Abstract
Epigenetic silencing of tumor suppressor gene promoters is one of the most common observations found in cancer. Despite the plethora of observed epigenetically silenced cancer related genes little is known about what is guiding the silencing to these particular loci. Two recent articles suggest that long antisense non-coding RNAs function as epigenetic regulators of transcription in human cells. These reports, along with previous observations that small antisense non-coding RNAs can epigenetically regulate transcription, imply that long antisense non-coding RNAs function as endogenous transcriptional regulatory RNAs in humans. Mechanistically, these long antisense non-coding RNAs may be involved in maintaining balanced transcription at bidirectionally transcribed loci as a method to modulate gene expression according to the selective pressures placed on the cell. The loss of this intricate bidirectional RNA based regulatory network can result in overt epigenetic silencing of gene expression. In the case of tumor suppressor genes, this silencing can lead to the loss of cellular regulation and be a contributing factor in cancer. This perspective will highlight the endogenous effector RNAs and mechanism of action whereby long antisense non-coding RNAs transcriptionally regulate gene expression in human cells.
Keywords: non-coding RNA, epigentics, antisense RNA, transcription, silencing
Introduction
The majority of the human genome is transcribed in both sense and antisense directionalities (reviewed in ref. 1). It is now known that much of the ~98% junk DNA is transcribed into non-coding RNAs, of which many are antisense to known coding mRNAs. Interestingly, there is limited conservation of long non-coding RNA sequence between various organisms, with the only noted commonality restricted to short stretches within the respective RNAs.1 This relative low sequence conservation between various species may in fact be the result of differing selective pressures placed on the organism and the selection of particular regions within the non-coding RNAs which are functional in regulating a particular locus in the genome. Theoretically distal changes in the long non-coding RNAs could also affect the respective secondary structure and thus alter particular target localization of the long non-coding RNA. However, to date there are few examples of phenotypic mutations resulting from alterations in long non-coding RNAs.2 This may be an attribute of selective pressures placed on the cell acting directly on the non-coding RNAs; changes that when positive are reinforced and selected for compared to when negative selected against and lost. It has indeed proven more difficult to assess the positive selection placed on non-coding RNAs due to the lack of proximal neutral sites (reviewed in ref. 3).
Many long non-coding RNAs are antisense to the coding region of protein producing genes. Notably, these sense/antisense or bidirectionally transcribed genes appear conserved throughout evolution suggesting a function as the retention of the cis acting antisense would be expected to diverge over evolutionary distance. However, this does not appear to be the case but rather that the sense/antisense RNAs are retained.4 Thus, the retention of sense mRNA with a corresponding antisense non-coding RNA and the vast number of long non-coding RNAs might be suggestive of a role for these RNAs in organismal complexity.5 Functionally, non-coding RNAs, both short6,7 and long,8,9 have been shown to be involved in the regulation of transcription by a mechanism that involves epigenetic changes in locus specific chromatin. Thus, emerging evidence suggests that non-coding RNAs function to bestow specificity in targeting epigenetic complexes to particular loci. Such control of transcription could be one possible avenue for how long non-coding RNAs would function to modulate organismal complexity. But how do long-coding RNAs regulate transcription?
Long Antisense Non-Coding RNAs Regulate Transcription in Human Cells
Imprinted genes have for several years been known to be controlled by non-coding RNAs and this mode of regulation is dependent on epigenetic changes at cis-acting imprint control regions (ICE) (reviewed in ref. 10). The ICE varies in position relative to the antisense non-coding RNA promoter and is thought to be involved in regulating the antisense non-coding RNA promoter and expression. The antisense non-coding RNA accordingly regulates the imprinted region. These imprinted regions cover large distances encompassing several genes. The long non-coding RNAs may thus be required to epigenetically regulate these imprinted regions to confer epigenetic memory that is not only nucleosome specific but also functional in nucleosome/nucleosome interactions allowing for epigenetic memory to be maintained at the imprinted cluster. In the case of imprinted genes, there is a correlation with imprinted gene expression and the non-coding RNA.11 But how might non-coding RNAs function to regulate non-imprinted genes?
Evidence for a mechanism of how long non-coding RNAs could regulate gene transcription emerged from studies using siRNAs targeted to human promoters (reviewed in ref. 12). Promoter targeted siRNAs, made de novo and introduced into human cells, were shown to be capable of instilling transcriptional gene silencing that was specific for the targeted promoter.13 Importantly, the observed transcriptional silencing appeared to function mechanistically through an epigenetic based mechanism.13 Indeed this initial observation has been disconcerting to many as tradition modes of RNAi based transcriptional silencing in plants and S. pombe required and RNA dependant RNA Polymerase, which is not known to be present or functional in humans.14 Nonetheless, several other groups have recapitulated this work using promoter targeted siRNAs.6,15-34 Despite the ability to generate promoter targeted siRNAs to control transcription of RNA polymerase II promoters little was known regarding to what extent this mechanism was being acted on in human cells, i.e., what if anything was the endogenous non-coding RNAs that actively utilized this pathway in human cells to regulate transcription?
During studies to uncover the mechanism whereby small RNAs regulate transcription in human cells it was learned that small antisense RNAs alone were sufficient for directing transcriptional silencing.35 Interestingly, in this same body of work it was learned that the small antisense RNAs were associated with the DNA methlytransferase 3A (DNMT3a, an enzyme involved in de novo DNA methylation36), and the silent state histone 3 lysine 27 tri-methylation mark at the targeted promoter.35 These observations were important as they suggested that a transcriptional regulatory pathway was active in human cells which relied on antisense non-coding RNAs and was mechanistically distinct from the RNAi pathways described in plants, S. pombe, Drosophila and C. elegans.14
How Antisense Non-Coding RNAs Guide Epigenetic Silencing Complexes to Target Loci?
Until recently it had remained unknown as to how the small antisense RNAs were able to find the particular promoter target site. Studies carried out to determine whether or not the small antisense RNAs were targeting either DNA or an RNA at the promoter demonstrated for the first time that RNA Polymerase II promoters were transcribed and that these low-copy promoter associated RNAs were in fact the target recognition/motif required for the small antisense RNAs to instill transcriptional silencing in humans.37 Since this discovery several others have reported pervasive transcription to be located at, upstream, or even overlapping 5' regions of protein coding genes, e.g., RNAPII promoters.5,15,38-41 Taken together these observations suggest that transcription found at RNAPII promoters could be the required target site for some yet to be discovered effector RNA involved in the regulation transcription. However, it had until recently unclear if there were any endogenous RNAs in human cells actively utilizing the emerging mechanism found to be involved in small antisense RNA directed transcriptional gene silencing.
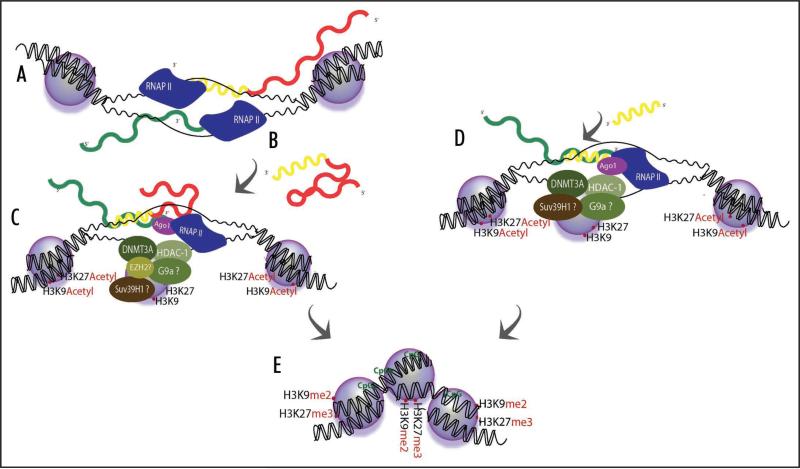
Interestingly, observations began to emerge suggesting promoter targeted siRNAs could also activate transcription.42-44 It was during studies into the mechanism involved in siRNA directed gene activation that a role for long antisense non-coding RNAs was discerned.8 These studies demonstrated that long antisense non-coding RNAs were the heretofore unknown endogenous RNA based transcriptional regulators that were actively utilizing the previously described mechanism to regulate transcription.8 This observation is supported by other observations in human cells whereby long antisense non-coding RNAs regulate transcription through targeting epigenetic silencing complexes to homology containing loci.9,45,46 The observations that small antisense non-coding RNAs, shown previously to modulate transcription in humans, as well as long antisense non-coding RNAs could function to regulate transcription in humans was further supported by reports of endogenous miRNAs capable of directing transcriptional silencing in humans.6,7 Taken together these data suggest that antisense non-coding RNAs, either short or long, are functional in regulating transcription in human cells. Interestingly, both forms of short and long antisense non-coding RNA mediated control of transcription require Argonaute 1 (Ago-1)8,23,24 and the enrichment of silent state histone 3 lysine 27 trimethylation at their respective target loci.8,20,30,35 As a result of these works a mechanism whereby antisense non-coding RNAs regulate transcription has begun to emerge (Fig. 1). Importantly, either long or short antisense non-coding RNAs can direct epigenetic remodeling complexes specifically to target loci and the mechanism of action appears to require Ago-1, DNMT3a and histone deacetylase 1 (HDAC-1)20,25,28,30 (Fig. 1).
Figure 1.
Model for both small and long antisense non-coding RNA directed transcriptional regulation in human cells. (A) Long antisense non-coding RNAs expressed at bidirectionally transcribed genes may fold into (B) secondary structured non-coding RNAs that can (C) interact with particular sites in the promoter of sense strand at the bidirectionally transcribed gene and also influence the recruitment of Ago-1, DNMT3a and HDAC-1 to this target site. (D) Small synthetic antisense non-coding RNAs can be designed to take advantage of the endogenous mechanism and also utilize the same pathway to transcriptionally silence gene expression. (E) The end result of either small or long antisense non-coding RNA transcription silencing is the targeted epigenetic remodeling of the particular RNA targeted loci.
Why Long Antisense Non-Coding RNAs?
It's beginning to seem that that the question of a function for “junk DNA” is no longer so enigmatic. Genome-wide analysis of the human transcriptome has suggested that virtually the entire genome is transcribed in both sense and antisense directions.47 Indeed much of the “junk DNA” is actively transcribed and roughly 40–50% of protein coding genes appear to exhibit long antisense non-coding RNAs.48 In fact virtually every protein coding gene we have looked at, using directional RT followed by PCR,8,49 appears to be expressing some level of a long antisense non-coding RNA. These antisense non-coding RNAs can be in cis or trans to their sense counterparts with a significant number found to lack poly-adenylation and subsequently retained in the nucleus. Such findings, taken together with recent observations, that both short and long antisense non-coding RNAs can function to epigenetically regulate gene transcription,8,9,12 implicates non-coding RNAs as functional endogenous regulators involved in epigenetic regulation of transcription in human cells.
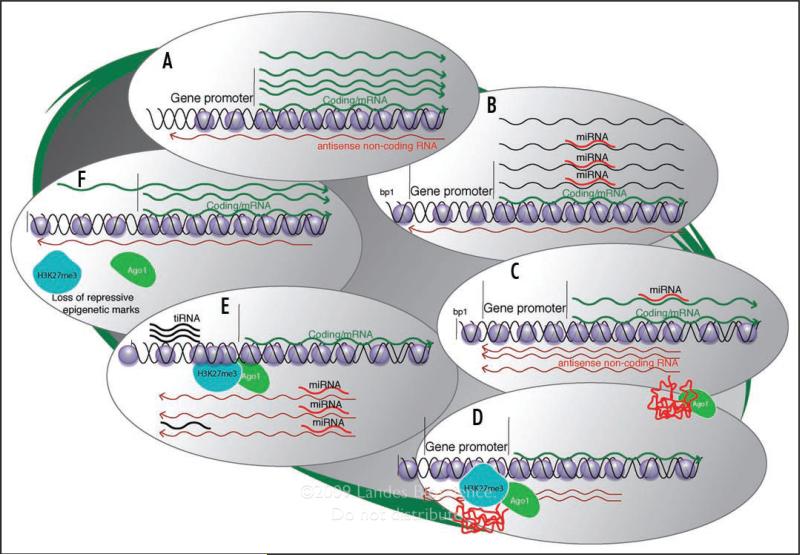
It is noteworthy that the extent of protein coding genes cannot account for the relative cellular and molecular complexity in human cells. Emerging evidence strongly suggests that biological complexity is to some extent linked to long non-coding RNAs.2,47,50 However, the form and function of non-coding RNAs between various organisms is of interest. While many organisms appear to depend on RNAi for particular functions that can drive cellular complexity, it is interesting to note some crucial differences may portend the reason long non-coding RNAs function in human cells to modulate chromatin and gene expression on the epigenetic level whereas siRNAs tend to do so in plants, yeast, C. elegans and Drosophila. Some aspects of the RNAi pathway are retained in human cells while others, such as an RNA dependant RNA polymerase (RdRP) are not. In fact the lack of an RdRP may be one key aspect of why long non-coding RNAs are found to function as epigenetic regulators of transcription in human cells. It would seem that humans may be utilizing long non-coding RNAs to control gene transcription via directing epigenetic complexes to particular loci. The advantage to utilizing long non-coding RNAs is that their ability to target a particular loci can be altered via subtle changes instilled in the long non-coding RNA. As only a small segment of the antisense non-coding RNA is required to direct transcription silencing in human cells9 and (reviewed in ref. 12) it seems reasonable to postulate that long antisense non-coding RNAs can undergo secondary structural changes that can modulate the folding of the long non-coding RNA and thus facilitate particular regions of the RNA to be capable of targeting particular chromatic regions with differing fidelities. Thus, changes in the long non-coding RNAs could affect the secondary structure and provide for either increased or reduced RNA/target affinity. Such a trait could allow for variation to be instilled in the cellular system which would allow for finer tuned adaptation to selective pressures. This could be one functional aspect of long non-coding RNAs that correlates with enhanced diversity and complexity found in humans. But why would humans need to retain miRNAs and RISC mediated regulation of RNAs? The retention of miRNAs, and also possibly the recently discovered tiny RNAs (tiRNAs),51 may in fact be linked to the regulation of long antisense non-coding RNAs, i.e., the miRNAs are present to control both the sense and antisense transcripts based on relative abundance while the tiRNAs are present to open upstream chromatin for regaining transcription and/or to suppress the long antisense non-coding RNAs (Fig. 2). An example for this ability of miRNAs to regulate bidirectional transcription can be found with miR373 which appeared to bind the observed antisense non-coding RNA for E-cadherin (reviewed in ref. 8). Examples of a function for tiRNAs has yet to be determined.
Figure 2.
How both tiRNAs and miRNAs might regulate long antisense non-coding RNAs at bidirectionally transcribed loci. (A) The endogenous state of a bidirectional transcribed gene is shown exhibiting low level antisense non-coding RNA expression relative to the highly expressed sense/mRNA. (B) This surplus of sense/mRNA could be targeted by particular sense/mRNA specific miRNAs. (C) The result of miRNA binding to sense/mRNA is a loss of sense/mRNA expression and unimpeded antisense non-coding RNA expression, which can lead to (D) Ago-1, and possibly several other yet-to-be determined proteins, associated interactions with the antisense non-coding RNA that results in the guiding of the epigenetic machinery necessary to transcriptionally silence the long antisense non-coding RNA homology containing region in the sense/mRNA promoter. (E) The result of increased silencing of sense/mRNA expression can be increased unimpeded antisense non-coding RNA expression and possibly a shift in transcription upstream of the long antisense non-coding RNA targeted site. This upstream transcription might then generate tiRNAs that together with the miRNAs would exhibit preferential binding, based on surplus substrate, to the antisense non-coding RNAs. (F) The result of tiRNA and/or miRNA binding the antisense non-coding RNA would be a loss of antisense non-coding RNA directed epigenetic repression leading to an increased potential for RNA polymerase II to intercalate into and transcribe the sense/mRNA gene promoter, ultimately regaining higher levels of sense/mRNA transcription.
Taken together there is mounting evidence implying a much overlooked role for long non-coding RNAs in transcriptional regulation and possibly cellular complexity. This role is argued to manifest via non-coding RNA guided epigenetic based modes of gene regulation. To date, there are only a few examples where long antisense non-coding RNAs have been shown to epigenetically control transcription at non-imprinted loci.8,9,45,46 Interestingly, these bidirectionally transcribed genes have been predominantly found to be tumor suppressor genes (reviewed in ref. 9). It is noteworthy that several tumor suppressor genes become epigenetically silenced during the progression to cancer. One has to wonder to what extent the loss of regulatory control of long antisense non-coding RNAs plays in the development of cancer? Indeed six long non-coding RNAs have been shown to be involved in carcinogenesis.52
Utilizing the Endogenous Long Non-Coding RNA Mechanism of Epigenetic Regulation for Therapeutic Gain
The evolutionary implications of long non-coding RNAs and their role in biological complexity and evolution is clearly of interest. However, equally interesting is the prospects of capitalizing on the molecular mechanism whereby antisense non-coding RNAs, be them small or long, regulate transcription for therapeutic benefit. Some of the key components of the mechanism whereby antisense non-coding RNAs regulate transcription have been worked out (reviewed in ref. 12) (Fig. 1). In essence small antisense non-coding RNAs can be designed to target particular genomic loci. These small RNAs associate with Ago-1 and the homology containing promoter associated RNAs during the first 12–24 hours following introduction into the cell24,37 (Fig. 1). After ~24 hours Ago-1 is lost at the targeted loci while silent state histone methylation begins to become pronounced.20,24,28 If the targeting lasts for ~3–4 days DNA methylation begins to appear at some of the targeted loci correlating with long-term stable transcriptional silencing.20,30 This form of RNA based silencing is distinctly different than traditional RNAi as (1) single stranded antisense non-coding and long antisense non-coding RNAs appear to be utilizing the same mechanism of action to epigenetically silence homology containing loci,8,12 (2) Ago-1, DNMT3a, and HDAC-1 are required for the establishment of silencing and DNMT1 and DNMT3a are required for the maintance of silencing,20,23,24,28 (3) this form of RNA mediated epigenetic silencing can be long-lasting if not permanent, following a relatively short exposure to the trigger antisense non-coding RNA.20,30 Thus, the endogenous machinery is operative in human cells to exert specific long-term transcriptional gene silencing, provided that the targeted promoter is in fact transcribed, i.e., transcription is required to establish transcriptional silencing.35
Equally interesting is the fact that this same molecular mechanism can be utilized to increase gene expression. To induce transcriptional activation of a particular gene the gene needs to exhibit bidirectional transcription, i.e., contain a long antisense non-coding RNA. One then designs traditional siRNAs or antisense phosphorothiate oligonucleotides to specifically target the degradation of the gene specific long antisense non-coding RNA. The loss of the regulatory long antisense non-coding RNA results in a loss of the epigenetic brake on the promoter expressing the sense strand and ultimately gene activation.8,53 As such the same long non-coding RNA based molecular mechanism can be utilized to specifically control transcription and either turn a gene on or off. The therapeutic potential to utilizing this mechanism to exert transcriptional control has only recently been realized with examples of transcriptional suppression demonstrated for HIV-1,28,30,54 CCR5,24 c-Myc,15 E-cadherin,29 prostate cancer27 and progesterone receptor.22 Clearly the potential discoveries with regards to the role of long non-coding RNAs in evolution and cellular complexity are just now beginning to emerge, as is the limitless potential to utilizing this emerging endogenous antisense non-coding RNA epigenetic regulatory pathway to exert therapeutic control over gene expression and avert disease progression.
Acknowledgements
This project is funded by R01 HL083473-02 to K.V.M. I thank Paula J. Morris at Seainsite http://seainsite.com/index.html for the generation of figures.
References
- 1.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Reviews Genetics. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS. The Genetic Signatures of Noncoding RNAs. PLoS Genet. 2009:5. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ponting CP, Lunter G. Signatures of adaptive evolution within human non-coding sequence. Hum Mol Genet. 2006;15:170–5. doi: 10.1093/hmg/ddl182. [DOI] [PubMed] [Google Scholar]
- 4.Dahary D, Elroy-Stein O, Sorek R. Naturally occurring antisense: Transcriptional leakage or real overlap? Genome Res. 2005;15:364–8. doi: 10.1101/gr.3308405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–99. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 6.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–5. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan Y, Zhang B, Wu T, Skogerbo G, Zhu X, Guo X, et al. Transcriptional inhibition of Hoxd4 expression by noncoding RNAs in human breast cancer cells. BMC Mol Biol. 2009;10:12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latos PA, Barlow DP. Regulation of imprinted expression by macro non-coding RNAs. RNA Biol. 2009:6. doi: 10.4161/rna.6.2.7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauler FM, Koerner MV, Barlow DP. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends in Genetics. 2007:23. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkins PG, Morris KV. RNA and transcriptional modulation of gene expression. Cell Cycle. 2008;7:602–7. doi: 10.4161/cc.7.5.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 14.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 15.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009 doi: 10.1038/emboj.2009.139. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buhler M, Mohn F, Stalder L, Muhlemann O. Transcriptional silencing of nonsense codon-containing immunoglobulin minigenes. Mol Cell. 2005;18:307–17. doi: 10.1016/j.molcel.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Castanotto D, Tommasi S, Li M, Li H, Yanow S, Pfeifer GP, Rossi JJ. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol Ther. 2005;12:179–83. doi: 10.1016/j.ymthe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–9. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez S, Pisano DG, Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7:2601–8. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- 20.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–95. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman AR, Hu JF. Directing DNA methylation to inhibit gene expression. Cell Mol Neurobiol. 2006;26:425–38. doi: 10.1007/s10571-006-9057-5. [DOI] [PubMed] [Google Scholar]
- 22.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, et al. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nature Chemical Biology. 2005;1:210–5. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 23.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–92. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 24.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–7. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 25.Lim HG, Suzuki K, Cooper DA, Kelleher AD. Promoter-targeted siRNAs induce gene silencing of simian immunodeficiency virus (SIV) infection in vitro. Mol Ther. 2008;16:565–70. doi: 10.1038/sj.mt.6300380. [DOI] [PubMed] [Google Scholar]
- 26.Park CW, Chen Z, Kren BT, Steer CJ. Double-stranded siRNA targeted to the huntingtin gene does not induce DNA methylation. Biochem Biophys Res Commun. 2004;323:275–80. doi: 10.1016/j.bbrc.2004.08.096. [DOI] [PubMed] [Google Scholar]
- 27.Pulukuri SM, Rao JS. Small interfering RNA directed reversal of urokinase plasminogen activator demethylation inhibits prostate tumor growth and metastasis. Cancer Res. 2007;67:6637–46. doi: 10.1158/0008-5472.CAN-07-0751. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, et al. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem. 2008;283:23353–63. doi: 10.1074/jbc.M709651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–10. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner AM, De La Cruz J, Morris KV. Mobilization-competent Lentiviral Vector-mediated Sustained Transcriptional Modulation of HIV-1 Expression. Mol Ther. 2009;17:360–8. doi: 10.1038/mt.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci USA. 2005;102:16055–9. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Feng Y, Pan L, Wang Y, Xu X, Lu J, Huang B. The proximal GC-rich region of p16(INK4a) gene promoter plays a role in its transcriptional regulation. Mol Cell Biochem. 2007;301:259–66. doi: 10.1007/s11010-007-9427-4. [DOI] [PubMed] [Google Scholar]
- 33.Yamagishi M, Ishida T, Miyake A, Cooper DA, Kelleher AD, Suzuki K, Watanabe T. Retroviral delivery of Promoter-targeted shRNA induces long-term silencing of HIV-1 transcription. Microbes Infect. 11:500–8. doi: 10.1016/j.micinf.2009.02.003. 200. [DOI] [PubMed] [Google Scholar]
- 34.Zhang MX, Zhang C, Shen YH, Wang J, Li XN, Chen L, et al. Effect of 27 nt small RNA on endothelial nitric-oxide synthase expression. Mol Biol Cell. 2008;19:3997–4005. doi: 10.1091/mbc.E07-11-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen Z, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2005:12. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–10. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 37.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. PNAS. 2007:104. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 40.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–4. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 41.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, et al. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–73. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 43.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–42. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cliffe AR, Garber DA, Knipe DM. Transcription of the Herpes Simplex Virus Latency-Associated Transcript Promotes the Formation of Facultative Heterochromatin on Lytic Promoters. J Virol. 2009 doi: 10.1128/JVI.00712-09. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33:462–71. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 47.Beiter T, Reich E, Williams RW, Simon P. Antisense transcription: A critical look in both directions. Cell Mol Life Sci. 2009;66:99–112. doi: 10.1007/s00018-008-8381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ge X, Rubinstein WS, Jung YC, Wu Q. Genome-wide analysis of antisense transcription with Affymetrix exon array. BMC Genomics. 2008;9:27. doi: 10.1186/1471-2164-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finocchiaro G, Carro MS, Francois S, Parise P, DiNinni V, Muller H. Localizing hotspots of antisense transcription. Nucleic Acids Res. 2007;35:1488–500. doi: 10.1093/nar/gkm027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 51.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–8. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 52.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Human Molecular Genetics. 2008;17:642–55. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. Journal of RNAi and Gene Silencing. 2005;1:66–78. [PMC free article] [PubMed] [Google Scholar]