleosomes alters the compaction of cellular chromatin and that in living cells, NSBP1 interacts with linker histones. We demonstrate that the negatively charged C-terminal domain of NSBP1 interacts with the positively charged C-terminal domain of H5 and that NSBP1 counteracts the linker histone-mediated compaction of a nucleosomal array. Dysregulation of the cellular levels of NSBP1 alters the transcription level of numerous genes. We suggest that mouse NSBP1 is an architectural protein that binds preferentially to euchromatin and modulates the fidelity of the cellular transcription profile by counteracting the chromatin-condensing activity of linker histones.
INTRODUCTION
Early studies revealed that at the global level, the chromatin fiber is organized into highly condensed, gene-poor, constitutive heterochromatin regions and into less-condensed, gene-rich euchromatin regions. Constitutive chromatin stains intensely and appears highly condensed throughout the cell cycle, while euchromatin condenses during mitosis, but during interphase decondenses and appears diffuse. An additional region of chromatin, named facultative heterochromatin, is enriched in silenced genes, which retain the potential for reactivation and conversion into euchromatin. Its cytological appearance is not well defined and depends on its state of activation (Fraser and Bickmore, 2007; Trojer and Reinberg, 2007). The dynamic properties of the chromatin fiber play a key role in the regulation of transcription and in the orderly progression of development and differentiation. In living cells, the chromatin fiber is continuously remodeled by regulatory complexes that modify specific amino acid residues in the nucleosomal histones or reposition nucleosomes along specific DNA sequences (Henikoff, 2008; Ng and Gurdon, 2008; Ruthenburg et al., 2007). In addition, structural proteins such as the linker histone H1 (Bustin et al., 2005; Woodcock et al., 2006) and members of the high-mobility group (HMG) protein superfamily (Bianchi and Agresti, 2005; Bustin, 1999), which bind to chromatin without any obvious specificity for DNA sequence or histone modification, dynamically alter the local and global architecture of the chromatin fiber and affect the binding of regulatory factors to their nucleosomal targets (Fan et al., 2005; Horn et al., 2002; Zlatanova et al., 2008). Although extensively studied, the cellular function and mechanism of action of these nucleosome-binding architectural proteins are still not fully understood.
Imaging analysis of living cells reveals that the interaction of most nuclear proteins including histone H1 and HMGs with chromatin is highly dynamic and that they continuously move throughout the nucleus, sampling the nucleosomes for optimal binding sites (Phair et al., 2004). Moreover, H1 variants and HMG proteins compete for nucleosome-binding sites, thereby creating a dynamic network of interdependent chromatin-binding activities in which the binding of one protein affects the binding of other members of the network (Bustin et al., 2005; Catez et al., 2004). The continuous turnover of the structural proteins on the surface of nucleosomes is part of the mechanism that provides functional and structural plasticity to the chromatin fiber. Thus, although H1 and HMGs are devoid of catalytic activity and of DNA sequence-binding specificity, these structural proteins do play important roles in optimizing DNA-related activities in the context of chromatin. Indeed, studies of human disease and of genetically modified mice with altered HMG or H1 expression indicate that these proteins affect the cellular phenotype and that changes in their expression levels may lead to developmental abnormalities and disease (Bianchi and Agresti, 2005; Fan and Skoultchi, 2004; Fusco and Fedele, 2007; Hock et al., 2006).
Although chromatin-binding proteins move rapidly throughout the nucleus and continuously turnover on nucleosomes, their global distribution in chromatin is not random. Thus, HP1 proteins are preferentially associated with condensed heterochromatin regions (Elgin and Grewal, 2003), HMGA and HMGB proteins are evenly distributed throughout the nucleus, while histone H1 and HMGNs colocalize with DNA, and these proteins seem to be enriched in constitutive heterochromatin where the nucleosomes are densely packed (Catez et al., 2004; Phair et al., 2004).
HMGNs are the only nuclear proteins shown to specifically bind to the 147-base-pair nucleosome core particle (CP), the building block of the chromatin fiber. HMGNs bind to nucleosomes through a highly conserved region known as the nucleosomal binding domain (NBD), which is the hallmark and the functional motif of the HMG protein family (Bustin, 2001; Ueda et al., 2008). Specific amino acid mutations in this region abolish the specific interaction of HMGNs with nucleosomes and inactivate most of their biological functions.
NSBP1 (originally named NBP-45 and now renamed HMGN5) is a novel member of the HMGN family (Shirakawa et al., 2000). It has been classified as an HMGN because it binds specifically to nucleosome cores and because it contains three important functional domains of the HMGN family: the nuclear localization signal (NLS), the NBD, and a conserved motif in the C-terminal domain (Figure 1A). However, NSBP1 differs significantly from HMGNs because it has four times as many amino acids, a third of which are negatively charged, resulting in a highly acidic protein (Shirakawa et al., 2000).
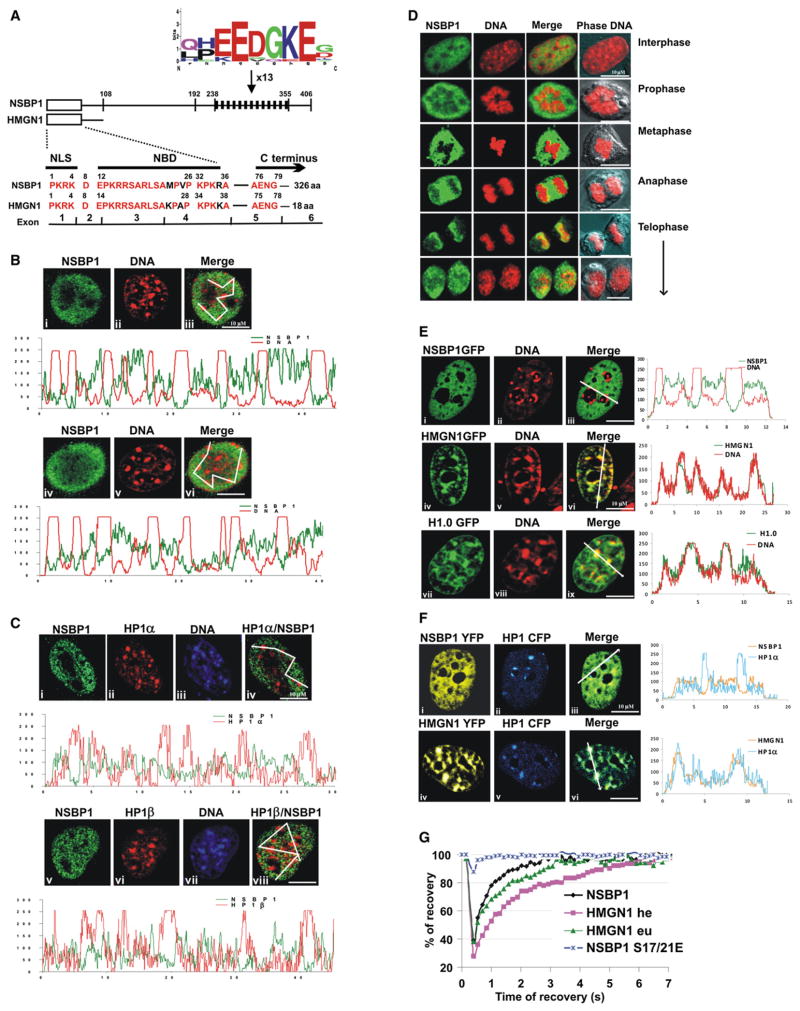
Figure 1. NSBP1 Protein Localizes to Nucleosomes in Euchromatin.
(A) Sequence similarity between HMGN1 and the N-terminal region of NSBP1. The schematic structure of NSBP1 and HMGN1 is shown above the sequence. The position of the exons is indicated below the sequence. NLS, nuclear localization signal; NBD, nucleosome binding domain. The location of the negatively charged sequence repeat is indicated by black bars; the consensus sequence of the repeat is shown above the diagram.
(B) NSBP1 does not localize to constitutive heterochromatin. Endogenous NSBP1 protein is visualized by immunofluorescence in green (i and iv) and DNA by Hoechst (ii and v). The merged images (iii and vi) and their localization profiles indicate that NSBP1 does not colocalize with constitutive heterochromatin (compact dots of DNA).
(C) NSBP1 does not colocalize with HP1 proteins. Immunostaining of endogenous NSBP1 protein is shown in green (i and v), HP1α (ii) or HP1β (vi) is shown in red, and corresponding DNA staining in blue (iii and vii). Merged images of NSBP1 and HP1 proteins (iv and viii) and their localization profiles are shown. Lack of yellow color indicates that NSBP1 does not colocalize with HP1α or HP1β.
(D) Endogenous NSBP1 (green) does not colocalize with metaphase chromosome DNA (red) throughout the mitosis.
(E) In live cells, NSBP1-GFP (i) is excluded from constitutive heterochromatin (ii, red), as shown by the absence of yellow (iii) and by the localization profiles. Controls of live cells expressing either HMGN1-GFP (iv) or H1-GFP (vii) indicate that these two proteins localize to heterochromatin.
(F) YFP-fused NSBP1 (i) or YFP-HMGN1 (iv) were cotransfected with CFP-fused HP1α (ii and v) and visualized in living cells. Merge images and colocalization profiles of NSBP1, HMGN1, and HP1α are shown. NSBP1 does not while HMGN1 does colocalize with HP1α in living cells.
(G) FRAP recovery curves for NSBP1-GFP and for HMGN1-GFP in either euchromatin (eu) or heterochromatin (he) indicate that, in living cells, both bind to chromatin. The extremely fast recovery of the FRAP signal for the NSBP1S17/21E indicates that this mutant does not bind to chromatin.
NSBP1 transcripts are present in many mouse (Shirakawa et al., 2000) and human (King and Francomano, 2001) tissues; however, the protein remains uncharacterized, and its biological function is unknown. Here, we report that the protein is ubiquitously present in all mouse tissues examined, suggesting that in differentiated tissues, it functions as a ubiquitously expressed nuclear protein. Interestingly, despite having an HMGN-canonical NBD, NSBP1 is depleted from constitutive heterochromatin and is preferentially targeted to nucleosomes in euchromatin by its acidic C-terminal domain. Alteration of the cellular levels of NSBP1 leads to widespread changes in the cellular transcription profile. We show that, both in vitro and in vivo, the interaction of NSBP1 with chromatin induces chromatin decondensation, and that NSBP1 binds to linker histones. We demonstrate that in vitro NSBP1 counteracts the chromatin-condensing activity of linker histone H5 and that the negatively charged C-terminal domain of NSBP1 interacts with the positively charged C-terminal domain of H5, a domain known to induce chromatin condensation. We suggest that the architectural changes in chromatin induced by the interaction of NSBP1 with nucleosomes in euchromatin are part of the cellular mechanism that fine-tunes the fidelity of the cellular transcription profile.
RESULTS AND DISCUSSION
NSBP1 Is a Ubiquitous Protein that Binds to Nucleosomes in Euchromatin
Previously, we reported that Nsbp1 transcripts are detectable in variable amounts in all the mouse tissues examined (Shirakawa et al., 2000). To test whether the protein is as widely expressed as the transcript, we prepared highly specific antibodies (Figure S1) and used western analysis to examine whether various mice tissues contain NSBP1 protein. We find that all the tissues examined contain NSBP1 protein (Figure S2), suggesting that the protein is widely expressed, perhaps acting as a ubiquitous chromatin architectural protein. In this respect, NSBP1 is similar to other members of the HMGN protein family, which are also ubiquitously present in most tissues. Furthermore, NSBP1 shares structural similarity with members of the HMGN protein family (Figure 1A). The N-terminal 86 amino acids, which are encoded by 5 distinct exons, contain the major functional hallmarks of the HMGN protein family: a highly conserved NLS encoded by the first exon, a highly conserved NBD encoded by exons 3 and 4, and the tetrapeptide AENG located in exon 5. However, the NSBP1 C-terminal region, encoded by exon 6, differs considerably from that of other members of the HMGN family, being 300 amino acids longer and significantly more acidic. The NSBP1 C-terminal domain is an array of multiple repetitive acidic sequences (Shirakawa et al., 2000); the most remarkable is the nanopeptide QP(LH)EEDGKEG(D), which is repeated 13 times in the C-terminal region spanning amino acids 238–355 (Figure 1A). Similar, but not identical, repeated acidic motifs are also present in the NSBP1 C-terminal region of other species (data not shown).
To examine whether the NSBP1 protein localizes to specific nuclear domains, we first analyzed its intranuclear localization by immunofluorescence in mouse embryonic fibroblasts (MEFs). We found that endogenous NSBP1 protein was not associated with a distinct nuclear subcompartment and was excluded from constitutive heterochromatic regions of chromatin, as judged by the lack of colocalization with DNA foci or with the heterochromatin-binding proteins HP1α and HP1β (Figures 1B, 1C, S5A, and S5B). NSBP1 did not colocalize with condensed DNA regions throughout the cell cycle and was not associated with mitotic chromosomes (Figure 1D). The salt concentrations at which the protein is released from nuclei are significantly lower than those necessary to extract histone H1 (Figure S3), suggesting that NSBP1 is not tightly associated with any nuclear structure.
Confocal microscopy of living cells expressing fluorescent fusion proteins verifies that, in vivo, NSBP1 is also not associated with constitutive heterochromatin regions, since its intranuclear location is clearly different from that of HMGN1 or H1, two proteins that are enriched in the constitutive heterochromatin regions identified by Hoechst staining of cellular DNA (Figures 1E and S5). Likewise, NSBP1 does not colocalize with HP1α, while HMGN1 does (Figure 1F). Furthermore, double immunofluorescence analyses reveal that NSBP1 is enriched in chromatin regions containing high levels of H3K9ac and H3K4me2,3, two histone modifications present in euchromatin, partially colocalized with H3K9me2 and H3K27me3, two modifications indicative of facultative heterochromatin (Figure S4), but is excluded from chromatin regions containing H3K9me3 (Figures 4B and S4), a modification associated with constitutive heterochromatin. Statistical analyses of multiple images such as shown in Figures 1, 2, S4, and S5, using heterochromatin-associated proteins Hp1α, linker histone H10, and HMGN1 as positive controls and H3K9Ac as a negative control for heterochromatin colocalization indicate that both the endogenous NSBP1 and the exogenously expressed NSBP1-GFP do not colocalize with constitutive heterochromatin. The Pearson correlation coefficients (see legend to Figure S5) obtained for NSBP1 and DNA staining were clearly different from those of the heterochromatin-binding protein for HP1 and H10 (Figure S5C). Thus, endogenous and exogenous NSPB1 is preferentially localized to euchromatin, a surprising finding since the protein contains the canonical HMGN NBD, and in vitro, it binds specifically to nucleosome CPs with an affinity constant similar to that of HMGN proteins (Shirakawa et al., 2000).
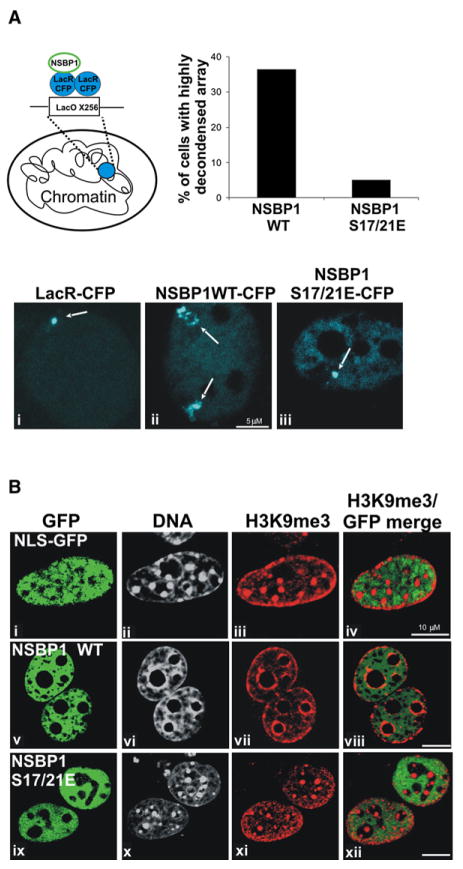
Figure 4. NSBP1 Induces Large-Scale Chromatin Decondensation.
(A) The interaction of NSBP1 with the nucleosomes decondenses a LacO chromatin array. Schematic drawing (top left) demonstrates the strategy for tethering NSBP1 protein to the array by fusion to LacR-CFP. The condensed array, visualized by tethering LacR-CFP (i), appears as a compact dot. Tethering of the wild-type NSBP1-CFP (ii) to the array induced formation of highly decondensed array, as judged by formation of extended structures with irregular shape. In contrast, tethering of NSBP1S17/21E mutant, which does not bind to nucleosomes (iii), did not unfold the condensed chromatin array. Bar graph above the images indicates the percent of cells containing highly decondensed arrays following tethering of either wild-type or NSBP1S17/21E mutant to the arrays. Thirty cells of each sample were examined.
(B) NSBP1 expression alters the global organization of chromatin. The global organization of the constitutive heterochromatin is visualized by DNA staining with Hoechst (ii, vi, and x) or H3K9me3 staining (iii, vii, and xi). Expression of NSBP1 leads to disappearance of the dense heterochromatic foci and condensation of heterochromatin around the nucleoli (vii). Merge images of GFP and H3K9me3 (iv, viii, and xii) demonstrate a lack of colocalization of NSBP1 protein with constitutive heterochromatin.
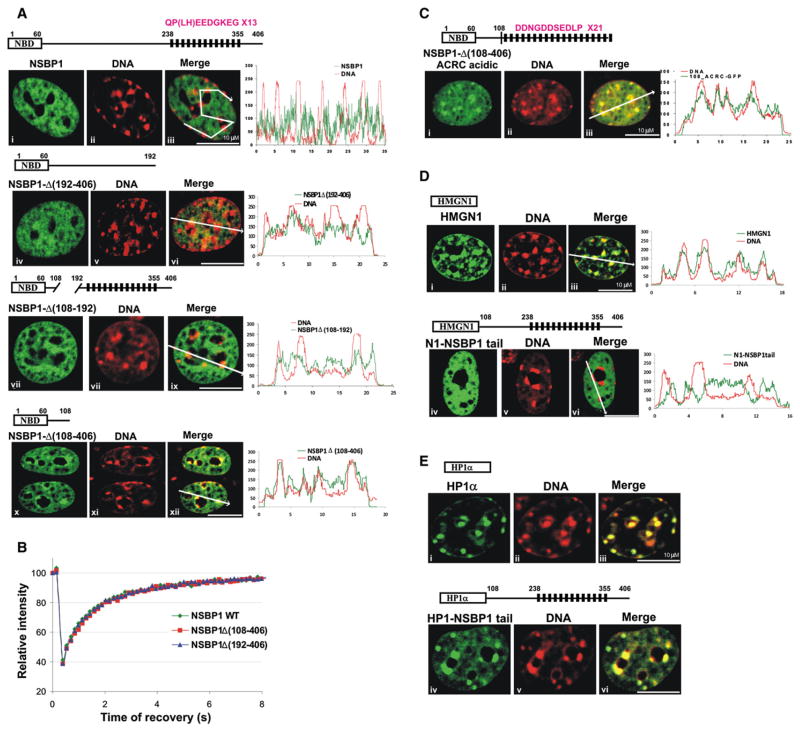
Figure 2. NSBP1 Is Specifically Targeted to Euchromatin by Its C-Terminal Tail.
(A) Intranuclear location of either NSBP1 protein (a–c) or various C-terminal deletion mutants (iv–vi, vii–ix, and x–xii), all expressed as GFP fusion proteins, compared to that of DNA (red), in living cells. A schematic diagram of the expressed proteins is shown above the corresponding images. Merge images (iii, vi, ix, and xii) and colocalization profiles demonstrate that progressive deletion of the C-terminal domain leads to a progressive relocation of the protein to heterochromatin.
(B) FRAP analyses of NSBP1-GFP and its C-terminal deletion mutants indicate the C-terminal tail does not affect the affinity of the protein to nucleosomes.
(C) A chimeric protein in which the C-terminal domain of NSBP1 protein was exchanged with the acidic C-terminal domain of human ACRC protein localized to heterochromatin. Schematic presentation of acidic portion of ACRC protein fused to tailless NSBP1 is shown above the images. Black rectangles represent the 21 repeats in ACRC. Consensus sequence of the repeat is shown (compare to Figure 2A).
(D) The NSBP1 C-terminal domain targets HMGN1 to euchromatin. Shown are the location of wild-type HMGN1 (i–iii) and the HMGN1 fused to the NSBP1 tail (iv–vi) in live cells. The merged images and the localization profile demonstrate that the wild-type HMGN1, but not the HMGN1-NSBP1 chimera, colocalizes with heterochromatin (DNA).
(E) The C-terminal domain of NSBP1 does not affect the chromatin interactions of HP1α. Wild-type HP1α (i) and HP1α fused to NSBP1 tail (iv–vi) were visualized in live cells. Colocalization of the proteins with Hoechst staining of DNA (ii and v) showed enrichment of both wild-type HP1α (iii) and HP1α fused to NSBP1 tail (v) in heterochromatin, as judged by the appearance of yellow color in merge images (iii and vi).
The difference in the nuclear localization of HMGN1 and NSBP1 raises the possibility that in living cells, the NSBP1 NBD is not functional and that the protein does not bind to nucleosomes. We previously demonstrated that fluorescence recovery after photobleaching (FRAP) is a sensitive tool to assess the interaction of HMGN protein with nucleosomes and that mutation of two conserved serine residues in the canonical NBD (see Figure 1A) significantly reduces the binding of the proteins to chromatin, both in vitro and in vivo (Ueda et al., 2008). The FRAP recovery curves of NSBP1-GFP were similar to those of HMGN1-GFP in euchromatin, although the chromatin residence time of NSBP1 was somewhat shorter, suggestive of a weaker binding to nucleosomes (Figure 1G). Most significantly, mutations in the conserved serine residues located in the NBD abolished the interaction of NSBP1 with chromatin, as evidenced by the large increase in intranuclear mobility, typical of nucleoplasmic proteins that do not bind to chromatin (Catez et al., 2004; Phair et al., 2004). Thus, like all HMGNs, the interaction of NSBP1 with chromatin is governed by the conserved NBD, an indication that the protein interacts with nucleosomes. Yet, despite these similarities with HMGNs that preferentially localize to heterochromatin, NSBP1 localizes to euchromatin. We therefore conclude that NSBP1 binds to nucleosomes in euchromatin.
The C-Terminal Domain of NSBP1 Localizes HMGNs to Euchromatin
The major structural difference between NSBP1 and other members of HMGN protein family is its long acidic C terminus, suggesting that it may affect its nuclear localization. Indeed, the NSBP1ΔC192–406 mutant lacking the C-terminal 214 amino acids partially colocalized with heterochromatin (Figure 2A, second row), while the NSBP1ΔC108–406 mutant fully colocalized with heterochromatin, and its distribution was similar to that of HMGN1 (compare bottom row in Figure 2A to top row in Figure 2D). An internal deletion of 84 amino acids (NSBP1ΔC108–192) partially increased the proportion of the protein associated with DNA (Figure 2A, third row), an indication that this region also affects the location of NSBP1 in chromatin. Although the C-terminal domain affected the intranuclear localization of the protein, the FRAP profiles of the wild-type protein are indistinguishable from that of the deletion mutant (Figure 2B). Thus, the C-terminal domain targets NSBP1 to euchromatin, but does not affect the affinity of the protein to nucleosomes, a finding that is in full agreement with the notion that the protein binds to nucleosomes through its NBD.
To test whether the negative charge in the C-terminal domain is the main element that targets NSBP1 to euchromatin, we replaced this domain with the highly acidic part of the acidic repeat-containing (ACRC) human protein (Nolte et al., 2001). ACRC resembles NSBP1, since its C-terminal domain contains 21 repeats of a decapeptide that is even more acidic than the repeat found in NSBP1 (compare peptide sequence in Figure 2A to that in Figure 2C). Although the C-terminal domain of the resulting NSBP-ACRC chimera protein is more acidic than that of the wild-type NSBP1, it binds to heterochromatin, and its nuclear location mimics that of the DNA, as evidenced by the confocal analysis of cells expressing the GFP-tagged mutant protein (Figure 2C). Thus, the specific sequence of the C-terminal domain, rather than its high negative charge, targets NSBP1 to euchromatin.
The NSBP1 C-terminal domain could serve either as a general euchromatin targeting module or as an HMGN-specific targeting module. To distinguish between these possibilities, we fused the entire C-terminal domain (residues 109–406) to either HMGN1 or to HP1α. Similar to wild-type NSBP1, the resulting HMGN1-NSBP1-tail chimera protein was preferentially localized to euchromatin (Figure 2D). In contrast, the HP1α-NSBP1-tail chimera still localized to heterochromatin, the cellular target of HP1α (Figure 2E). The results indicate that NSBP1 C-terminal domain per se does not change the protein localization in the chromatin and does not change the specificity of the chromatin-interacting site. Thus, since HP1α has binding sites for heterochromatin, but not for euchromatin, the HP1α-NSBP1 chimera also localizes to heterochromatin. HMGNs, however, bind to all nucleosomes through their highly conserved NBD, and in the context of this NBD, the C-terminal domain of NSBP1 specifically targets NBD-bearing proteins to euchromatin. These results indicate that the C-terminal domain of NSBP1 is HMGN specific, rather than a “universal” euchromatin-targeting module.
NSBP1 Modulates the Cellular Transcription Profile
The location of NSBP1 in euchromatin, taken together with its potential to activate transcription (Shirakawa et al., 2000), raises the possibility that NSBP1 may affect the cellular transcription profile. In support, immunofluorescence analyses indicate that NSBP1 is partially colocalized with RNAPII (Figure S6). We selected AtT20 cells, a mouse pituitary cell line (Ooi et al., 2004), to test the role of NSBP1 in modulating the cellular transcription profile, because an inquiry of the GEO database (NCBI) indicated high Nsbp1 expression in mouse pituitary gland and because immunofluorescence analyses indicate that also in AtT20 cells, NSBP1 is not enriched in heterochromatin (Figure 3A).
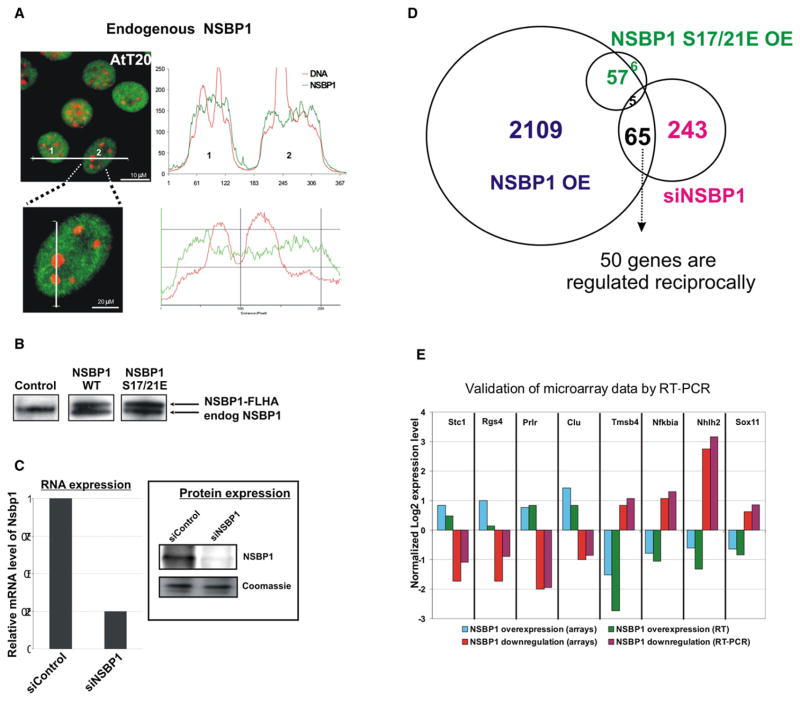
Figure 3. NSBP1 Modulates the Transcription Profile of AtT20 Cells.
(A) Fluorescent images of mouse pituitary AtT20 cells demonstrate that NSBP1 is depleted from constitutive heterochromatin. Localization profiles are shown for two cells, labeled 1 and 2, and a magnification of cell 2.
(B) Western blot analysis of AtT20 cells stably transfected with vectors expressing FLAG-HA (FLHA)-tagged wild-type (WT) NSBP1 or NSBP1S17/21E. Note that the levels of the exogenous protein are similar to the endogenous NSBP1 (endog NSBP1).
(C) Downregulation of NSBP1 expression by siRNA. Shown are RT-PCR and western blot analysis of AtT20 cells treated with siRNA against NSBP1 or control siRNA.
(D) Venn diagram depicting the overlap of genes whose expression is altered by either overexpressing wild-type NSBP1 (NSBP1 OE) or the NSBP1S17/21E mutant (NSBP1S17/21E OE) or by siRNA-mediated downregulation of NSBP1 (siNSBP1).
(E) Validation of microarray data. Shown is normalized log 2 expression of eight selected genes whose expression was reciprocally up- and downregulated by the overexpression or deletion of NSBP1 in AtT20 cells.
By quantitative western analyses, we estimated that there are approximately 12,000 NSBP1 molecules per AtT20 cell (Figure S7). We used a retroviral-based expression system to stably overexpress and approximately double the levels of NSBP1. As controls, we overexpressed similar amounts of the NSBP1S17/21E mutant that does not bind to chromatin. The resulting AtT20 clones expressed the FLAG- and HA-tagged exogenous proteins at levels comparable to those of the endogenous NSBP1 protein (Figure 3B). Additionally, we efficiently downregulated Nsbp1 expression with siRNA (Figure 3C) and tested whether loss of the protein affects the cellular transcription profile. The transcription profile of the various cells was analyzed using Affymetrix expression arrays (see Experimental Procedures).
Class comparison of significant changes in transcription profiles (p < 0.001) indicated that overexpression of wild-type NSBP1 altered the expression of over 2000 genes (Table S1 and Figure 3D). Significantly, overexpression of similar levels of the NSBP1S17/21E mutant protein had a considerably lower effect on the transcription profile, as the expression of only 68 genes was affected. Thus, the changes in the transcription profile are due to the interaction of NSBP1 with chromatin. Downregulation of NSBP1 altered the expression of 308 genes. The expression of 65 genes was affected by both the upregulation and the downregulation of NSBP1 protein levels; 50 of these genes were reciprocally affected. The results of the expression arrays were validated by RT-PCR of 8 genes whose expression was most significantly and reciprocally changed by either over-expression or downregulation of NSBP1 (Figure 3E). All the changes in transcription levels were highly significant; however, the expression levels of most of the affected genes changed only moderately (between 1.5- and 2-fold); less than 10% of the genes were changed by more than 2-fold. Gene ontology analysis did not suggest that NSBP1 preferentially affects a specific pathway or a distinct set of biological functions.
The effects of NSBP1 on the cellular transcription profile are similar to those resulting from altering the levels of histone acetylation (Chittur et al., 2008; Fenic et al., 2008). They also coincide with previously reported transcriptional changes following altered expression of structural chromatin binding proteins, such as H1 (Fan et al., 2005), the methyl-CpG binding protein (MeCP2) (Chahrour et al., 2008), and the genome organizing protein SATB1 (Han et al., 2008). Loss of H1 or of MeCP2, two structural proteins that bind to multiple sites and are known to condense chromatin, leads to both up- and downregulation of gene expression. Histone acetylation leads to global chromatin decompaction, and HDAC treatment alters the expression of numerous genes, yet in some instances, more genes are down-regulated then upregulated. The emerging picture suggests that perturbation of chromatin structure leads to complex changes in the cellular transcription profile. The fact that multiple genes were up- and downregulated by altered NSBP1 expression is in full agreement with the role of NSBP1 as a chromatin architectural protein rather than specific transcription factor.
In a fashion similar to that of other chromatin-binding proteins, the NSBP1-mediated global changes in gene expression could be due to a direct interaction of the protein with genomic regulatory sequences, to secondary effects on the function of chromatin-regulatory factors, to direct changes in chromatin, or to a combination of these. While the exact mechanism whereby NSBP1 affects the expression of specific genes remains to be elucidated, it is clear that the effects are due to the interaction of NSBP1 with nucleosomes, since forced expression of the NSBP1S17/21E mutant that does not bind to chromatin affects the expression of only 68 genes. Thus, the interaction of NSBP1 with nucleosomes participates in the cellular mechanisms that modulate the fidelity of the cellular transcription profile.
NSBP1 Decompacts Cellular Chromatin
NSBP1 is classified as a member of the HMGN protein family; these proteins are known to bind specifically to nucleosomes, alter the structure of chromatin, and affect transcription (Bustin, 2001). Moreover, the structure of the NSBP1 C-terminal tail is reminiscent of acidic activators that have been shown to induce changes in the large-scale chromatin structure (Carpenter et al., 2005). Thus, the effects of NSBP1 on the cellular transcription profile are most likely the result of NSBP1-induced alterations in chromatin architecture.
To test the ability of NSBP1 to affect chromatin architecture, we used NIH2/4 cells that are stably transfected with an array of 256 repeats of Lac operon sequence (LacO) that can bind multiple copies of Lac repressor (LacR) fused to CFP (LacR-CFP). Fusion proteins containing the LacR-CFP are tethered to the LacO sites, inducing local structural changes that can be visualized in living cells (Soutoglou and Misteli, 2008; Tumbar et al., 1999). In the absence of LacO, the LacR-CFP forms a small, compact dot in the nucleus of the cells (Figure 4Ai). Tethering of wild-type NSBP1 to the array leads to significant chromatin decondensation, as evidenced by the formation of significantly extended structures in more than 35% of the transfected cells (Figure 4Aii). Significantly, the tethering of the double mutant NSBP1S17/21E, which does not bind specifically to nucleosomes, did not lead to array decondensation (Figure 4Aiii), thereby indicating that chromatin decondensation is contingent on the correct binding of NSBP1 to nucleosomes. The chromatin unfolding is not due to the loss of histone H3 or linker histone H1 and is not accompanied by increased histone acetylation (Figure S8), suggesting that the altered chromatin architecture is not accompanied by significant changes in chromatin composition. The degree of array decondensation is reminiscent of that induced by the acidic domain of the VP16 activator (Tumbar et al., 1999), implying that NSBP1 induces large-scale higher-order chromatin unfolding.
In the LacO array experiments, NSBP1 is strongly tethered to a defined chromatin location by the high-affinity interaction between LacO and LacR. In living cells, however, the binding of NSBP1 to chromatin is weaker, as the protein rapidly shuttles between chromatin-binding sites (Figure 1G). Confocal microscopy of live cells overexpressing NSBP1-GFP reveals that the interaction of NSBP1 with nucleosomes also alters the large-scale structure of cellular chromatin. In cells expressing wild-type NSBP1, but not in cells expressing the mutant NSBP1S17/21E, the number of heterochromatic foci, identified by their intense staining with Hoechst, was significantly reduced, and the heterochromatic DNA regions were clustered around nucleoli (compare Figure 4Bvii to 4Bii or 4Bx). Immunofluorescence analyses with antibodies to the heterochromatic-specific histone modification H3K9me3 verified that the interaction of NSBP1 with chromatin leads to a significant reorganization of cellular heterochromatin. In control cells expressing only GFP or in cells expressing the NSBP1S17/21E-GFP double mutant that does not bind to nucleosomes, H3K9me3-containing chromatin regions are clustered into several dense heterochromatin foci and around the nuclear periphery (Figures 4Bi–iv and 4Bix–xii), while in cells expressing wild-type NSBP1 protein, these regions are significantly depleted, and H3K9me3-containing chromatin regions are now clustered around the nucleoli and in the nuclear periphery (Figure 4B, e–h). Thus, similar to the NSBP1-induced chromatin unfolding of the LacO arrays, NSBP1, but not the NSBP1S17/21E mutant, induces large-scale reorganization of native chromatin fibers.
NSBP1-mediated changes in large-scale chromatin organization are also evident by micrococcal nuclease digestion assays. The chromatin of cells overexpressing NSBP1 was digested faster than the chromatin of cells overexpressing the NSBP1S17/21E mutant, as evidenced by the accelerated reduction in the proportion of large oligonucleosomes and a concomitant increase in the relative proportion of di- and mononucleosomes (Figure S11). Taken together, all the data are consistent with the notion that the interaction of NSBP1 with nucleosomes reduces the compaction of the higher-order chromatin structure.
NSBP1 Counteracts the Linker Histone-Mediated Stabilization of Chromatin Compaction
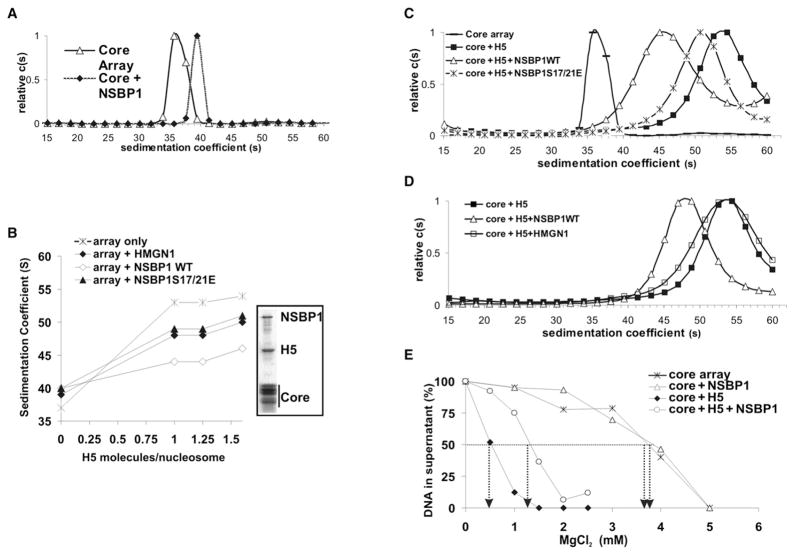
In considering possible mechanisms whereby NSBP1 reduces the compaction of the chromatin fiber, we focused on linker histone H1, the most abundant nucleosome-binding protein, whose ability to stabilize the compact structure of the chromatin fiber is well documented (Carruthers et al., 1998; McBryant et al., 2006; Robinson and Rhodes, 2006; Woodcock et al., 2006). In these studies, we tested the effect of purified NSBP1 protein on the structure of a defined nucleosome array, which either did or did not contain the linker histone H5, a linker histone variant whose interaction with chromatin has been previously studied (Carruthers et al., 1998; Robinson et al., 2008). In these arrays, nucleosomes are tightly positioned on the DNA, and therefore changes in chromatin compaction reflect spatial reorganization of chromatin architecture rather than nucleosome sliding. Following assembly, compaction of chromatin was measured by analytical ultracentrifugation and assessed by the sedimentation coefficient (S20,w) (Carruthers et al., 2000; Schuck, 2000).
In the absence of H5, the S20,w of the unfolded array was 37S, and addition of NSBP1 to the array increased the S20,w to 40S, most likely reflecting the increased mass of the NSBP1-containing arrays (Figure 5A). Addition of H5, at a ratio of one H5 molecule per CP or higher, increased the S20,w to 54S (Figure 5B), reflecting the increased compaction of the array. NSBP1 antagonizes the H5-induced chromatin compaction, as evidenced by the lower S20,w values obtained after the addition of both proteins. In the presence of NSBP1, the S20,w of the arrays was 45S, even when the H5-to-core-particle ratio was 1.6. Significantly, substitution of the wild-type NSBP1 with either the NSBP1S17/21E mutant, which does not bind to chromatin, or with HMGN1 protein, which lacks a long acidic domain, had a significantly lower effect on the chromatin compaction ability of H5. In the presence of these proteins, addition of H5 resulted in S20,w of 50S, an indication that both the chromatin-binding activity and the C-terminal domain of NSBP1 are required to fully antagonize the H5-mediated chromatin condensing activity. Mg2+-precipitation of the fully assembled array (Huynh et al., 2005; Robinson and Rhodes, 2006) reveals that the array contains both NSBP1 and H5 in amounts comparable to core histones (Figure 5B, box), indicating that NSBP1 prevents chromatin compaction without displacing H5 from the array.
Figure 5. NSBP1 Counteracts the Linker-Histone-Dependent Chromatin Compaction.
(A) NSBP1 does not decrease the compaction of a nucleosome array in the absence of linker histone H5. Shown are the sedimentation profiles of core arrays and core arrays to which NSBP1 was added at a molar ratio of ~2 NSBP1 molecules per core particle (CP).
(B) Efficient prevention of H5-mediated chromatin compaction by NSBP1. Shown are sedimentation coefficients of nucleosome arrays incubated with increasing amounts of H5 in the presence of the indicated proteins. NSBP1 and HMGN1 were added at a ratio of two molecules per nucleosome. The Coomassie-stained picture of the SDS-PAGE gel in the boxed area shows that both histone H5 and NSBP1 are bound to the assembled array.
(C) NSBP1, but not the NSBP1S17/S21E mutant, unfolds H5-containing arrays. NSBP1 or NSBP1S17/21E proteins were added to a folded array already containing H5 at a molar ratio of ~2 NSBP1 molecules per CP.
(D) HMGN1 does not unfold H5-containing arrays. To account for relative protein levels in the cell, 2 HMGN1 but only 0.57 NSBP1 molecules per each H5-containing CP were added.
(E) NSBP1 specifically counteracts H5-dependent self-association of the chromatin arrays. Self-association assays were performed with increased concentrations of magnesium chloride and measured as described in the Experimental Procedures. +H5 arrays contained two molecules of histone H5 per CP; +NSBP1 arrays contained two molecules NSBP1 per CP. Arrows indicate magnesium chloride concentration required for 50% of arrays to precipitate. In all samples, the concentration of the core arrays (12 × 207 repeats, see Experimental Procedures) was 25 ng/μl (183 nM mononucleosomes).
In an additional series of experiments, we tested whether NSBP1 unfolds a compacted array already containing H5. The results clearly indicated that wild-type NSBP1 decreased the S20,w of the compacted array significantly better than the NSBP1S17/21E mutant (Figure 5C). Thus, an efficient counteraction of H5-induced chromatin compaction is contingent on the proper binding of the full-length NSBP1 protein to nucleosomes. Furthermore, to compare the effect of NSBP1 with HMGN1 protein, which lacks a long acidic domain but is more abundant in the nucleus, we conducted an experiment with 2 HMGN1 but only 0.57 NSBP1 per CP so as to approximate the relative ratios of the protein levels in the cell. Once again, NSBP1 reduced the sedimentation velocity of the H5-containing nucleosome array, while HMGN1 did not (Figure 5D). These experiments confirm that both the chromatin-binding activity and the C-terminal domain of NSBP1 are required to fully antagonize the H5-mediated chromatin condensing activity.
In addition to causing chromatin fiber folding, linker histones promote stabilization of oligomeric tertiary chromatin structures (Carruthers et al., 1998; McBryant et al., 2006). Reversible chromatin self-association assays in the presence or absence of linker histone H5 provide additional evidence that NSBP1 counteracts the H5-mediated chromatin structural transitions. In such an assay, increased MgCl2 concentration induces formation of tertiary chromatin structures, leading to reversible self-association and precipitation of the array from the solution (Carruthers et al., 1998; Huynh et al., 2005). The degree of array oligomerization can be assessed by measuring the amount of DNA left in solution at various MgCl2 concentrations. Arrays lacking H5 reach a 50% condensation at around 4 mM MgCl2, and this value is not affected by NSBP1 addition (Figure 5E). Addition of increasing amounts of linker histone H5 reduces the concentration of MgCl2 required for precipitation of 50% of the arrays to 0.5 mM (at 2 H5 molecules per CP), reflecting the high degree of the array oligomerization. Addition of NSBP1 to these H5-containing arrays counteracts chromatin oligomerization, as indicated by the 3-fold increase (from 0.5 mM to 1.5 mM) (Figure 5E) in the MgCl2 concentration required to precipitate 50% of the array. We conclude that the interaction of NSBP1 with chromatin counteracts the H5-dependent secondary and tertiary chromatin structures.
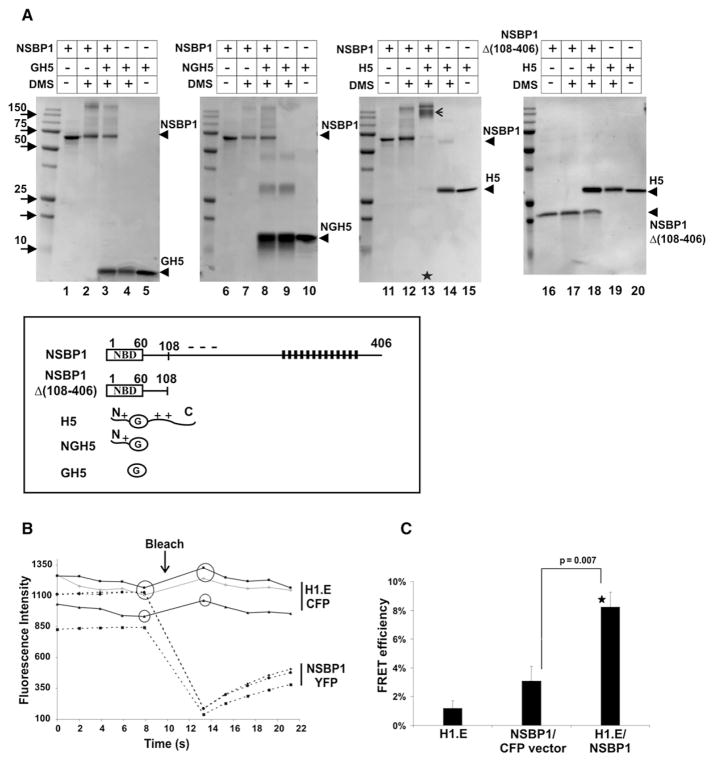
It is well documented that the C-terminal region of linker histones plays a major role in stabilizing chromatin compaction (Lu et al., 2008; Woodcock et al., 2006). Since NSBP1 counteracts the chromatin condensing activity of H5, while HMGN1 does not, it is likely that the highly negative C terminus of NSBP1 interacts directly with H5. To test this possibility, we first treated either purified HMGN1, NSBP1, HMGB1, and H5 or equimolar molar mixtures of each of these proteins and H5 with the crosslinker Dimethyl suberimidate (DMS), as previously described for HMGB1 (Cato et al., 2008). These experiments clearly indicate that NSBP1 formed a specific complex with the linker histone H5 (asterisk, Figure S9, lane 9). The individual proteins at the same conditions gave no specific products (Figure S9). The complex formation was accompanied by depletion of both NSBP1 and H5. Under the same conditions, HMGB1 (positive control) did crosslink with H5, but HMGN1 did not (Figure S9), suggesting that, indeed, NSBP1 crosslinks to H5 through its negatively charged C terminus.
Like all linker histones, H5 contains a globular domain (GH5) flanked by two positively charged domains. To gain more detailed insights into the interactions among the various H5 and NSBP1 domains, we crosslinked wild-type NSBP1 or the deletion mutant NSBP1Δ(108–409) lacking negatively charged C-terminal domain with the native H5, the purified globular domain (GH5) lacking both positively charged flanking domains, or an H5 fragment lacking only the C-terminal domain (NGH5). The results clearly indicate that only the wild-type H5 and wild-type NSBP1 form a complex (Figure 6A, arrow in lane 13). Deletion of either only the C terminus of NSBP1 (Figure 6A, lanes 16–20) or only the C terminus of H5 (Figure 6A, lanes 6–10) abolishes the interaction between the proteins. Thus, the negatively charged C-terminal region of NSBP1 targets the positively charged C-terminal region of H5 in a fashion similar to that shown for HMGB1 (Cato et al., 2008). Conceivably, this interaction interferes with the chromatin condensing activity of linker histones that is mediated by the intrinsic disorder and amino acid composition of its C-terminal region (Lu et al., 2008).
Figure 6. NSBP1 Interacts Directly with Linker Histone H5 Both In Vitro and In Vivo.
(A) In vitro crosslinking experiments. Full-length NSBP1 and histone H5 or their deletion mutants (see the schematic representation on the bottom of panel A; in this panel, NBD is the nucleosomal NBD of NSBP1, and G is the globular region of H5) were crosslinked by Dimethyl suberimidate (DMS) and analyzed by 15% SDS-PAGE. Lanes 2, 4, 7, 9, 12, 14, 17, and 19 represent the proteins crosslinked individually, whereas lanes 3, 8, 13, and 18 shows the products of crosslinking between H5 and NSBP1 proteins or various deletion mutants. Arrow in line 13 indicates the specific NSBP1-H5 complex formed after crosslinking. Note the disappearance of monomers of NSBP1 and H5 only in lane 13.
(B) Detection of linker histone:NSBP1 interaction in living cells by fluorescence resonance energy transfer (FRET). Mouse NIH 3T3 were cotransfected with plasmids expressing H1.E-CFP and NSBP1-YFP proteins. Acceptor photobleaching FRET was performed as described in the Experimental Procedures. Shown is a graphic presentation of fluorescence intensity in three representative ROIs following the bleach of the NSBP1-YFP acceptor. Note the increased fluorescence intensity of H1.E-CFP donor (arrow).
(C) Summary of FRET efficiency. The H1.E bar alone is a control for false FRET; NSBP1/CFP vector bar measures random FRET; H1.E/NSBP1 measures the interactions between H1.E-CFP donor and NSBP1-YFP acceptor. Error bars represent standard error (n = 15). P value was calculated using Student’s t test. Interaction between H1.E and NSBP1 was found to be highly statistically significant (*).
Analysis of the NSBP1 structure with the FoldIndex program (http://bip.weizmann.ac.il/fldbin/findex) (Prilusky et al., 2005) predicts that the C-terminal region of NSBP1 is also intrinsically disordered (Figure S10). Intrinsically disordered proteins are predicted to bind with low affinity, but specifically and rapidly, to their interacting partners (Hansen et al., 2006), a prediction consistent with the observed high mobility of NSBP1 (Figure 1G). Since both H1 and NSBP1 have disordered C-terminal domains and move rapidly in the nucleus, we reasoned that they would not form stable complexes and therefore used fluorescence resonance energy transfer (FRET) to test for their possible interaction in living cells. FRET analysis by the acceptor photobleaching technique measures the ability of a donor fluorophore (CFP) to transfer the energy to an adjacently positioned acceptor (YFP). This energy transfer is reduced following YFP bleaching, thereby yielding an increase in CFP fluorescence (Figure 6B). FRET will be observed only if two fluorescent proteins are in the nanometer scale proximity: in other words, interact with each other. By comparing FRET efficiency with false FRET and random FRET controls (see Experimental Procedures), we determined that there is a specific and highly significant interaction between NSBP1 and linker histone H1.E (Figures 6B and 6C). In summary, our data clearly demonstrate a direct interaction between NSBP1 and linker histones both in vitro and in vivo.
Of the three domains contained in the linker histones (Bustin and Cole, 1970), the central globular region serves to position the protein in the major groove near the dyad axis of the nucleosome (Brown et al., 2006), while the C-terminal domain, which is highly positive, is essential for chromatin compaction (Lu et al., 2008; Woodcock et al., 2006). At the nucleosomal dyad axis, the binding sites of HMGN partially overlap with those of H1 (Alfonso et al., 1994). HMGNs bind to nucleosomes through their conserved NBD; however, this binding does not seem to significantly affect the H5-dependent chromatin compaction (Figure 5), perhaps because HMGN1 competes with the globular domain of H5 without interacting with the positively charged H5 C-terminal domain. Our finding that the ability of NSBP1 to counteract the chromatin condensing activity of H5 is contingent on chromatin binding raises the possibility that NSBP1 also affects the interaction of the globular domain of H5 with chromatin. Indeed, in living cells, where the interaction of linker histones with chromatin is highly dynamic, HMGNs, as well as other HMGs, compete with H1 for nucleosomal binding and weaken the binding of H1 to chromatin without significantly affecting chromatin condensation (Catez et al., 2004).
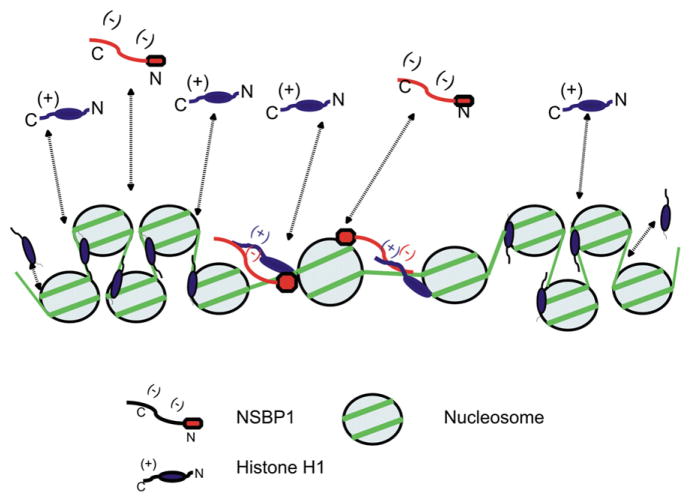
In summary, we report that NSBP1 is a nucleosome-binding protein that is widely expressed in mouse tissues, affects the binding of linker histones to chromatin, and destabilizes the compact structure of the chromatin fiber. Most likely, these changes in chromatin architecture lead to alteration in the cellular transcription profile. We envision (Figure 7) that in living cells, where the interaction of chromatin-binding protein with nucleosomes is highly dynamic (Bustin et al., 2005; Catez et al., 2004), the temporary binding of NSBP1 to nucleosomes places its negatively charged C-terminal domain in the vicinity of the positively charged C-terminal domain of H1. The juxtaposition of these domains diminishes the ability of the H1 C-terminal domain to stabilize a compact chromatin structure. Our studies do not exclude the possibility that the specific interaction of NSBP1 and other HMGN variants with chromatin also impact the chromatin condensing activities of the core histone tails (Kan et al., 2008). Since the chromatin residence time of NSBP1 is short, and the protein rapidly migrates among nucleosomes, its effects on chromatin structure and on the cellular transcription profile are larger than would be expected from a protein of such low cellular abundance.
Figure 7. Model of the Effect of NSBP1 on Chromatin Compaction.
In living cells, H1 and NSBP1 bind dynamically to chromatin (dotted lines). The binding of NSBP1 to H1-containing nucleosomes juxtaposes the C-terminal domains of the proteins and counteracts the H1-induced stabilization of a compact chromatin structure.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and siRNA Treatment
SV-40-transformed MEFs were purchased from American Type Culture Collection and used for microscopy and FRAP experiments. NIH 3T3 cells were used for micrococcal nuclease digestion. NIH2/4 stable cell line, which carries an array of 256 copies of the LacR binding sequence and 96 copies of the tetracycline response element sequence (Soutoglou and Misteli, 2008), was a gift from Evi Soutoglou (T. Misteli, NCI). AtT20 mouse pituitary-derived cell line was a gift from the laboratory of G. Hager (NCI). Cells were maintained in DMEM (Invitrogen) with 10% FCS (GIBCO-BRL). Stable AtT20 cell lines were prepared using retrovirus, generated by transient transfection of ecotropic helper Phoenix cells with pHAN vector bearing either wild-type or mutated NSBP1 protein, tagged with FLAG and HA.
For siRNA-mediated downregulation of NSBP1, AtT20 cells were transfected with specific siRNA (L-044143-00) or control siRNA (D-001810-01-05; ON-TARGETplus SMARTpool) using Dharmafect 4 transfection reagent (T-2004-01, Dharmacon [Thermo Scientific]; Lafayette, CO). Following initial transfection, cells were grown for 48 hr, split, and retransfected. Cells were collected 72 hr after the second round of siRNA treatment.
Antibodies
Polyclonal anti-NSBP1 and anti-H1 antibodies were generated in rabbits and affinity-purified. Antibodies against histone H3 and histone modifications were from Abcam (Cambridge, MA) (ab1791-H3, ab12179-H3K9Ac, ab6000-H3K4Me2+3, ab6002-H3K27Me3, ab1220-H3K9Me2). Anti-acetyl-lysine, clone 4G12, and anti-Hp1α antibodies were from Millipore (Billerica, MA) (#05-515 and #05-689). Anti-actin clone AC-74 antibodies were from Sigma (A5316), and anti-Hp1β antibodies were from Millipore (Billerica, MA) (MAB3448). Anti-RNAPII 8WG16 antibody was from Covance (Gaithersburg, MD) (MMS-126R). Secondary antibodies for microscopy were from Jackson Laboratory. HRP-conjugated secondary antibodies for western blots were from Pierce (Rockford, IL).
Plasmids
NSBP1-GFP and HMGN1-GFP fusion proteins were cloned in GFPN2 vector (Clontech; Mountain View, CA). LacR-CFP plasmid, HP1α-CFP, and H10-GFP were a gift from the laboratory of T. Misteli (NCI). For tethering NSBP1 to LacR array, the protein was fused in frame to the N terminus of LacR-CFP construct. pHAN NSBP1 FLHA retroviral vector bearing NSBP1 proteins, tagged with FLAG and HA at C terminus under the control of CMV promoter, was used for generation of stable clones. For FRET nuclear CFP vector, pEYFPN1 (Clontech), CFP-YFP fusion (Karpova et al., 2003), NSBP1-YFP, and hH1.E-CFP were used.
RNA and cDNA Preparation
RNA was prepared with TRIzol reagent according to the manufacturer’s protocol. It was additionally cleaned by QIAGEN RNeasy kit. For RT-PCR analysis, 500 ng of RNA were used for cDNA synthesis by iScript kit (Bio-Rad). An equal amount of cDNA was applied for RT-PCR reactions with Power SYBR Green mix (Applied Biosystems). Reactions and measurements were performed using AB 7900HT Fast Real-Time PCR System and software.
Confocal Microscopy
For live cell imaging, cells were plated on MatTek glass-bottom culture dishes 24 hr before transfection, and constant temperature of 37°C was maintained during the experiment. For fixed cells, imaging cells were plated on poly-D-lysine-coated coverslips on 6-well plates.
Cells were fixed by freshly prepared 4% paraformaldehyde for 5–15 min at RT and then either permeabilized by 0.5% Triton X-100 for 5 min at RT or post-fixed in cold methanol for 5 min at −20°C. Following fixation, cells were blocked in 1% BSA for 30 min at RT and incubated with indicated primary and secondary Abs for 1 hr at RT. Standard dilution for primary and secondary antibodies was 1:200 in blocking buffer. DNA was counterstained with Hoechst dye. Confocal images were collected using a Zeiss LSM 510 system mounted on a Zeiss Axiovert 200M microscope (Carl Zeiss, Inc.; Thornwood, NY) using an oil-immersion Plan-Apochromat 63×/1.4 DIC objective lens. Colocalization of proteins with DNA was analyzed by the ImageJ program using the Colocalization Finder Plugin.
Fluorescence Resonance Energy Transfer
For FRET experiments, NIH 3T3 cells were cotransfected with H1.E-CFP and NSBP1-YFP proteins by FuGENE 6 (Roche) for 24 hr and then fixed with 4% paraformaldehyde for 20 min at RT. We used a Zeiss LSM 510 META confocal microscope (Carl Zeiss) operating with a 30 mW argon laser tuned to 458 nm, 488 nm, and 514 nm laser lines. Cells were examined with a 60× 1.4 NA Zeiss oil-immersion objective and 6×zoom. FRET was measured using the acceptor photobleaching method, with a CFP/YFP filter set as described (Karpova et al., 2003). According to this protocol, three circular regions of interest × 75 pixels in diameter were bleached in the nucleus of a fixed cell by scanning them in the YFP channel with the 514 laser line at 70% intensity. Before and after the bleach, five images were collected in the CFP channel to access changes in donor fluorescence. To minimize the effect of photobleaching due to imaging, images in the CFP channel were collected at 1.4% of the 458 nm laser line intensity. Average intensity was measured for each region of interest in the CFP channel before and after the bleach, and the background average intensity was subtracted from this value. The FRET energy transfer efficiency (EF) was calculated as EF = (I6 − I5)/I6, where I is the background-corrected average CFP intensity at the indicated time point. I6 and I5 correspond to the average intensity values of the postbleach and prebleach time points. The following controls were tested in the same way: negative control CFP-H1.E (no acceptor, only donor present), positive control expressing a CFP-YFP fusion protein (Karpova et al., 2003), and a random FRET control (cells cotransfected with both NSBP1-YFP and CFP-expressing vector). The FRET efficiency for the positive control (CFP-YFP fusion protein) was approximately 30%. P value was calculated using Student’s t test.
Nucleosome Array Conformation Studies
Nucleosome arrays were based on clone 601 nucleosome positioning sequence (Lowary and Widom, 1998), which was used to construct 12 × 207 nucleosome repeats and reconstituted with histone octamers as described (Wang et al., 2009). Linker histone H5 was isolated from chicken erythrocytes and reconstituted as described (Springhetti et al., 2003) at a ratio of one molecule per nucleosome. The input histone H5 concentration was determined from its sequence-based predicted absorbance of 0.217 for 1 mg/ml H5 protein solution in water at 280 nm (ProtParam at http://www.expasy.ch). Pure recombinant mouse NSBP1 protein, HMGN1 protein, and/or H5 were added as described in the figure legends. To monitor physical compactness of assembled nucleosome arrays, sedimentation velocity analysis was performed using Beckman Optima XL-A ultracentrifuge, as described (Carruthers et al., 2000; Wang et al., 2009) in 150 mM NaCl at 20,000 rpm and 20°C for ~3 hr. Sedimentation velocity analysis was conducted using the continuous c(s) distribution model with SEDFIT software (Schuck, 2000) (http://www.analyticalultracentrifugation.com).
For magnesium-dependent self-association experiments (Carruthers et al., 1998), arrays containing 0, 1, or 2 molecules of histone H5 per nucleosome were incubated with increasing magnesium chloride concentrations, as indicated on the graph. The DNA from pellets (self-associated material) and supernatants (non-self-associated material) was separated on agarose gels. The gel band intensities were quantified using OptiQuant software, and the percent of total DNA that remained in the supernatant was plotted on the graph.
Western Blots
Whole-cell lysates were prepared in 1× SDS-PAGE sample buffer (Bio-Rad) supplemented with protease inhibitors. Tissue lysates from 2-month-old C57BL mouse were prepared as follows. A tissue specimen was collected in 500 μl of RIPA buffer (20 mM HEPES [pH 7.0], 300 mM NaCl, 10 mM KCl, 1 mM MgCl2, 20% glycerol, 0.5% Triton X-100, 0.5 mM DTT, complete protease inhibitor cocktail, 1 mM orthovanadate) and homogenized on ice by electricity-driven homogenizer. The samples were fractionated on 10% precast Criterion gels, transferred by semidry method to PVDF membrane, blocked with nonfat milk in PBS, and probed with antibodies as in the text. Chemiluminescent detection using ECL Plus has been performed according to Amersham (GELifesciences; Piscataway, NJ) recommendations.
Salt Extraction
SV-40-transformed MEFs were washed in 1× PBS and then in washing buffer (20 mM HEPES [pH 7.0], 10 mM KCl, 10 mM MgCl2, 20% glycerol, 0.1% Triton X-100, 0.5 mM DTT, 1 mM orthovanadate, complete protease inhibitor, 0.25 mM PMSF). Finally, cells were resuspended in 200 μl of the washing buffer, and NaCl was added to final concentration of 0, 75, 125, 250, 350, and 500 mM. Extraction was performed on ice for 15 min, with over-the-top mixing every 2–3 min. Cells were centrifuged at 15,000 g for 5 min. The pellet and the supernatant fraction were collected. Laemmli buffer supplemented with protease inhibitors was added to both fractions, followed by brief sonication and boiling.
Micrococcal Nuclease Digestion
Three 10 cm plates of 40% confluent NIH 3T3 cells were transfected with 6 μg of either NSBP1-GFP or NSBP1S17/21E plasmids by FuGENE 6 (Roche). According to GFP fluorescence measurements, transfection efficiency was close to 80%. After 48 hr, the cells were washed and the nuclei isolated and digested with 30 units of MNase I (Sigma) for 1, 2, 4, 8, 16, and 24 min. Isolated DNA were analyzed by 1.5% TAE agarose gel and ethidium bromide staining.
Microarray Expression Analysis
Stable AtT20 clones overexpressing NSBP1-FLHA, NSBP1S17/121E FLHA, or control empty retroviral vector and AtT20 cells treated with specific or control siRNA were collected for RNA extraction. RNA was prepared as described previously. Expression analysis has been done using Affymetrix GeneChips 430 2.0 mouse array at LMT (Laboratory of Molecular Technology, NCI-Frederick). Raw data were preprocessed using BRB Array Tools (Biometric Research Branch, NCI, NIH) to filter out genes with p values > 0.001.
Assessment of NSBP1 Copy Number
Whole-cell extracts (WCEs) from AtT20 cells have been prepared using Nuc-Buster Protein Extraction Kit by Novagen (EMD Chemicals, Merk; Darmstadt, Germany). Recombinant wild-type NSBP1 and mouse thymus histone H1 have been purified, and their concentration has been determined using BCA assay. Fixed number of cells and purified proteins were run on SDS-PAGE, and the amount of histone H1 and NSBP1 per fixed number of cells has been calculated as described below. WCE from 4 × 105 cells contains approximately 250 ng of histone H1 (scan measurements of the gel), which equals 625 fg/cell. One mole of H1 equals 25,000 g; therefore, there are 25 Attomoles (25 × 10−18 moles) of H1 per 1 cell. One mole equals 6 × 1023 molecules (Avogadro number); therefore, 25 × 10−18 moles equal to 15,000,000 molecules per cell. Note that calculated copy number of linker histone robustly corresponds to predicted number of nucleosomes per cell. NSBP1 amount was calculated by comparing signal intensities of fixed number of cells with that of purified NSBP1 protein by western blot. Content NSBP1 per 1 cell was calculated as 1 fg or 0.02 AtTomoles (2 × 10−20 moles). One mole of NSBP1 equals 45,000 g or 6 × 1023 molecules; therefore, 2 × 10−20 mole is equal to 12,000 molecules per 1 cell.
Chemical Crosslinking
The NSBP1 and HMGN1 proteins were cloned and expressed in bacteria as previously described (Shirakawa et al., 2000). HMGB1 protein has been isolated from mouse thymus by perchlorate extraction and purified by size-exclusion HPLC. Histone H5 was purified from chicken erythrocytes by acid extraction and Mono S ion-exchange chromatography. GH5 and NGH5 have been obtained by limited proteolysis with either trypsin or endoproteinase Asp-N (P3303) and purified using reverse-phase C18 column HPLC. Chemical cross-linking between purified HMGN1, HMGB1, NSBP1 and its mutants, and histone H5 and H5 truncated polypeptides was performed as described (Cato et al., 2008). Briefly, the purified proteins were mixed at a final concentration of 2 μM in 150 mM NaCl and Triethanolamine-HCl buffer (pH 8.0), cross-linked by 100 μM DMS (Aldrich #179523) for 60 min at RT, and fractionated on precast 15% SDS-PAGE Criterion gel (Bio-Rad).
Supplementary Material
Acknowledgments
We thank Valarie Barr for technical assistance with microscopy. We thank Doctor F. Catez for the initial observation that HMGN5 does not localize to heterochromatin. This project is supported by the Center for Cancer Research, intramural program of the NCI, NIH, by contract number N01-CO-12400 granted by NCI, NIH and by NSF grant MCB-0615536 to S.A.G.
Footnotes
ACCESSION NUMBERS
Microarray expression data were analyzed by BRB Array Tools. Data were deposited to GEO database, accession number GSE16564.
Supplemental Data include 11 figures and one table and can be found online at http://www.cell.com/molecular-cell/supplemental/S1097-2765(09)00476-6.
References
- Alfonso PJ, Crippa MP, Hayes JJ, Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J Mol Biol. 1994;236:189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem Sci. 2001;26:431–437. doi: 10.1016/s0968-0004(01)01855-2. [DOI] [PubMed] [Google Scholar]
- Bustin M, Cole RD. Regions of high and low cationic charge in a lysine-rich histone. J Biol Chem. 1970;245:1458–1466. [PubMed] [Google Scholar]
- Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Memedula S, Plutz MJ, Belmont AS. Common effects of acidic activators on large-scale chromatin structure and transcription. Mol Cell Biol. 2005;25:958–968. doi: 10.1128/MCB.25.3.958-968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- Carruthers LM, Schirf VR, Demeler B, Hansen JC. Sedimentation velocity analysis of macromolecular assemblies. Methods Enzymol. 2000;321:66–80. doi: 10.1016/s0076-6879(00)21187-7. [DOI] [PubMed] [Google Scholar]
- Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato L, Stott K, Watson M, Thomas JO. The Interaction of HMGB1 and Linker Histones Occurs Through their Acidic and Basic Tails. J Mol Biol. 2008;384:1262–1272. doi: 10.1016/j.jmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittur SV, Sangster-Guity N, McCormick PJ. Histone deacetylase inhibitors: a new mode for inhibition of cholesterol metabolism. BMC Genomics. 2008;9:507. doi: 10.1186/1471-2164-9-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SC, Grewal SI. Heterochromatin: silence is golden. Curr Biol. 2003;13:R895–R898. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Fan Y, Skoultchi AI. Genetic analysis of H1 linker histone subtypes and their functions in mice. Methods Enzymol. 2004;377:85–107. doi: 10.1016/S0076-6879(03)77005-0. [DOI] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Fenic I, Hossain HM, Sonnack V, Tchatalbachev S, Thierer F, Trapp J, Failing K, Edler KS, Bergmann M, Jung M, et al. In vivo application of histone deacetylase inhibitor trichostatin-a impairs murine male meiosis. J Androl. 2008;29:172–185. doi: 10.2164/jandrol.107.003848. [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2006;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Carruthers LM, Logie C, Hill DA, Solomon MJ, Wade PA, Imbalzano AN, Hansen JC, Peterson CL. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol. 2002;9:263–267. doi: 10.1038/nsb776. [DOI] [PubMed] [Google Scholar]
- Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- Kan PY, Caterino TL, Hayes JJ. The H4 tail domain participates in intra- and inter-nucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol. 2008;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Baumann CT, He L, Wu X, Grammer A, Lipsky P, Hager GL, McNally JG. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J Microsc. 2003;209:56–70. doi: 10.1046/j.1365-2818.2003.01100.x. [DOI] [PubMed] [Google Scholar]
- King LM, Francomano CA. Characterization of a human gene encoding nucleosomal binding protein NSBP1. Genomics. 2001;71:163–173. doi: 10.1006/geno.2000.6443. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Lu X, Hamkalo B, Parseghian MH, Hansen JC. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder (dagger) Biochemistry. 2008;48:164–172. doi: 10.1021/bi801636y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBryant SJ, Adams VH, Hansen JC. Chromatin architectural proteins. Chromosome Res. 2006;14:39–51. doi: 10.1007/s10577-006-1025-x. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- Nolte D, Ramser J, Niemann S, Lehrach H, Sudbrak R, Muller U. ACRC codes for a novel nuclear protein with unusual acidic repeat tract and maps to DYT3 (dystonia parkinsonism) critical interval in xq13.1. Neurogenetics. 2001;3:207–213. doi: 10.1007/s100480100120. [DOI] [PubMed] [Google Scholar]
- Ooi GT, Tawadros N, Escalona RM. Pituitary cell lines and their endocrine applications. Mol Cell Endocrinol. 2004;228:1–21. doi: 10.1016/j.mce.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Rhodes D. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa H, Landsman D, Postnikov YV, Bustin M. NBP-45, a novel nucleosomal binding protein with a tissue-specific and developmentally regulated expression. J Biol Chem. 2000;275:6368–6374. doi: 10.1074/jbc.275.9.6368. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springhetti EM, Istomina NE, Whisstock JC, Nikitina T, Woodcock CL, Grigoryev SA. Role of the M-loop and reactive center loop domains in the folding and bridging of nucleosome arrays by MENT. J Biol Chem. 2003;278:43384–43393. doi: 10.1074/jbc.M307635200. [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Catez F, Gerlitz G, Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol. 2008;28:2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li M, Stadler S, Correll S, Hayama R, Leonelli L, Li P, Han H, Nathan C, Grigoryev SA, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184:205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Seebart C, Tomschik M. The linker-protein network: control of nucleosomal DNA accessibility. Trends Biochem Sci. 2008;33:247–253. doi: 10.1016/j.tibs.2008.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.