Abstract
MicroRNAs (miRNAs) are short, non-coding RNAs that regulate gene expression and are aberrantly expressed in human cancer. The ERBB-2 tyrosine kinase receptor is frequently overexpressed in prostate cancer and is associated with disease progression and poor survival. We have identified two specific miR-331-3p target sites within the ERBB-2 mRNA 3′-untranslated region and show that miR-331-3p expression is decreased in prostate cancer tissue relative to normal adjacent prostate tissue. Transfection of multiple prostate cancer cell lines with miR-331-3p reduced ERBB-2 mRNA and protein expression and blocked downstream phosphatidylinositol 3-kinase/AKT signaling. Furthermore, miR-331-3p transfection blocked the androgen receptor signaling pathway in prostate cancer cells, reducing activity of an androgen-stimulated prostate-specific antigen promoter and blocking prostate-specific antigen expression. Our findings provide insight into the regulation of ERBB-2 expression in cancer and suggest that miR-331-3p has the capacity to regulate signaling pathways critical to the development and progression of prostate cancer cells.
Prostate cancer (PCa)3 is the second leading cause of cancer death among men in the United States. In 2007, 218,890 new cases and 27,050 prostate cancer-related deaths had occurred in the United States (1). Initially, prostate tumors express the androgen receptor (AR) and are dependent on androgens for their growth, providing the basis of androgen ablation therapy; however, some of these tumors will eventually recur in an androgen-independent form, with a significantly worse prognosis. There are no effective therapies for androgen-independent prostate cancer. Overexpression of the human epidermal growth factor receptor 2 (ERBB-2/HER2) and activation of its downstream signaling cascades, including the PI3K/AKT pathway, which promotes cell proliferation, metastasis, apoptosis resistance, and angiogenesis, has been identified in a range of tumors, including those of the breast, prostate, and pancreas (2). Studies in patients with localized PCa have shown that ERBB-2 expression is significantly associated with disease progression, metastasis, and survival (3). In addition, it is thought that elevated ERBB-2 expression and AKT signaling facilitates the development of androgen-independent PCa by activating AR signaling in the absence of androgen (4–7). These studies emphasize the important functional role of ERBB-2 and its signaling pathways in the progression of PCa, in part through interactions with AR signaling, and highlight its potential as a therapeutic target.
MicroRNAs (miRNAs) are a class of short, endogenous, non-coding RNA molecules that bind with imperfect complementarity to the 3′-untranslated regions (3′-UTRs) of target mRNAs, causing translational repression or message degradation (8, 9). MiRNAs have important roles in normal cellular development and function (10, 11), and altered expression of miRNAs is associated with cancer (12). Many miRNA genes are located at fragile genomic regions that are amplified, deleted, or rearranged in cancer (13), whereas aberrant expression of other miRNAs in cancer can be attributed to alterations in miRNA biogenesis or miRNA promoter methylation (14–16), or to transcription factors such as MYC and p53 that directly regulate miRNA transcription (17, 18). It has been suggested that some miRNAs may act as oncogenes or tumor suppressor genes (12, 19). For example, decreased expression of the let-7 miRNAs is associated with RAS oncogene overexpression and reduced survival in non-small cell lung cancer (20, 21), whereas increased miR-21 expression in a range of cancers, including those of the breast, prostate, lung, colon, pancreas, and stomach (22), is associated with reduced apoptosis, chemoresistance, and increased tumor growth (23). Several studies have reported aberrant patterns of miRNA expression in PCa (24, 25), whereas others have implicated specific miRNAs in the development of androgen-independent PCa. These include miR-125b, which regulates expression of the pro-apoptotic factor Bak1 and promotes androgen-independent growth (26) and miR-146a, which regulates expression of the oncogenic ROCK1 kinase and modulates tumorigenicity (27). Interestingly, the tumor suppressor miRNAs miR-125a and miR-125b (22) have been shown to directly regulate ERBB-2 expression in breast cancer (28), but not in PCa (26). To date it has been unclear whether miRNAs regulate ERBB-2 expression in PCa.
In this study, we show that miR-331-3p directly regulates ERBB-2 mRNA and protein expression in multiple PCa cell lines via two specific ERBB-2 3′-UTR target binding sites. We found that miR-331-3p expression is down-regulated in ERBB-2-overexpressing PCa tissue relative to normal adjacent prostate tissue, and that miR-331-3p reduces downstream ERBB-2 signaling via phosphorylated AKT in PCa cells. Furthermore, miR-331-3p blocked the AR signaling pathway by reducing transcriptional activity and expression of prostate-specific antigen (PSA), an AR target gene. Our data show a new mechanism by which reduced miR-331-3p expression in PCa promotes elevated ERBB-2 expression and signaling; through cross-talk this facilitates AR signaling and has implications for the development, progression, and treatment of PCa.
EXPERIMENTAL PROCEDURES
Cell Culture, miRNA Precursors, and LNA Inhibitors, and Normal/Tumor Tissue RNA
LNCaP-FGC, CWR-22RV1, and DU145 cell lines were obtained from the American Type Culture Collection (ATCC) and cultured at 37 °C in 5% CO2 with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. For ligand treatments, cells were starved overnight in 1% charcoal-stripped serum and stimulated with heregulin (β-subunit, Sigma; 50 ng/ml for 20 min), or DHT and/or bicalutamide at 10 nm and 10 μm, respectively, for 24 h. Synthetic miRNA precursor molecules corresponding to human miR-331-3p (precursor miR miRNA product ID PM10881) and a negative control miRNA (miR-NC; precursor miR miRNA negative control number 1, product ID AM17110) were obtained from Ambion. Synthetic LNA precursor molecules corresponding to hsa-miR-331-3p (miRCURY knockdown number 138573-00) and a negative control scramble-LNA (miRCURY knockdown number 199002-00) were obtained from Exiqon. Total RNA from normal adjacent tissue (NAT) and prostate tumor were purchased from Ambion (FirstChoice; product ID AM7288, acinar adenocarcinoma, moderately differentiated, stage II, T2N0M0, Gleason score 6 (3 + 3)).
Luciferase Plasmid Construction
pmiR-REPORT-miR-331- 3p-target was generated by ligating annealed DNA oligonucleotides corresponding to a perfect hsa-miR-331-3p target site (forward, 5′-CAA CAA AAT CAC TAG TCT TCC A-3′ and reverse, 5′-TGG AAG ACT AGT GAT TTT GTT G-3′) to unique SpeI and HindIII sites that were inserted 3′ of the luciferase open reading frame of pmiR- REPORT (Promega) firefly luciferase reporter vector. Full-length ERBB-2 3′-UTR reporter plasmid was generated by cloning the PCR-amplified, full-length ERBB-2 3′- UTR (nucleotides 4006–4624 of GenBankTM accession number NM_004448) into the pmiR-REPORT luciferase plasmid backbone (Ambion). Wild type and mutant ERBB-2 target reporter plasmids pmiR-REPORT-ERBB-2-A and -B were generated by cloning annealed oligonucleotides corresponding to nucleotides 4093–4115 and 4513–4535, respectively, of ERBB-2 (GenBank accession number NM_004448) mRNA 3′-UTR into SpeI and HindIII sites in pmiR- REPORT. Mutant vectors contained mutations in the miR-331-3p seed binding regions. Oligonucleotide sequences were: target A wild type, 5′-ACT AGT GCC CTC CGA CCA CTT CCA GGG GAA AGC TT; target A mutant, 5′-ACT AGT GCC CTC CGA CCA CTT CGA CGC GAA AGC TT; target B wild type, 5′-ACT AGT AGA TGA AAT AAA GAC CCA GGG GGA AGC TT; and target B mutant, 5′-ACT AGT AGA TGA AAT AAA GAG CGA CGC GGA AGC TT. Mutated bases are underlined. All plasmid DNA sequences were verified by DNA sequencing.
Transfections and Luciferase Assays
Cells were seeded 24 h prior to transfection and transfected using Lipofectamine 2000 (Invitrogen) with miRNA precursor molecules at final concentrations ranging from 1 to 30 nm. Cells were harvested at 12–24 h (for RNA extraction) or 3 days (for protein extraction). For reporter gene assays, cells were seeded in 24-well plates and co-transfected using Lipofectamine 2000 (Invitrogen) with 100 ng of firefly luciferase reporter DNA and either 20 ng of pRL-CMV or 100 ng of pRL-thymidine kinase Renilla luciferase reporter DNA as a transfection control. Cell lysates were assayed for firefly and Renilla luciferase activities 24 h after transfection using the Dual Luciferase Reporter Assay System (Promega) and a Fluostar OPTIMA luminometer (BMG Labtech), and firefly luciferase activities normalized to Renilla luciferase activities.
RNA Extraction and RT-PCR
Total RNA was extracted from cell lines with TRIzol reagent (Invitrogen) and treated with DNase I (Promega) to eliminate contaminating genomic DNA. For qRT-PCR analysis of ERBB-2, GAPDH, and pri-miR-331-3p expression, 125 ng of total RNA was reverse transcribed to cDNA with random hexamers and Thermoscript (Invitrogen). Real-time PCR for ERBB-2, GAPDH, and pri-miR-331-3p was performed using a Corbett 3000 RotorGene instrument (Corbett Research) with Platinum® SYBR® Green (Invitrogen) and ERBB-2 and GAPDH primers from Primer Bank (29) and pri-miR-331-3p primers (30): ERBB-2-F, 5′-TGA CAC CTA GCG GAG CGA T-3′; ERBB-2-R, 5′-GGG GGA TGT GTT TTC CCT CAA-3′; GAPDH-F, 5′-ATG GGG AAG GTG AAG GTC G-3′; GAPDH-R, 5′- GGG GTC ATT GAT GGC AAC AAT A-3′; miR-331-3p-F, 5′-GAG CTG AAA GCA CTC CCA A-3′; miR-331-3p-R, 5′-CAC ACT CTT GAT GTT CCA GGA-3′. Expression of ERBB-2 or pri-miR-331 RNA relative to GAPDH mRNA was determined using the 2−ΔΔCT method (31). For analysis of miR-331-3p expression by qRT-PCR, reverse transcription and PCR were carried out using TaqMan miRNA assay kits (Applied Biosystems) for hsa-miR-331 (part number 4373046), U44 small nuclear RNA (part number 4373384), and U6 small nuclear RNA (part number 4373381) with a Corbett 3000 RotorGene thermocycler (Corbett Research) according to the manufacturer's instructions.
Western Blotting
Cytoplasmic protein extracts were prepared as described (32), resolved on NuPAGE 4–12% BisTris gels or NuPAGE 10% BisTris gels (Invitrogen), and transferred to polyvinylidene difluoride membranes (Roche). Membranes were blocked in 5% skim milk/Tris-Buffered-Saline Tween and probed with anti-β-actin mouse monoclonal antibody (1:10000, Abcam ab6276-100), anti-ERBB-2 (CB-11) mouse monoclonal antibody (1:1000, Abcam ab8054-1), anti-phospho-ERBB-2 rabbit monoclonal antibody (1:1000, Abcam ab47755-100), anti-AKT rabbit monoclonal antibody (1:1000, Cell Signaling Technology number 9272), anti-phospho-AKT (Ser-473) rabbit monoclonal antibody (1:500, Cell Signaling Technology number 4060), anti-AR (H-280) rabbit monoclonal antibody (1:1000, Santa Cruz Biotechnology sc-13062), or anti-PSA rabbit polyclonal antibody (1:1000, DakoCytomation A0562). Secondary horseradish peroxidase-linked anti-mouse IgG (NA931V) and anti-rabbit IgG (NA934V) antibodies were used at 1:10000 (GE Healthcare), prior to detection with ECL Plus detection reagent and ECL-Hyperfilm (GE Healthcare).
Statistical Analysis
Statistical analysis of qRT-PCR data were performed using GenEx software (MultiD). All analyses were performed at a minimum confidence interval of 95% (CI = 0.95) and normality of data were confirmed by the Kolmogorov-Smirnov test (KS test).
RESULTS
The ERBB-2 3′-UTR Contains Two Putative Target Sites for miR-331-3p
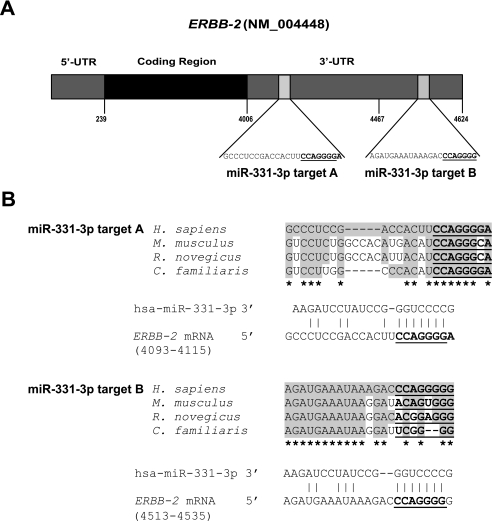
TargetScan (33) analysis (release 5: December 2008) predicted that the ERBB-2 3′-UTR contains two putative miR-331-3p binding sites at nucleotide 4108–4115 (labeled A) and a second site at nucleotide 4528–4534 (labeled B, Fig. 1A). The A target site has a context score of 97% on TargetScan, the B site has a lower score of 88%, and is less well conserved within the seed region (34). However, both miR-331-3p sites had some degree of sequence conservation between human, mouse, rat, and dog (Fig. 1B). Although there were three other miRNAs with two predicted putative binding sites in the ERBB-2 mRNA 3′-UTR (miR-1197, miR-1207-5p, and miR-1252) the TargetScan context score for each site was significantly lower than for both of the miR-331-3p sites (70 and 27, 42 and 54, and 35 and 41%, respectively).
FIGURE 1.
Identification of two specific miR-331-3p target sites within the ERBB-2 mRNA 3′-UTR. A, schematic representation of the ERBB-2 mRNA with two 3′-UTR miR-331-3p binding sites (A and B) predicted by TargetScan. The miR-331-3p seed sequence is underlined. B, sequence alignment of the predicted ERBB-2 3′-UTR miR-331-3p target sites showing conservation between human, mouse, rat, and dog. The miR-331-3p seed sequence (CCAGGGG) is shown in bold and underlined, and conserved nucleotides are shaded. Stars indicate nucleotides conserved across all four species.
miR-331-3p Down-regulates ERBB-2 Gene Expression in Human Prostate Cancer Cells
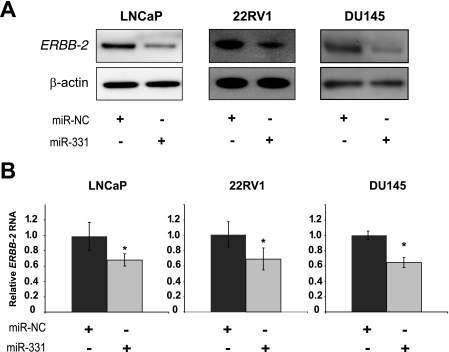
Given our interest in ERBB-2 expression and signaling in PCa, we investigated the potential for miR-331-3p to regulate ERBB-2 expression in three different PCa cell lines (LNCaP, AR+; 22RV1, AR+; and DU145, AR). We found low miR-331-3p expression across the cell lines, with the highest levels in DU145 cells and lower levels in the two AR+ cell lines (supplemental Fig. S1A). ERBB-2 mRNA expression was inversely correlated with miR-331-3p expression, as DU145 cells express the least, whereas LNCaP and 22RV1 cells contain higher levels of ERBB-2 mRNA (supplemental Fig. S1B). To evaluate the effects of miR-331-3p on each of these cell lines, we transfected either a negative control or miR-331-3p precursor into the cells and determined the level of ERBB-2 gene expression at 72 h post-transfection. Compared with a negative control miRNA precursor, miR-331-3p significantly down-regulated expression of both ERBB-2 protein and RNA in each of the PCa cell lines, independent of AR status (Fig. 2, A and B).
FIGURE 2.
miR-331-3p regulates ERBB-2 expression in prostate cancer cell lines. A, immunoblotting detection of ERBB-2 and β-actin expression using protein extracts harvested from LNCaP, 22RV1, and DU145 cells 3 days after transfection with miR-331-3p or the miR-NC precursor. B, qRT-PCR analysis of ERBB-2 mRNA expression in LNCaP, 22RV1, and DU145 cells 24 h after transfection with miR-331-3p or miR-NC. ERBB-2 RNA expression was normalized to GAPDH RNA expression, and is shown as a ratio of miR-331-3p-transfected cells to miR-NC-transfected cells using the 2−ΔΔCT method and GenEx statistical software. Data are representative of three independent experiments. Asterisk indicates a significant difference from miR-NC-transfected control cells (p < 0.03). Error bars represent confidence intervals (CI = 0.95).
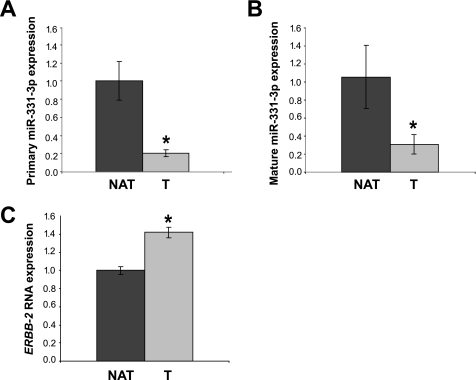
We next examined the expression of miR-331-3p in PCa versus normal adjacent tissue. The expression of miR-331-3p was substantially lower in the tumor tissue compared with normal prostate, for both miR-331-3p, as well as the pri-miR-331-3p transcript (Fig. 3, A and B). We also noted elevated ERBB-2 expression in the same tumor sample relative to normal adjacent tissue (Fig. 3C), suggesting a reciprocal relationship between the levels of miR-331-3p and ERBB-2 in human PCa.
FIGURE 3.
miR-331-3p expression is reduced in prostate tumor relative to normal adjacent tissue and is inversely correlated with ERBB-2 mRNA expression. A, qRT-PCR analysis of the pri-miR-331-3p expression in normal adjacent prostate tissue (NAT) RNA versus prostate tumor (T) RNA. Total RNA was reverse transcribed and miR-331-3p expression determined by qRT-PCR. Data were normalized to GAPDH expression and relative tumor miR-331-3p expression was calculated. Asterisk indicates a significant difference between pri-miR-331-3p expression in NAT versus tumor (p < 0.0001). B, qRT-PCR analysis for mature miR-331-3p in NAT versus tumor. Total RNA was reverse transcribed and miR-331-3p expression determined by the TaqMan miRNA qRT-PCR assay. Data were normalized to U44 and U6 small nuclear RNA expression and relative miR-331-3p expression was calculated. Asterisk indicates a significant difference between mature miR-331-3p expression in tumor versus NAT (p < 0.00001). C, qRT-PCR analysis of ERBB-2 mRNA expression in NAT versus tumor RNA. Total RNA was reverse transcribed and ERBB-2 and GAPDH expression determined by qRT-PCR. Data were normalized to GAPDH RNA expression and tumor ERBB-2 was expressed relative to NAT ERBB-2. Asterisk indicates a significant difference between ERBB-2 expression in tumor versus NAT (p < 0.0001). Error bars are as described in the legend to Fig. 2.
The 3′-UTR of ERBB-2 Is a Direct Target of miR-331-3p in Prostate Cancer Cells
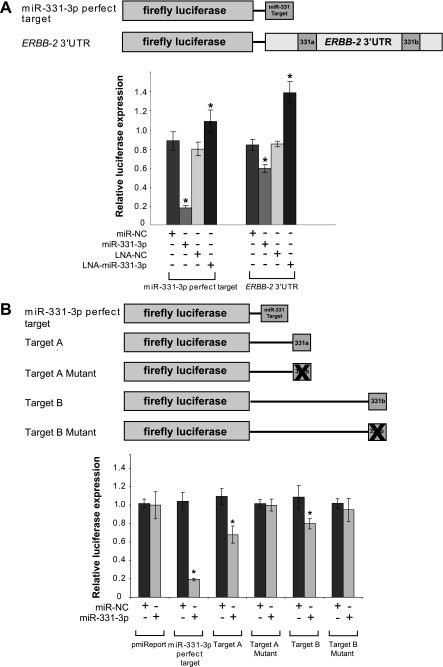
TargetScan analysis suggested that the 3′-UTR of ERBB-2 would be a direct target for miR-331-3p, via one or both predicted binding sites. To test this hypothesis, we generated miR-331-3p perfect target and ERBB-2 3′-UTR reporter constructs (Fig. 4A). PCa cells (22RV1) were transfected with either a negative control miRNA (miR-NC) or miR-331-3p precursor to up-regulate miR-331-3p expression, or a negative control or miR-331-3p antagonist (locked nucleic acid-modified oligonucleotide, LNA-anti-miR) to block miR-331-3p activity. In 22RV1 cells co-transfected with miR-331-3p precursor and a perfect miR-331-3p target site reporter, there was a significant reduction in reporter activity, compared with cells with the miR-NC precursor (Fig. 4A). Similarly, with the full-length ERBB-2 3′-UTR, the miR-331-3p precursor induced a significant decrease in reporter activity. In contrast, cells transfected with LNA-miR-331-3p showed an increase in reporter activity, which was not evident in LNA NC-transfected cells (Fig. 4A).
FIGURE 4.
The 3′-UTR of ERBB-2 mRNA is a direct target of miR-331-3p via two miR-331-3p target sites. A, schematic representation of firefly luciferase reporter constructs for full-length, wild type ERBB-2 3′-UTR and perfect miR-331-3p target. 22RV1 cells were co-transfected with pmiR-REPORT constructs and miR-NC (1 nm), miR-331-3p (1 nm), LNA-NC (10 nm), or LNA-miR-331-3p (10 nm). B, schematic representation of firefly luciferase reporter constructs of wild type and mutant ERBB-2 3′-UTR miR-331-3p-A and -B target sites and perfect -3p target. 22RV1 cells were co-transfected with pmiR-REPORT and CMV-Renilla constructs, and miR-NC or miR-331-3p (1 nm), and assayed for firefly and Renilla luciferase activities after 24 h. Relative luciferase expression values are expressed as a ratio of miR-NC to LNA. Asterisk indicates significant difference between miR-NC transfected control cells (p < 0.05). All data are representative of at least three independent experiments. Error bars are as described in the legend to Fig. 2.
We next determined the contribution of each of the two miR-331-3p sites to the regulation of reporter activity. Using the miR-331-3p perfect target reporter, and wild type ERBB-2 3′-UTR site A and B reporter constructs, we found that there was a similar contribution to reporter activity from both sites with miR-331-3p transfection, when compared with transfection with an unrelated miRNA (supplemental Fig. S2). Furthermore, smaller 3′-UTR ERBB-2 constructs were tested (see Fig. 4B). In 22RV1 cells transfected with mutant A and B site reporters, the effect of miR-331-3p was eliminated, consistent with miR-331-3p binding directly to each of the two target sites (Fig. 4B).
Taken together, these data confirm that each of the two miR-331-3p target sites in the ERBB-2 3′-UTR is a direct and specific target for miR-331-3p, and that both sites contribute to the regulation of ERBB-2 expression by miR-331-3p. These effects are observed in a range of human PCa cell lines, independent of AR status.
MiR-331-3p Regulates ERBB-2 and AR Signaling in Prostate Cancer Cells
To evaluate the effects of miR-331-3p on PCa cell signaling, we initially treated 22RV1, DU145, and LNCaP cells with heregulin to confirm activation of the ERBB-2 pathway. In 22RV1, DU145, and LNCaP cells treated with heregulin for 20 min, transfection with the miR-331-3p precursor significantly reduced the level of phosphorylated ERBB-2 (Fig. 5A and supplemental Fig. S3, A and B). Furthermore, we examined the effects of miR-331-3p on phosphorylated AKT levels in prostate cancer cells to determine whether miR-331-3p could regulate the PI3K/AKT signaling pathway downstream of ERBB-2. Cells transfected with miR-331-3p had substantially lower phosphorylated AKT levels, consistent with miR-331-3p regulating the PI3K/AKT kinase pathway.
FIGURE 5.
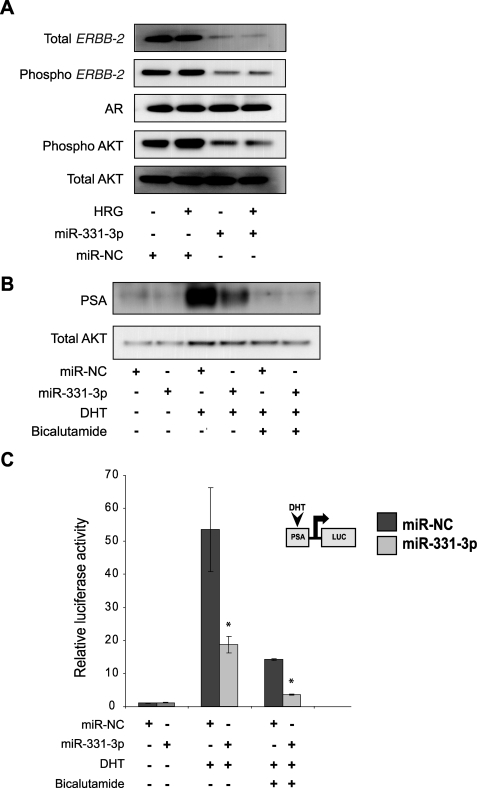
miR-331-3p decreases ERBB-2 protein expression and signaling, and blocks PSA expression and promoter activity in LNCaP cells. A, LNCaP cells were transfected with miR-NC or miR-331-3p (30 nm) for 48 h and serum starved for 24 h thereafter, followed by stimulation ± heregulin (HRG; 50 ng/ml) for 20 min. Cell lysates were analyzed for total ERBB-2, phospho-ERBB-2, AR, total AKT, and phospho-AKT expression by immunoblotting. B, LNCaP cells were transfected with miR-331-3p for 48 h and treated ± DHT (10 nm) and ± bicalutamide (10 μm). Total PSA expression was determined by immunoblotting. C, LNCaP cells were co-transfected with a PSA-luciferase vector (PSA-LUC) and thymidine kinase-Renilla vector and with miR-NC or miR-331-3p (1 nm). Relative luciferase expression (firefly normalized to Renilla) values are expressed as a ratio of miR-NC-transfected cells (±S.D.). Asterisk indicates significant difference between miR-NC transfected control cells (p < 0.05). Error bars represent confidence intervals (CI = 0.95).
It has been proposed that elevated levels of ERBB-2 expression and PI3K/AKT signaling promote the progression of prostate cancer by activating the AR signaling pathway. Cross-talk between the ERBB-2 and AR pathways can activate transcription of AR pathway genes, even in the absence of androgen stimulation (4, 6). To investigate whether miR-331-3p modifies AR signaling via its blockade of ERBB-2 signaling, we transfected LNCaP cells with miR-331-3p or the miR-NC precursor and, after 24 h, stimulated the cells with DHT, with or without bicalutamide, an AR antagonist. As expected, DHT induced PSA protein expression and this effect was blocked by either bicalutamide or miR-331-3p treatment (Fig. 5B), suggesting that miR-331-3p represses AR signaling indirectly by regulating ERBB-2 expression and signaling, because miR-331-3p did not regulate AR expression directly (Fig. 5A), and the AR mRNA 3′-UTR does not contain the predicted miR-331-3p target sites (data not shown).
To define the mechanism by which miR-331-3p blocks AR signaling, we co-transfected LNCaP cells with a DHT-responsive PSA-LUC reporter construct and either miR-331-3p or miR-NC (Fig. 5C). Reporter activity was increased following DHT treatment, and this effect was significantly reduced by miR-331-3p transfection. Furthermore, the combination of miR-331-3p and bicalutamide was more effective at reducing DHT-induced PSA-LUC reporter activity than either treatment alone.
Taken together, these data suggest that by regulating ERBB-2 expression and PI3K/AKT signaling, miR-331-3p can regulate AR signaling in PCa. miR-331-3p blocked DHT-induced PSA expression and PSA-LUC reporter gene activity in LNCaP cells but it did not alter AR expression, suggesting that miR-331-3p indirectly reduces transcription and expression of AR pathway target genes such as PSA via cross-talk between ERBB-2 and AR signaling pathways.
DISCUSSION
We have demonstrated that the ERBB-2 mRNA 3′-UTR contains two specific, direct miR-331-3p target sites, and that miR-331-3p down-regulates ERBB-2 mRNA and protein expression and signaling in multiple PCa cell lines. The observed reduction in ERBB-2 mRNA levels by miR-331-3p is consistent with recent reports indicating that many miRNAs regulate target gene expression by promoting mRNA decay (35, 36). miR-331-3p expression is decreased in ERBB-2 overexpressing PCa tissue relative to normal adjacent tissue, suggesting a role for miR-331-3p in the development and progression of this disease. Furthermore, we have shown that miR-331-3p blocked AR signaling in PCa cells, without reducing AR expression, by decreasing androgen-induced PSA promoter activity and PSA protein expression. These data suggest that loss of miR-331-3p expression could promote the increased ERBB-2 expression and signaling seen in many prostate cancers, and that this may promote the AR signaling pathway.
Although ERBB-2 gene amplification is common in ERBB-2 overexpressing breast tumors, it is a very rare event in PCa (37), suggesting that post-transcriptional mechanisms, such as regulation by miRNAs, may determine its expression in this disease. Recent evidence suggests that expression of the ERBB receptor family in cancer cells can be mediated in part by miRNAs (28, 38, 39). There is poor conservation of the two ERBB-2 3′-UTR miR-331-3p target sites (Fig. 1B, A and B) between human and rodent. However, TargetScan analysis (33) indicates that these sites are among the highest ranking predicted miR-331-3p target sites, with context score percentiles of 97 and 88%, respectively, when taking into account criteria that are associated with miRNA target site functionality, such as 3′ compensatory base pairing, local AU sequence content, and position within the 3′-UTR (34). Our reporter gene assays indicate that target sites A and B contribute to the direct repression of ERBB-2 expression by miR-331-3p despite their weak evolutionary conservation. This is consistent with our previous findings with miR-7 and the epidermal growth factor receptor, where miR-7 targets two poorly conserved sites within the epidermal growth factor receptor 3′-UTR to regulate epidermal growth factor receptor expression and signaling (39), emphasizing that strong miRNA target site conservation between species is not essential for a site to be functional. Importantly, mutation of each ERBB-2 3′-UTR miR-331-3p target site seed binding region impaired repression of reporter gene activity by miR-331-3p, whereas in contrast, transfection with unrelated miRNA did not repress reporter activity. This confirmed the direct and specific interaction of miR-331-3p with each ERBB-2 mRNA 3′-UTR site.
Anti-androgen therapy (bicalutamide (CasodexTM)) is widely used for locally advanced PCa; however, after an initial response to treatment some tumors recur in an androgen-independent form. The molecular mechanisms underlying androgen independence are not fully understood, however, elevated ERBB-2 expression and signaling is thought to promote the progression of PCa from an androgen-dependent to an androgen-independent state by producing constitutive activation of AR signaling despite androgen withdrawal or blockade (4, 6). This may be achieved by enhanced recruitment of the AR to androgen-responsive promoters (40). Thus, ERBB-2 has emerged as a therapeutic target in PCa, with preclinical and clinical studies investigating the efficacy of anti-ERBB-2 monoclonal antibodies (e.g. trastuzumab or pertuzumab) for the treatment of the disease (41–44). These studies have yielded disappointing results with little or no anti-tumor activity observed. However, these trials have involved patients with advanced PCa and patients have not been stratified or recruited according to tumor ERBB-2 expression, suggesting that better selection may be required in future studies (37). It is possible that blocking ERBB-2 signaling will hinder the progression of PCa to androgen independence, or that ERBB-2 blockade will restore androgen dependence, suggesting that co-administration of an ERBB-2 inhibitor with an anti-androgen (e.g. bicalutamide) might have clinical benefits (45). Our transfection studies are consistent with this hypothesis, with the combination of miR-331-3p and bicalutamide giving stronger repression of androgen-stimulated PSA promoter activity than either miR-331-3p or bicalutamide alone (Figs. 5C and 6). It has also been suggested that resistance to second line therapies, including ERBB-2 inhibitors, may result from loss of the tumor suppressor PTEN and subsequent activation of the PI3K/AKT pathway, a common event in advanced PCa that is associated with androgen independence (46–48). Indeed, PTEN loss is associated with trastuzumab resistance in breast cancer (49). Furthermore, bicalutamide monotherapy is associated with increased expression of ERBB-2 and phosphorylated AKT and reduced expression of PTEN in PCa (5). Interestingly, our work suggests that miR-331-3p has the capacity to block PI3K/AKT signaling in PCa cells independent of their PTEN status (LNCaP, PTEN-negative; 22Rv1 and DU145, PTEN-positive) (50), possibly by targeting other molecules involved in this pathway downstream of PTEN, and thus miR-331-3p could be used to overcome the inherent resistance of PTEN-negative prostate cancers to anti-ERBB-2 and other second line therapies (e.g. chemotherapy).
FIGURE 6.
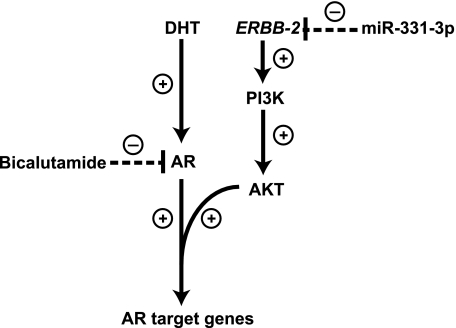
miR-331-3p blocks AR signaling via inhibition of ERBB-2 expression and AKT activity in prostate cancer cells. AR antagonists such as bicalutamide bind to the AR and prevent its activation and expression of AR target genes, such as PSA. Nevertheless, AR signaling may persist in prostate cancer cells despite AR blockade, in part via increased expression of the ERBB-2 receptor tyrosine kinase and subsequent activation of the PI3K/AKT pathway, which causes AR phosphorylation and promotes expression of AR target genes. miR-331-3p directly targets the ERBB-2 mRNA 3′-UTR to regulate ERBB-2 protein expression, thereby reducing PI3K/AKT signaling and AR signaling. The combination of an AR antagonist (bicalutamide) and miR-331-3p effectively blocks AR signaling (PSA expression and PSA promoter activity) in LNCaP prostate cancer cells. (+) indicates activation step of pathway and (−) indicates inhibition of pathway component.
Our study is the first to implicate miR-331-3p in PCa. Deregulated expression of miRNAs in cancer is often associated with gain or loss of chromosomal regions, with many miRNA genes located in fragile sites that are frequently altered in cancer (51–53). Alternatively, aberrant miRNA expression can occur through epigenetic mechanisms or by defects in miRNA biogenesis (14–16). We observed decreased expression of both primary and mature miR-331-3p in PCa tissue relative to normal adjacent prostate tissue (Fig. 3, A and B), suggesting that reduced miR-331-3p expression in PCa cells does not result from abnormal processing of miR-331-3p. The human miR-331 gene is located at 12q22, a chromosomal region that is not commonly altered in PCa. One possible explanation for decreased miR-331-3p expression in PCa is that there is reduced miR-331-3p gene transcription, resulting in less primary, and therefore mature, miR-331-3p being produced. Other studies have reported altered transcription of miRNA genes in cancer (17, 54). In this regard, it will be interesting to characterize the miR-331-3p promoter and its activity in PCa cells.
In summary, we have identified ERBB-2 as a direct and specific target of miR-331-3p in PCa cells. miR-331-3p expression is reduced in PCa and promotes elevated ERBB-2 expression and signaling, which increases AR signaling. Transfection of PCa cell lines with miR-331-3p reduced ERBB-2 expression, PI3K/AKT signaling, and blocked AR signaling. This suggests that miR-331-3p has the capacity to regulate activity of critical signaling pathways in PCa cells. Ongoing studies will investigate the potential for miR-331-3p to mediate cross-talk between the AR and ERBB-2 signaling pathways and modulate the sensitivity of PCa cells to anti-androgen and anti-ERBB-2 therapies.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Thomson, Dianne Beveridge, and Kavitha Iyer for helpful discussions regarding this manuscript.
This work was supported by the National Health and Medical Research Council of Australia and the Cancer Council of Western Australia.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PCa
- prostate cancer
- AR
- androgen receptor
- miRNA
- microRNA
- pri
- primary
- qRT-PCR
- quantitative reverse transcriptase-PCR
- UTR
- untranslated region
- NAT
- normal adjacent tissue
- DHT
- dihydrotestosterone
- LNA
- locked nucleic acid
- miR-NC
- negative control miRNA
- CMV
- cytomegalovirus
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- PSA
- prostate-specific antigen
- PI3K
- phosphatidylinositol 3-kinase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PTEN
- phosphatase and tensin homolog.
REFERENCES
- 1.Jemal A., Siegel R., Ward E., Murray T., Xu J., Thun M. J. (2007) CA Cancer J. Clin. 57, 43–66 [DOI] [PubMed] [Google Scholar]
- 2.Vernimmen D., Gueders M., Pisvin S., Delvenne P., Winkler R. (2003) Br. J. Cancer 89, 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isharwal S., Miller M. C., Epstein J. I., Mangold L. A., Humphreys E., Partin A. W., Veltri R. W. (2008) Int. J. Cancer 123, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craft N., Shostak Y., Carey M., Sawyers C. L. (1999) Nat. Med. 5, 280–285 [DOI] [PubMed] [Google Scholar]
- 5.Festuccia C., Gravina G. L., Muzi P., Pomante R., Ventura L., Vessella R. L., Vicentini C., Bologna M. (2007) Endocr. Relat. Cancer 14, 601–611 [DOI] [PubMed] [Google Scholar]
- 6.Mellinghoff I. K., Vivanco I., Kwon A., Tran C., Wongvipat J., Sawyers C. L. (2004) Cancer Cell 6, 517–527 [DOI] [PubMed] [Google Scholar]
- 7.Shi X. B., Ma A. H., Tepper C. G., Xia L., Gregg J. P., Gandour-Edwards R., Mack P. C., Kung H. J., deVere White R. W. (2004) Prostate 60, 257–271 [DOI] [PubMed] [Google Scholar]
- 8.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 9.Humphreys D. T., Westman B. J., Martin D. I., Preiss T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16961–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng A. M., Byrom M. W., Shelton J., Ford L. P. (2005) Nucleic Acids Res. 33, 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B., Pan X., Cobb G. P., Anderson T. A. (2007) Dev. Biol. 302, 1–12 [DOI] [PubMed] [Google Scholar]
- 13.Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bueno M. J., Pérez de Castro I., Gómez de Cedrón M., Santos J., Calin G. A., Cigudosa J. C., Croce C. M., Fernández-Piqueras J., Malumbres M. (2008) Cancer Cell 13, 496–506 [DOI] [PubMed] [Google Scholar]
- 15.Schmittgen T. D. (2008) J. Cell Mol. Med. 12, 1811–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viswanathan S. R., Daley G. Q., Gregory R. I. (2008) Science 320, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang T. C., Yu D., Lee Y. S., Wentzel E. A., Arking D. E., West K. M., Dang C. V., Thomas-Tikhonenko A., Mendell J. T. (2008) Nat. Genet. 40, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007) Nature 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esquela-Kerscher A., Slack F. J. (2006) Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 20.Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. (2005) Cell 120, 635–647 [DOI] [PubMed] [Google Scholar]
- 21.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Mitsudomi T., Takahashi T. (2004) Cancer Res. 64, 3753–3756 [DOI] [PubMed] [Google Scholar]
- 22.Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Si M. L., Zhu S., Wu H., Lu Z., Wu F., Mo Y. Y. (2007) Oncogene 26, 2799–2803 [DOI] [PubMed] [Google Scholar]
- 24.Ambs S., Prueitt R. L., Yi M., Hudson R. S., Howe T. M., Petrocca F., Wallace T. A., Liu C. G., Volinia S., Calin G. A., Yfantis H. G., Stephens R. M., Croce C. M. (2008) Cancer Res. 68, 6162–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozen M., Creighton C. J., Ozdemir M., Ittmann M. (2008) Oncogene 27, 1788–1793 [DOI] [PubMed] [Google Scholar]
- 26.Shi X. B., Tepper C. G., White R. W. (2008) J. Cell. Mol. Med. 12, 1456–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin S. L., Chiang A., Chang D., Ying S. Y. (2008) RNA 14, 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott G. K., Goga A., Bhaumik D., Berger C. E., Sullivan C. S., Benz C. C. (2007) J. Biol. Chem. 282, 1479–1486 [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Seed B. (2003) Nucleic Acids Res. 31, e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang J., Lee E. J., Gusev Y., Schmittgen T. D. (2005) Nucleic Acids Res. 33, 5394–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 32.Giles K. M., Daly J. M., Beveridge D. J., Thomson A. M., Voon D. C., Furneaux H. M., Jazayeri J. A., Leedman P. J. (2003) J. Biol. Chem. 278, 2937–2946 [DOI] [PubMed] [Google Scholar]
- 33.Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 34.Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filipowicz W., Jaskiewicz L., Kolb F. A., Pillai R. S. (2005) Curr. Opin. Struct. Biol. 15, 331–341 [DOI] [PubMed] [Google Scholar]
- 36.Liu T., Papagiannakopoulos T., Puskar K., Qi S., Santiago F., Clay W., Lao K., Lee Y., Nelson S. F., Kornblum H. I., Doyle F., Petzold L., Shraiman B., Kosik K. S. (2007) PLoS ONE 2, e804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solit D. B., Rosen N. (2007) J. Clin. Oncol. 25, 241–243 [DOI] [PubMed] [Google Scholar]
- 38.Kefas B., Godlewski J., Comeau L., Li Y., Abounader R., Hawkinson M., Lee J., Fine H., Chiocca E. A., Lawler S., Purow B. (2008) Cancer Res. 68, 3566–3572 [DOI] [PubMed] [Google Scholar]
- 39.Webster R. J., Giles K. M., Price K. J., Zhang P. M., Mattick J. S., Leedman P. J. (2009) J. Biol. Chem. 284, 5731–5741 [DOI] [PubMed] [Google Scholar]
- 40.Liu Y., Majumder S., McCall W., Sartor C. I., Mohler J. L., Gregory C. W., Earp H. S., Whang Y. E. (2005) Cancer Res. 65, 3404–3409 [DOI] [PubMed] [Google Scholar]
- 41.Bradbury J. (2007) Lancet Oncol. 8, 287. [DOI] [PubMed] [Google Scholar]
- 42.Albanell J., Codony J., Rovira A., Mellado B., Gascón P. (2003) Adv. Exp. Med. Biol. 532, 253–268 [DOI] [PubMed] [Google Scholar]
- 43.Small E. J., Bok R., Reese D. M., Sudilovsky D., Frohlich M. (2001) Semin. Oncol. 28, Suppl. 15, 71–76 [DOI] [PubMed] [Google Scholar]
- 44.Bianco A. R. (2004) J. Chemother. 16, Suppl. 4, 52–54 [DOI] [PubMed] [Google Scholar]
- 45.Gravina G. L., Festuccia C., Millimaggi D., Tombolini V., Dolo V., Vicentini C., Bologna M. (2009) Urology 74, 452–457 [DOI] [PubMed] [Google Scholar]
- 46.Malik S. N., Brattain M., Ghosh P. M., Troyer D. A., Prihoda T., Bedolla R., Kreisberg J. I. (2002) Clin. Cancer Res. 8, 1168–1171 [PubMed] [Google Scholar]
- 47.Murillo H., Huang H., Schmidt L. J., Smith D. I., Tindall D. J. (2001) Endocrinology 142, 4795–4805 [DOI] [PubMed] [Google Scholar]
- 48.Whang Y. E., Wu X., Suzuki H., Reiter R. E., Tran C., Vessella R. L., Said J. W., Isaacs W. B., Sawyers C. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5246–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagata Y., Lan K. H., Zhou X., Tan M., Esteva F. J., Sahin A. A., Klos K. S., Li P., Monia B. P., Nguyen N. T., Hortobagyi G. N., Hung M. C., Yu D. (2004) Cancer Cell 6, 117–127 [DOI] [PubMed] [Google Scholar]
- 50.Festuccia C., Gravina G. L., D'Alessandro A. M., Millimaggi D., Di Rocco C., Dolo V., Ricevuto E., Vicentini C., Bologna M. (2008) Int. J. Oncol. 33, 381–388 [PubMed] [Google Scholar]
- 51.Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selcuklu S. D., Yakicier M. C., Erson A. E. (2009) Cancer Genet. Cytogenet. 189, 15–23 [DOI] [PubMed] [Google Scholar]
- 53.Wei J. S., Song Y. K., Durinck S., Chen Q. R., Cheuk A. T., Tsang P., Zhang Q., Thiele C. J., Slack A., Shohet J., Khan J. (2008) Oncogene 27, 5204–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddy S. D., Ohshiro K., Rayala S. K., Kumar R. (2008) Cancer Res. 68, 8195–8200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.