Summary
Prostate cancer biomarkers are enriched in urine after prostatic manipulation, suggesting whole cells might also be detectable for diagnosis. We tested multiplex staining of urinary sediments as a minimally-invasive method to detect prostate cancer. Urine samples were collected after prostatic massage (attentive digital rectal examination) from 35 men in Urology clinic, and without massage from 15 control men without urologic disease, for a total of 50 specimens (27 cancer positive cases, 23 cancer negative cases). LNCaP prostate cancer cells spiked into urine were used for initial marker optimization. Urine sediments were cytospun onto glass slides and stained. Multiplex urine cytology was compared to conventional urine cytology for cancer detection: anti-alpha-methylacyl CoA racemase (AMACR) antibody was used as a marker of prostate cancer cells, anti-Nkx3.1 as a marker of prostate epithelial cells, anti-nucleolin as a marker of nucleoli, and DAPI to highlight nuclei. Prostate cancer cells were successfully visualized by combined staining for AMACR, Nkx3.1, and nucleolin. Of 25 informative cases with biopsy-proven prostate cancer, 9 were diagnosed as suspicious or positive by multiplex immunofluorescence urine cytology, but only 4 were similarly judged by conventional cytology. All cases without cancer were read as negative by both methods. Multiplex cytology sensitivity for cancer detection in informative cases was 36% (9/25) and specificity was 100% (8/8). In conclusion, we have successfully achieved multiple-staining for AMACR, Nkx3.1, Nucleolin, and DAPI to detect prostate cancer cells in urine. Further refinements in marker selection and technique may increase sensitivity and applicability for prostate cancer diagnosis.
Keywords: prostate cancer, urine cytology, biomarkers, diagnosis, multiplex staining
1. Introduction
A prostate cancer (PC) diagnosis is suspected when prostate specific-antigen (PSA) and/or digital rectal examination (DRE) findings are abnormal. Prostate biopsy is currently the only way to confirm the diagnosis of clinically localized prostate cancer following abnormal PSA and/or DRE, and is not without risks, including local infection, systemic sepsis, and hemorrhage. Prostate biopsy itself suffers from inherent inadequacies in tissue sampling resulting in a sensitivity deficit (false negatives), ie. many cancers are missed. Nevertheless, prostate biopsy remains the gold standard for prostate cancer detection, and is the basis for preoperative pathologic grading and cancer volume estimation. A noninvasive and definitive test for prostate cancer would be most welcome to patients and clinicians, whether by imaging or molecular biofluid analysis. More effective tools to discriminate patients who harbor undetected cancer from those who do not, and who harbor clinically significant cancer are needed.
It is known that prostate cancer cells are shed into biological fluids, particularly when the prostate is subjected to physical manipulation, thus creating the potential for their non-invasive detection in either urine or expressed prostatic fluid (EPF)[1, 2]. However, unlike bladder cancer, where urine cytology has proven utility in cancer detection and surveillance, past attempts at detecting prostate cancer cells in urine by traditional cytology were thwarted by unacceptably low sensitivities, although specificities were consistently high. This sensitivity deficit was due primarily to the low numbers of prostate cancer cells present in urine cytology preparations, plus difficulty in differentiating malignant prostate cells from the other cells and debris present in Pap stained urine cytology slides[2–6]. This problem became even worse following the widespread use of PSA testing, which contributed to downward stage migration such that most prostate cancers detected in the modern era are much smaller in volume, lower in grade, and probably less likely to shed malignant cells into the urine. Consequently, detecting prostate cancer cells via urine cytology has been largely abandoned. Rather than whole cells, recent attention has focused on detecting various molecular markers linked to prostate cancer including proteins, enzymatic activities, coding and non-coding RNA species, and modified DNA (e.g. gene promoter region hypermethylation), each displaying various degrees of sensitivity and specificity for prostate cancer detection[7–12]. None of these tests have replaced prostate biopsy for diagnosis, though each confers differing levels of diagnostic importance. Indeed, much progress has recently been made in identifying and detecting novel molecular alterations present in prostate cancer cells [13, 14]. Considering the expanding pool of candidate biomarkers, plus current methods for sensitive marker visualization unavailable to previous cytologists, we revisited the cell-based approach to prostate cancer detection in urine samples. In this pilot study, we used simultaneous fluorescent co-staining of multiple prostate cancer biomarkers to allow the detection of even a few cancer cells if present in the urine. If definitively identified, prostate cancer cells shed in urine in conjunction with biomarkers and improved imaging modalities may one day supplant the modern 12-core biopsy for actual prostate cancer diagnosis.
2. Materials and Methods
2.1 Cell culture
LNCaP prostate cancer cells and T24 bladder cancer cells were obtained from ATCC and maintained as adherent cultures in RPMI-2640 medium supplemented with 10% FBS (plus 1nM R1881 for LNCaP). To mimic clinical urine samples, 5×103 LNCaP cells were spiked into 10ml of urine freshly obtained from a normal male volunteer.
2.2 Patients and urine specimens
Urine specimens were prospectively collected from 35 men, under an IRB-approved protocol. This study cohort comprised 12 men about to undergo diagnostic prostate needle core biopsy for standard indications (abnormal DRE, elevated PSA or elevated PSA velocity), one patient prior to trans-urethral resection (TUR) for symptomatic lower urinary tract symptoms, 2 men on expectant management for known small volume, low grade PC, and 20 men with known prostate cancer referred for a second opinion regarding definitive treatment Clinicopathologic characteristics of this cohort are listed in Table1. Ages ranged from 40–76 years and serum PSA levels ranged from 0.6–20.5 nl/ml. Prior to the biopsy, an “attentive” (approximately 30 second) DRE was performed, and the first 50 milliliters of voided urine were collected. An additional 15 control specimens were collected, without prostate massage, from men without suspicion of prostate cancer or with no evidence of disease (NED) following definitive treatment for prostate cancer (11 male volunteers without standard indications for prostate cancer, 3 patients NED following radical prostatectomy for prostate cancer, 1 patient NED following radiation therapy for prostate cancer). All urine specimens were stored refrigerated until processed, which typically occurred within 3 hours of collection.
Table 1.
Clinicopathologic information on the study cohort
| Case# | Prostate Biopsy Status | Biopsy Gleason | Clinical stage | Pathol. Stage | Standard urine cytology | Molecular urine cytology | Tumor volume | Age | Race | PSA (ng/ml) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | -- | -- | -- | -- | N | N | -- | 53 | W | 4.2 |
| 2 | -- | -- | -- | -- | N | N | -- | 65 | W | 9.1 |
| 3 | -- | -- | -- | -- | N | N | -- | 62 | W | 0.6 |
| 4 | -- | -- | -- | -- | N | N | -- | 54 | W | 3.4 |
| 5 | -- | -- | -- | -- | N | N | -- | 57 | W | 4.4 |
| 6 | -- | -- | -- | -- | N | N | -- | 72 | W | 7.1 |
| 7 | -- | -- | -- | -- | N | N | -- | 74 | W | 5.9 |
| 8 | -- | -- | -- | -- | N | N | -- | 40 | W | 0.8 |
| 9 | + | 4+3 | cT1c | pT3bN0 | N | N | Ext. | 56 | W | 5.3 |
| 10 | + | 3+3 | cT1c | pT2 | N | N | Mod. | 50 | B | 6.8 |
| 11 | + | 3+3 | cT1c | pT2 | N | N | Mod. | 50 | B | 6.2 |
| 12 | + | 3+4 | cT1c | pT2cN0 | Uninform | Uninform. | Ext. | 45 | W | 4.5 |
| 13 | + | 3+3 | cT1c | N/A | N | N | Small | 57 | W | 2.4 |
| 14 | + | 3+3 | cT1c | pT3aN0 | N | N | Ext. | 54 | W | 4.9 |
| 15 | + | 3+3 | cT1c | pT2aN0 | N | N | Small | 50 | W | 4.8 |
| 16 | + | 3+3 | cT1c | N/A | N | N | Ext. | 45 | W | 9.6 |
| 17 | + | 3+3 | cT2a | N/A | N | N | Mod. | 58 | W | 6.6 |
| 18 | + | 3+3 | cT2a | N/A | N | N | Ext. | 64 | W | 6.0 |
| 19 | + | 3+3 | cT1c | N/A | N | N | Small | 76 | W | 4.9 |
| 20 | + | 3+3 | cT1c | N/A | N | N | Small | 49 | W | 3.7 |
| 21 | + | 4+4 | cT2b | N/A | N | Cancer | Ext. | 76 | W | 20.5 |
| 22 | + | 3+3 | cT1c | N/A | N | Suspicious | Small | 61 | W | 6.6 |
| 23 | + | 3+3 | cT1c | pT2 | N | Suspicious | Mod. | 53 | W | 2.5 |
| 24 | + | 3+3 | cT1c | N/A | Cancer | Cancer | Mod. | 71 | W | 8.1 |
| 25 | + | 3+4 | cT1c | N/A | N | Cancer | Mod. | 60 | B | 11.3 |
| 26 | + | 3+3 | cT2b | pT3a | N | Suspicious | Ext. | 56 | B | 5.5 |
| 27 | + | 3+3 | cT2b | pT2cN0 | Suspicious | N | Ext. | 59 | W | 4.8 |
| 28 | + | 3+3 | cT1c | N/A | N | N | Small | 54 | W | 4.6 |
| 29 | + | 3+3 | cT1a | N/A | N | Suspicious | Small | 70 | W | 2.4 |
| 30 | + | 3+4 | cT1c | pT2 | Suspicious | Suspicious | Ext. | 59 | W | 5.2 |
| 31 | + | 3+3 | cT1c | pT2cN0 | N | Uninform. | Mod. | 58 | W | 17.0 |
| 32 | + | 3+3 | cT1c | N/A | N | N | Mod. | 68 | W | 4.4 |
| 33 | + | 3+3 | cT2b | pT3bN0 | Suspicious | N | Ext. | 56 | W | 7.9 |
| 34 | + | 3+3 | cT1c | N/A | N | N | Small | 68 | W | 5.4 |
| 35 | + | 3+3 | cT1c | pT2cN0 | N | Cancer | Small | 45 | W | 6.9 |
2.3 Sample preparations
In order to develop a method for the isolation of intact prostate cancer cells from urine and their deposition onto microscope slides suitable for multiplex staining, we utilized LNCaP prostate cancer tissue culture cells added to freshly collected urine in order to simulate the behavior of prostate cancer cells in patient urine specimens. For urine spiked with tissue culture cells and one-half volume of each post-DRE urine sample, the specimens were centrifuged at 800g for 5min, and pellets were suspended in PBS buffer. Cell suspensions were then applied to PrepStain Density Reagent (Tripath Imaging, Burlington, NC) and centrifuged at 200g for 2min, and the top layers were removed and centrifuged at 800g for 10min. The pellets were then resuspended in PBS and were applied to charged surface microscope slides by centrifugation in a cytocentrifuge at 1000 rpm for 3min (Shandon Cytospin IV, Thermo Scientific, Waltham, MA). By this procedure, two slides were made from each urine sample. On separate areas of the same cytospin slides T24 bladder carcinoma cells were deposited, serving as negative controls for antibody staining. While optimizing multiplex staining, several different fixatives were assayed including acid alcohols, acetone, zinc, paraformaldehyde and phosphate-buffered neutral formalin. Formalin fixation was eventually chosen for use, as it displayed good compatibility for staining with multiple different markers. Slides were kept at 4 degrees Centigrade in PBS until used.
2.4. Antibodies and immunostaining
2.4.1. Slide pre-treatment
Of several different target retrieval buffers assessed for heat-induced epitope retrieval (HIER) prior to antibody staining, we found that immersion of the slides for one minute in 2% Tween-20 detergent followed by steam-heating in high pH target retrieval solution (Cat.# S3307; DAKO Cytomation, Carpinteria, CA) for 20 minutes, then PBS with 0.05% Tween 20 (PBST) for 5 minutes, produced good results for the particular marker combinations used in these studies.
2.4.2. Immunofluorescence
Primary antibodies were incubated overnight at 4°C. The following antibodies were used in these studies: rabbit monoclonal anti-AMACR (Cat.# 13H4; Zeta, Sierra Madre, CA), rabbit polyclonal anti-Nkx3.1[15], mouse monoclonal anti-hTERT antibody (Cat.# NCL-hTERT; Novocastra , UK; shown to be specific for nucleolin rather than hTERT)[16]. For detection of the rabbit polyclonal anti-AMACR and anti-Nkx3.1 antibodies, signal amplification was performed using the CSA detection system (DAKO Cytomation, Carpinteria, CA), followed by application of 1:100 diluted streptavidin-Alexa Fluor 488 conjugated fluorescent secondary reagent (Cat.# S-32354; Molecular Probes, Eugene, OR) for 45 minutes at room temperature. For detection of the mouse monoclonal anti-nucleolin antibody, Alexa Fluor 568 goat anti-mouse IgG (Cat.#_A11061; Molecular Probes, Eugene, OR) was applied at a dilution of 1:100 in PBS for 45 minutes at room temperature. Slides were then stained with the nuclear counter stain 4’-6-diamidino-2-phenylindole (DAPI; Cat # D-8417, Sigma Chemicals, St. Louis, MO) and mounted with ProlongTM Anti-fade Mounting Media (Cat# P-7481;Molecular Probes, Eugene, Oregon) and cover slipped.
2.5. Evaluation of Fluorescently stained slides
Fluorescence microscopic evaluation of multiplex stained urine cytospin slides was performed in a blinded fashion, by two urologic pathologists (A.D.M., G.N.) using an epifluorescence microscope with appropriate fluorescence filter sets (Nikon Eclipse 400; Nikon Instruments, Inc., Melville, NY). The Slides were categorized for molecular cytology into one of 3 categories: (i) Malignant = presence of cells displaying simultaneous markers of both prostatic origin and prostate cancer, (ii) Suspicious = presence of cells displaying one or more of the cancer-associated markers but lacking definitive markers of prostatic origin, or cells with positive markers but of sub-optimal quality, thus precluding definitive categorization as malignant, (iii) Negative = lacking cells displaying markers of the malignant phenotype, although cells of prostate origin may be present. When both slides from each case were diagnosed as negative, the cases were considered negative for prostate cancer.
2.6. Evaluation of PAP stained slides
Following complete evaluations of the cytospin slides stained for immunofluorescence, the subset of slides from patients with prospective urine samples collected prior to biopsy or TUR (35 cases) were processed for standard cytopathologic examination. This was accomplished by cover slip removal, washing to remove anti-fade mounting medium and staining by the technique of Papanicolau[17]. The PAP stained slides were evaluated by a cytopathologist with expertise in urine cytopathology (S.A.) and scored for the presence of glandular epithelial cells which were characterized as benign, atypical/suspicious, or malignant per standard cytologic convention.
2.7. Statistical analysis
Differences between age and PSA between study subjects with or without prostate cancer were analyzed by the Mann-Whitney test. The immunofluorescence staining results were analyzed by the Fisher exact test.
3. Results
3.1. Validation of multiplex staining with LNCaP cells in urine
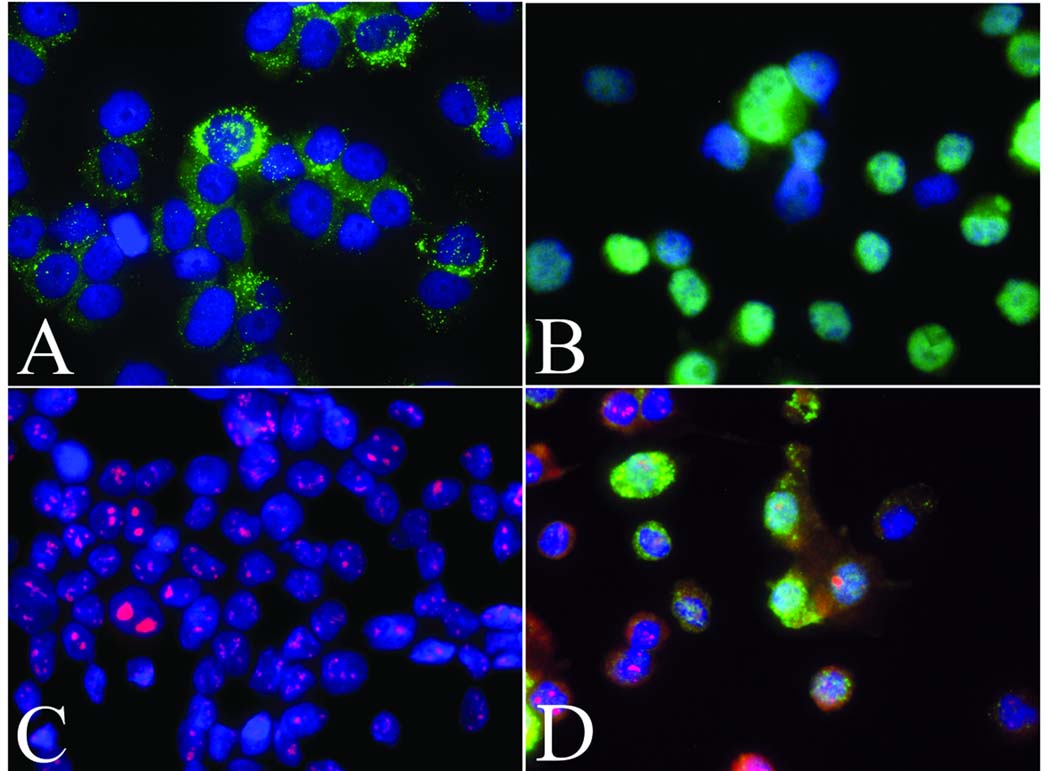
Figure 1 shows representative staining of cytospin slides derived from LNCaP-spiked urine fluorescently stained with individual antibodies directed against (i) the prostate-specific marker Nkx3.1, (ii) AMACR, which is over-expressed in prostate cancer, and (iii) nucleolin, which serves to highlight abnormal nucleoli - a hallmark of prostate cancer cells. Combined staining for multiple markers was also achieved. For example, a combination of AMACR positivity, prominent and multiple nucleoli (nucleolin staining), plus nuclear Nkx3.1 positivity (Figure 1D) allowed LNCaP prostate cancer cells to be readily distinguished from other cell types present in the sample. Rare normal cells were positive for single markers, typically staining weakly for either nucleolin or AMACR, but no normal cells stained positively for two or more of the biomarkers used.
Figure 1. Immunofluorescence staining of LNCaP prostate cancer cell cytospins.
LNCaP cytospin preps were stained with: (A) anti-Alpha-methylacyl-CoA racemase (AMACR) antibody detected in green; (punctuate cytoplasmic staining pattern); (B) anti-Nkx3.1 (nuclear pattern) and (C) anti-Nucleolin (nucleolar pattern); (D) Combined fluorescent staining for AMACR, Nkx3.1, and nucleolin. In all cases nuclei are co-stained blue with the DNA-binding dye DAPI. Original magnification=400X.
When observed, autofluorescence was primarily cytoplasmic and exhibited sample-to-sample variability as well as variability between differing cell types. Although bright at times, it did not interfere with the detection of our marker panel due to the facts that two of the markers, nucleolin and Nkx3.1, are nuclear, while AMACR positivity manifests as a distinctive punctate cytoplasmic staining pattern restricted to the single color channel used for the fluorescent secondary detection reagent, in contrast, cellular autofluorescence tended to exhibit a more homogeneous pattern and was also present in more than one color channel.
Regarding cell recovery and reproducibility, by varying the numbers of LNCaP cells added into the urine we found that we could reproducibly detect LNCaP cells in the cytospin slides 100% of the time from a input number of 1,000 LNCaP cells however, cells were only detected on the cytospin slides 50% of the time when 100 LNCaP cells were introduced and no cells were detected when only 10 LNCaP cells were added to the urine samples.
3.2. Staining of urine samples
On the cytospin slides made using clinical urine samples, the number of cellular elements (including all cell types; see below) present varied markedly from case to case. Of the 35 study samples, 19 cases displayed low to very low cell numbers (a few hundred cells or less), 8 cases displayed moderate cell numbers (a few thousand cells), and 6 cases displayed high cell numbers (several thousand cells). The presence of corpora amylacea was noted on 11 of the 35 cases (31%), indicating that the prostate was contributing material to at least a subset of the urine sediments. 6 of the 11 specimens with corpora also had moderate to high cellularity. In two cases (cases #12 and #31) the cytospin slides contained virtually no cells, thus were considered inadequate for evaluation by multiplex fluorescence analysis and were classified as uninformative.
In 27 of the 35 study cases, prostate cancer was detected on needle biopsy (Table 1). There were no significant differences in age or PSA between the cancer group and the negative-biopsy group. Table 2 compares the molecular cytology results to the pathologic findings on needle biopsy. Among the 25 informative cases with biopsy-proven prostate cancer, 4 cases were classified as malignant by molecular cytology, 5 cases were considered suspicious, and 16 cases were considered negative. Positive cases typically contained small clumps of suspicious or malignant cells, although solitary positive cells were sometimes observed (Figure 2). Suspicious cases generally had cells with very weak or absent Nkx3.1 staining, although they displayed staining for AMACR and multiple or large nucleoli. The group of 15 control patients and NED patients were all negative by molecular cytology.
Table 2.
Performance of molecular cytology in clinical urine specimens
| Cancer status | NED voided urines | ||
|---|---|---|---|
| Molecular cytology | Positive (n=25) | Negative (n=8) | (n=15) |
| Positive | 4 | 0 | 0 |
| Suspicious | 5 | 0 | 0 |
| Negative | 16 | 8 | 15 |
| Uninformative | 2 | ||
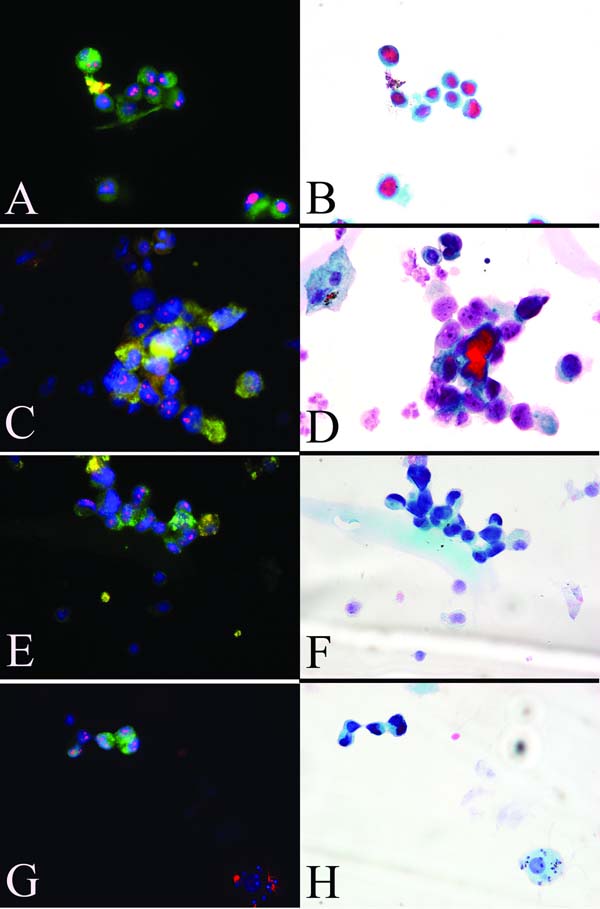
Figure 2. Multiplex staining of urine sediments obtained from clinical urine specimens.
Panels A,C,E and G: combined fluorescent staining for AMACR (green, cytoplasmic), Nkx3.1 (green, nuclear), and nucleolin (red, sub-nucleolar) strongly suggests the presence of exfoliated prostate cancer cells in these urine specimens. Panels B,D,F and H: same specimens stained by the Papanicolau procedure.
By combining the two abnormal categories (suspicious and malignant), the overall sensitivity of multiplex staining to detect prostate cancer in urine specimens following prostatic manipulation was 33% (9/27), with a specificity of 100% (8/8). Notably, of the 9 cases judged to be abnormal, 7 were from slides having cellularity in the moderate-high range. No significant differences were found between the 3 molecular cytology categories and any of the clinicopathologic variables in this cohort.
The study cohort consisted of 27 men with PC and 8 without. Pathologically, six of the PC cases had Gleason pattern 4 disease while the other 21 had only Gleason pattern 3 disease. Four of the 6 (67%) cases with pattern 4 produced molecular cytology results that were either suspicious or positive on multiplex immunofluorescence cytology. In one of the other cases the specimen was deemed inadequate in terms of cellularity (likely due to improper collection), and in the other a pattern 4+3 case was missed (negative urine sediment). While the yield of molecular cytology for cases of high grade PC (Gleason pattern 4) was much higher than for the pattern 3 cases, the subset under analysis was smaller.
3.3. Comparison of molecular cytology and standard cytopathology
We further wished to determine how these same slides would be judged by standard cytopathology. In an attempt to address this question, we carefully removed the cover slips and submitted the slides for PAP staining and blinded cytopathology review. A range of cell types, typical of those seen in urinary cytology specimens, were noted on the PAP-stained slides including; urothelial cells, umbrella cells, squamous cells, inflammatory cells, renal tubule cells and mature sperm. Of the 27 biopsy-confirmed cancer cases, 3 were diagnosed as having atypical or suspicious glandular epithelial cells and 1 case was diagnosed as containing frankly malignant epithelial cells consistent with prostate adenocarcinoma, giving a sensitivity of 15% (4/27) for prostate cancer detection by standard cytology when suspicious and malignant categories were combined. The single case deemed malignant by standard cytology had also been deemed malignant by molecular cytology and, upon review, this assessment turned out to be based on the same group of abnormal cells (Figure 2C&D). All 4 cases deemed atypical/suspicious or malignant by PAP staining were from cases that had moderate to high cellularity on the cytospin slides. Of the two cases considered uninformative by multiplex fluorescence staining due to low cellularity, one (case# 12) was likewise deemed uninformative by standard cytology, while the other (case# 31), although scant, was nonetheless characterized as normal. As with molecular cytology, all cases negative for cancer on biopsy were deemed benign by cytopathology.
Following the blinded cytopathology review, we re-examined the slides from the discordant cases that had first been deemed abnormal by molecular cytology, but later diagnosed as benign by standard cytopathology. In 5 of these cases we were able to find the identical cells on the PAP stained slides that had previously been judged abnormal on the molecular cytology slides, and these specific cells were presented to the cytopathologist for review. In one case, the cells had been classified previously as benign appearing epithelial glandular cells, likely of prostatic origin. In the remaining 4 cases the cells had not been noted during the initial review of the PAP stained slides. In 3 of these cases, the cells were described, on review, as small cells with high N/C ratios with poorly preserved, smudgy chromatin without visible or prominent nucleoli (for example, see Figure 2G&H). It was further noted that while the small cell sizes and high N/C ratios would be consistent with prostate adenocarcinoma, the morphological preservation was sub-optimal and precluded such a diagnosis. In the remaining case, the cells were described as a three dimensional glandular fragment with smudgy chromatin and difficult to discern nucleoli.
4. Discussion
Over 600,000 diagnostic prostate biopsy procedures are performed annually in the United States. Prostate biopsies are not without risk, and inherently undersample the prostate. The false negative rate of initial biopsy has been estimated as high as 34%. Indeed, some 20%-35% of patients sent for repeat biopsy are diagnosed with cancer not detected on initial biopsy [18, 19]. On the other hand, many men lacking cancer on first biopsy do not have clinically detectable prostate cancer (true negatives) despite the continued presence of suspicious DRE or PSA results. Thus, patients whose initial biopsy results are negative present a serious clinical dilemma due to our inability to differentiate those patients at high risk for harboring undetected cancer who might benefit from immediate repeat biopsy, from those unlikely to have cancer who could be spared unnecessary additional biopsies and their associated morbidity and cost. Men in this situation understandably experience much anxiety due to these uncertainties, and many will ultimately undergo multiple repeat biopsy procedures. In this regard, a non-invasive test to help augment current screening and testing modalities would be of enormous benefit.
Following the successful demonstration by Papanicolaou in 1942 of his cytologic staining method for detecting exfoliated malignant cells of the uterine cervix, clinical researchers eagerly sought to apply the technique to detect cancer in bodily fluids[17]. With respect to the male GU tract, although successful in detecting bladder cancer cells in urine sediments, the method failed to yield similar results for other types of GU cancer, including prostate cancer. Despite consistently high specificities, attempts at detecting prostate cancer cells by urine cytology were plagued by low sensitivities due to the relatively low numbers of prostate cancer cells present in urine cytology preps, plus difficulty in differentiating malignant prostate cells from the other cells and debris present [3–6, 20, 21]. Despite the inability of routine cytology to reliably detect prostate cancer cells in the urine, it was clear that, at least in a subset of cases, intact, exfoliated prostate cancer cells could sometimes be recovered from urine samples or expressed prostatic fluid (EPF).
The recent application of molecular techniques to the study of prostate cancer has led to the identification of several novel molecular alterations present in prostate cancer cells; for example, increased expression of the enzyme alpha-methylacyl-CoA racemase (AMACR), abnormally short telomeres, and recurrent chromosomal translocations involving members of the ETS family of transcription factors, to name just a few [13, 22–26]. With the abandonment of the cytologic approach, current efforts are now focused on detecting such molecular changes in the urine or EPF [7–12, 15].
Paralleling the advances in biomarker discovery, significant advances in antibody and in situ nucleic acid detection methods have also been made, such as tyramide signal amplification, rolling circle amplification and the advent of fluorescent quantum dot secondary detection reagents [14, 27, 28]. Considering the expanding pool of candidate prostate cancer biomarkers, plus these modern tools for sensitive marker visualization that were unavailable to previous researchers attempting urine cytology, we felt that re-visiting a cell-based approach, using molecular cytopathology to detect prostate cancer cells in urine samples could be rewarding [29]. Specifically, we hypothesized that prostate cancer cells isolated from the urine could be stained simultaneously for a panel of molecular markers and imaged using fluorescence microscopy, thus providing a sensitive, non-invasive and unambiguous method for their detection. Such a method would include; (i) a clear delineation of prostate cells from other cell types present in urine samples, such as lymphocytes and cells originating from other portions of the GU tract (e.g. bladder, urethra or kidney) and (ii) identification of the subset of prostate epithelial cells that are malignant.
In this study we have demonstrated the feasibility of using molecular markers to detect apparently intact prostate cancer cells in urine sediments. We first demonstrated the staining of LNCaP in urine with our marker set, then analyzed clinical urine samples in a cohort of men enriched with prostate cancer cases, some men who were biopsy negative, and some normal controls. For this pilot study, we specifically attempted to exclude men with conditions that might confound our urine sediment assay: Men with high grade prostate intraepithelial neoplasia (PIN) or urothelial carcinoma of the bladder, which could show positive AMACR or prominent nucleolin staining, respectively. Men with these conditions, as well as patients undergoing radical cystectomy for urothelial carcinoma without coexistent prostate adenocarcinoma would be interesting subsets to study cytologically in the future with our multiplex technique, for more accurate sensitivity and specificity determinations. Certainly, more prostate cancer-specific markers will be developed over the coming years; it will be of interest to optimize and study the most specific of these in such cohorts as well as in larger groups of men being screened and subsequently biopsied.
Several groups have demonstrated the ability to detect prostate cancer-specific biomarkers (e.g. methylated DNA, protein and RNA) in urine samples after manipulation of the prostate [7–12]. We, therefore, collected our urine specimens following trans-rectal prostatic massage, or “attentive DRE”, from men scheduled to undergo needle biopsy for suspicion of cancer. We then optimized methods for cell preservation, isolation, cytospin slide preparation, and multiplex staining for a panel of prostate cancer biomarkers. The specific markers and fluorescent secondary reagents used for their detection were chosen based upon consideration of the following: (1) cell type specificity, (2) species of origin of the primary antibody (restricts which secondary may be used), and (3) specific sub-cellular localization and staining character of the marker. These considerations presented some restrictions on which markers could be used together without interfering with one another, but also allowed for economy in the numbers of colored fluorescent secondaries used. For example, the anti-Nkx3.1 antibody homogeneously labels the nuclei of prostate epithelial cells, while the anti-racemase antibody yields strong punctate cytoplasmic staining in prostate cancer cells. Given their restriction to different, spatially distinct cellular compartments (nucleus versus cytoplasm) the same colored secondary antibody (green in this example) can be used for the detection of both of these rabbit-derived primary antibodies without interference. Furthermore, this same red channel may also be utilized to detect a nucleolar marker because, despite its nuclear localization, the much larger nucleolus is readily distinguishable from the much smaller and more numerous telomere signals. There is no question that more accurate and more specific markers will be developed over the coming years – ours represents an attempt at developing a diagnostic panel given contemporary data and available reagents.
By assaying various combinations of antibodies and staining conditions we decided upon a set of 3 antibodies (anti-Nkx3.1, anti-nucleolin and anti-AMACR) for use in evaluating the feasibility of detecting prostate cancer cells by simultaneous fluorescent staining of cytospins from clinical urine specimens. Table 3 contrasts the expected staining phenotype of prostate cancer cells versus normal cells using this particular combination of markers. Elevated staining of AMACR, prominent nucleoli, and nuclear morphologic abnormalities revealed by DAPI staining were used as markers of prostate cancer cells, while Nkx3.1 positivity confirmed their prostatic epithelial origin [15, 22, 25, 30–32]. Other potentially useful markers of prostate epithelial cells such as Prostate Specific Membrane Antigen (PSMA), Prostate Specific Antigen (PSA), and telomere FISH were also assayed but did not perform well in combination with the other antibodies or lacked specificity in our assays (data not shown).
Table 3.
Expected staining characteristics of prostate cancer versus normal cells in urine sediments for combined immunofluorescence
| Marker | Subcellular localization | Cell staining phenotype: Normal | Cell staining phenotype: Prostate cancer |
|---|---|---|---|
| Anti-AMACR | Cytoplasm | Negative-weak | Strong punctuate pattern |
| Anti-nucleolin | Nucleolus | Absent or weak, single nucleolus | Strong, prominent and/or multiple nucleoli |
| Anti-Nkx3.1 | Nucleus | Homogeneous nuclear stain in normal prostate epithelial cells, other cell types negative. | Homogeneous nuclear stain |
Regarding assay performance, there were no false positives and thus specificity was 100%. Sensitivity for prostate cancer detection was 36% by molecular cytology which, although somewhat low, provides a proof of principle for this new approach. A recent multi-center study with multiplexed quantitative methylation-specific polymerase chain reaction assay demonstrated 53–55% sensitivity and 76–80% specificity for prostate cancer detection with urine obtained similarly (after DRE) [33]. Four of six (67%) cases with higher grade (Gleason pattern 4) were deemed suspicious or positive by our assay, with one of the other six specimens having been judged acellular and thus inadequate from a collection standpoint. We hypothesize that in the rare instances of a virtually acellular sediment, patients either provided a midstream rather than an initial urine sample after attentive DRE, or the DRE was not vigorous enough. Alternately, in some cases prostatic material may not be shed in sufficient quantity to be detectable in initial urine samples (at least by our current assays). Indeed the main problem we encountered with the clinical specimens was low cellularity. Only 37% of samples yielded moderate to high cellularity on the cytospin slides. The fact that 78% of the samples displaying suspicious or malignant cells were cases with moderate to high cellularity supports the importance of obtaining adequate numbers of cells. This is likely to be especially important in cases where cancer cell numbers are limited; a situation reasonably to be expected with the small volume cancers currently typically detected in the PSA-era. Currently we are pursuing methods for stabilizing the pH and osmotic balance of the urine specimens in an attempt to help overcome this problem [34]. More thorough prostatic massage techniques, faster sample processing, accurate instruction to patients on providing initial post-DRE samples, and alternate fixation strategies may improve cell yields. In addition, alternative cell isolation strategies, such as the use of immunomagnetic beads (e.g. Dynabead Epithelial Enrich or CELLextion beads, Invitrogen, Carlsbad, CA), or filtration in combination with automated fluorescence microscopy may also lead to improvements [35].
In summary, we have provided a proof of principle for the direct detection of prostate cancer cells from urine samples collected in the clinic. The assay was able to detect very small numbers of cancer cells and was 100% specific. Although the sensitivity of the molecular cytology assay was relatively low, it did outperform standard cytology on the same samples, and it is reasonable to expect an increase in sensitivity with further refinement of the assay and development of better molecular targets. Molecular cytology, perhaps coupled with assays for soluble cancer biomarkers in the urine currently under development, should prove useful in solving the clinical dilemma of how best to proceed with men who are suspected of harboring prostate cancer but whose initial needle biopsy result is negative. In such patients, the finding of prostate cancer cells in post-biopsy urine would strongly imply undetected cancer and indicate the need for immediate re-biopsy. In addition to addressing this significant clinical problem, our assay could conceivably prove useful in other ways, such as for primary cancer detection following DRE, for monitoring disease status in prostate cancer patients who opt for expectant management (e.g. “watchful waiting” or “active surveillance”), or for monitoring disease status in men who have undergone non-extirpative treatment approaches such as cryotherapy or radiotherapy. An assay such as this might also be useful to diagnose prostate cancer in patients who are unsuitable for biopsy due to hemorrhagic diathesis, need for constant anti-coagulation, or severe medical comorbidities. Furthermore, it is conceivable that some qualitative or quantitative measure of prostate cancer cells found in the urine (e.g. absolute numbers of cancer cells present) may correlate with tumor aggressiveness and thus also be of use in prognostication (like Gleason grade), though this remains highly speculative. We believe the current study provides a compelling rationale for the continued development of methods for detecting prostate cancer in the urine by the same standard that is currently used for definitive diagnosis by pathologists in needle biopsies – direct visualization of the cancer cells themselves.
Acknowledgements
The authors wish to express their gratitude to Karen Plowden of the Johns Hopkins Pathology Department for her assistance in the use of the Surepath centrifugal technique for the isolation of cells from urine specimens. We also thank Tsuyoshi Iwata of the Johns Hopkins Pathology Department for technical advices.
This work was supported by the following research funding mechanisms: NIH/NIDDK grant 1K23DK071262 (C.P.P.), Dept. of Defense grant W81XWH-05-1-0167 (C.P.P.) and a Patrick C. Walsh Foundation grant (A.K.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure/Conflict of Interest
The authors have no disclosures to declare nor any competing interests that would constitute a conflict of interest regarding the work described in this manuscript.
References
- 1.Bologna M, Vicentini C, Festuccia C, et al. Early diagnosis of prostatic carcinoma based on in vitro culture of viable tumor cells harvested by prostatic massage. Eur Urol. 1988;14:474–476. doi: 10.1159/000473012. [DOI] [PubMed] [Google Scholar]
- 2.Garret M, Jassie M. Cytologic examination of post prostatic massage specimens as an aid in diagnosis of carcinoma of the prostate. Acta Cytol. 1976;20:126–131. [PubMed] [Google Scholar]
- 3.Albers DD, Mc DJ, Thompson GJ. Carcinoma cells in prostatic secretions. J Am Med Assoc. 1949;139:299–303. doi: 10.1001/jama.1949.02900220025005. [DOI] [PubMed] [Google Scholar]
- 4.Guinan P, Gilham N, Nagubadi SR, Bush I, Rhee H, McKiel C. What is the best test to detect prostate cancer? CA Cancer J Clin. 1981;31:141–145. doi: 10.3322/canjclin.31.3.141. [DOI] [PubMed] [Google Scholar]
- 5.Sharifi R, Shaw M, Ray V, Rhee H, Nagubadi S, Guinan P. Evaluation of cytologic techniques for diagnosis of prostate cancer. Urology. 1983;21:417–420. doi: 10.1016/0090-4295(83)90170-x. [DOI] [PubMed] [Google Scholar]
- 6.Koss LG, Deitch D, Ramanathan R, Sherman AB. Diagnostic value of cytology of voided urine. Acta Cytol. 1985;29:810–816. [PubMed] [Google Scholar]
- 7.Cairns P, Esteller M, Herman JG, et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res. 2001;7:2727–2730. [PubMed] [Google Scholar]
- 8.Gonzalgo ML, Pavlovich CP, Lee SM, Nelson WG. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin Cancer Res. 2003;9:2673–2677. [PubMed] [Google Scholar]
- 9.Hessels D, Klein Gunnewiek JM, van Oort I, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8–15. doi: 10.1016/s0302-2838(03)00201-x. discussion -6. [DOI] [PubMed] [Google Scholar]
- 10.Laxman B, Tomlins SA, Mehra R, et al. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia. 200;6(8):885–888. doi: 10.1593/neo.06625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers CG, Yan G, Zha S, et al. Prostate cancer detection on urinalysis for alpha methylacyl coenzyme a racemase protein. J Urol. 2004;172:1501–1503. doi: 10.1097/01.ju.0000137659.53129.14. [DOI] [PubMed] [Google Scholar]
- 12.Jeronimo C, Usadel H, Henrique R, et al. Quantitative GSTP1 hypermethylation in bodily fluids of patients with prostate cancer. Urology. 2002;60:1131–1135. doi: 10.1016/s0090-4295(02)01949-0. [DOI] [PubMed] [Google Scholar]
- 13.Tricoli JV, Schoenfeldt M, Conley BA. Detection of prostate cancer and predicting progression: current and future diagnostic markers. Clin Cancer Res. 2004;10:3943–3953. doi: 10.1158/1078-0432.CCR-03-0200. [DOI] [PubMed] [Google Scholar]
- 14.Smith AM, Dave S, Nie S, True L, Gao X. Multicolor quantum dots for molecular diagnostics of cancer. Expert Rev Mol Diagn. 2006;6:231–244. doi: 10.1586/14737159.6.2.231. [DOI] [PubMed] [Google Scholar]
- 15.Bethel CR, Faith D, Li X, et al. Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with gleason score and chromosome 8p deletion. Cancer researc. 2006;66:10683–10690. doi: 10.1158/0008-5472.CAN-06-0963. [DOI] [PubMed] [Google Scholar]
- 16.Wu YL, Dudognon C, Nguyen E, et al. Immunodetection of human telomerase reverse-transcriptase (hTERT) re-appraised: nucleolin and telomerase cross paths. J Cell Sci. 2006;119:2797–2806. doi: 10.1242/jcs.03001. [DOI] [PubMed] [Google Scholar]
- 17.Papanicolaou GN. A New Procedure for Staining Vaginal Smears. Science. 1942;95:438–439. doi: 10.1126/science.95.2469.438. [DOI] [PubMed] [Google Scholar]
- 18.Djavan B, Zlotta A, Remzi M, et al. Optimal predictors of prostate cancer on repeat prostate biopsy: a prospective study of 1,051 men. J Urol. 2000;163:1144–1148. discussion 8–9;. [PubMed] [Google Scholar]
- 19.Keetch DW, Catalona WJ, Smith DS. Serial prostatic biopsies in men with persistently elevated serum prostate specific antigen values. J Urol. 1994;151:1571–1574. doi: 10.1016/s0022-5347(17)35304-1. [DOI] [PubMed] [Google Scholar]
- 20.Foot NC, Holmquist ND. Supravita staining of sediments of serous effusions a simple technique for rapid cytological diagnosis. Cancer. 1958;11:151–157. doi: 10.1002/1097-0142(195801/02)11:1<151::aid-cncr2820110127>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 21.Garret M, Hamm FC. Atypical cells of orgin from the seminal vesicles, complicating cytologic evaluation of prostatic secretions. Am J Clin Pathol. 1963;39:265–272. doi: 10.1093/ajcp/39.3.265. [DOI] [PubMed] [Google Scholar]
- 22.De Marzo AM, DeWeese TL, Platz EA, et al. Pathological and molecular mechanisms of prostate carcinogenesis: implications for diagnosis, detection, prevention, and treatment. J Cell Biochem. 2004;91:459–477. doi: 10.1002/jcb.10747. [DOI] [PubMed] [Google Scholar]
- 23.Luo J, Zha S, Gage WR, et al. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer research. 2002;62:2220–2226. [PubMed] [Google Scholar]
- 24.Meeker AK, Hicks JL, Platz EA, et al. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer research. 2002;62:6405–6409. [PubMed] [Google Scholar]
- 25.Rubin MA, Zhou M, Dhanasekaran SM, et al. alpha-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. Jama. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 26.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson M. Lock and roll: single-molecule genotyping in situ using padlock probes and rolling-circle amplification. Histochem Cell Biol. 2006;126:159–164. doi: 10.1007/s00418-006-0213-2. [DOI] [PubMed] [Google Scholar]
- 28.Speel EJ, Hopman AH, Komminoth P. Tyramide signal amplification for DNA and mRNA in situ hybridization. Methods Mol Biol. 2006;326:33–60. doi: 10.1385/1-59745-007-3:33. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt FC, Longatto-Filho A, Valent A, Vielh P. Molecular techniques in cytopathology practice. Journal of clinical pathology. 2008;61:258–267. doi: 10.1136/jcp.2006.044347. [DOI] [PubMed] [Google Scholar]
- 30.Ali TZ, Epstein JI, Bieberich CM, De Marzo AM. Nkx3.1 as a new tissue marker of prostate adenocarcinoma. Modern Pathology. 2006;19:128A. [Google Scholar]
- 31.Kelemen PR, Buschmann RJ, Weisz-Carrington P. Nucleolar prominence as a diagnostic variable in prostatic carcinoma. Cancer. 1990;65:1017–1020. doi: 10.1002/1097-0142(19900215)65:4<1017::aid-cncr2820650429>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 32.Sturgis CD, Box M, D'Costa R, Forgue B, McGuire MS, Dieterich M. Ancillary alpha-methylacyl-CoA racemase immunocytochemistry in the diagnosis of adenocarcinoma of the prostate in urinary cytology: a case report. Acta Cytol. 2006;50:335–338. doi: 10.1159/000325965. [DOI] [PubMed] [Google Scholar]
- 33.Vener T, Derecho C, Baden J, et al. Development of a multiplexed urine assay for prostate cancer diagnosis. Clinical chemistry. 008;54:874–882. doi: 10.1373/clinchem.2007.094912. [DOI] [PubMed] [Google Scholar]
- 34.Kern WH, Bales CE, Webster WW. Cytologic evaluation of transitional cell carcinoma of the bladder. J Urol. 1968;100:616–622. doi: 10.1016/s0022-5347(17)62583-7. [DOI] [PubMed] [Google Scholar]
- 35.Ntouroupi TG, Ashraf SQ, McGregor SB, et al. Detection of circulating tumour cells in peripheral blood with an automated scanning fluorescence microscope. British journal of cancer. 2008;99:789–795. doi: 10.1038/sj.bjc.6604545. [DOI] [PMC free article] [PubMed] [Google Scholar]