Abstract
There is little information on how neuropeptide Y (NPY) proteolysis by peptidases occurs in serum, in part because reliable techniques are lacking to distinguish different NPY immunoreactive forms and also because the factors affecting the expression of these enzymes have been poorly studied. In the present study, LC-MS/MS was used to identify and quantify NPY fragments resulting from peptidolytic cleavage of NPY1–36 upon incubation with human serum. Kinetic studies indicated that NPY1–36 is rapidly cleaved in serum into 3 main fragments with the following order of efficacy: NPY3–36 ≫ NPY3–35 > NPY2–36. Trace amounts of additional NPY forms were identified by accurate mass spectrometry. Specific inhibitors of dipeptidyl peptidase IV, kallikrein, and aminopeptidase P prevented the production of NPY3–36, NPY3–35, and NPY2–36, respectively. Plasma kallikrein at physiological concentrations converted NPY3–36 into NPY3–35. Receptor binding assays revealed that NPY3–35 is unable to bind to NPY Y1, Y2, and Y5 receptors; thus NPY3–35 may represent the major metabolic clearance product of the Y2/Y5 agonist, NPY3–36.
Neuropeptide Y (NPY)2 is a 36-amino acid peptide involved in the central and peripheral control of blood pressure (1–4) and in feeding behavior and obesity (5–9). NPY stimulates at least 6 types of receptors, called Y1, Y2, Y3, Y4, Y5, and y6 (10–12). The Y1 receptor has high affinity for full-length NPY, while Y2 and Y5 receptors bind and are stimulated by full-length and N-terminally truncated NPY. The physiological effects associated to the Y1 and Y2 receptors are the best known; exposure to a Y1 agonist causes an increase in blood pressure and potentiates postsynaptically the action of other vasoactive substances (1, 4, 13), whereas Y2 receptors are mainly located presynaptically, and upon stimulation mediate the inhibition of neurotransmitter release (14, 15). NPY is a prototype of peptide whose function can be altered by proteases. Among peptidases displaying a high affinity for NPY, the primary role appears to be played by dipeptidyl peptidase IV (DPPIV, EC 3.4.14.5), a serine-type protease, also known as CD26, that releases an N-terminal dipeptide, Xaa-Xab- -Xac, preferentially when Xab is a proline or an alanine residue (16). By cleaving the Tyr-Pro dipeptide off the NPY N-terminal extremity, DPPIV generates NPY3–36, a truncated form that loses its affinity for the Y1 receptor and becomes a Y2/Y5 receptor agonist (17, 18).
NPY can also be degraded by aminopeptidase P (AmP, EC 3.4.11.9), a metalloprotease that hydrolyzes the peptide bond between the first and the second amino acid residue at the N terminus of proteins, if the second amino acid is a proline (19). AmP removes the N-terminal tyrosine from NPY to generate NPY2–36, a selective Y2 agonist (18, 20). There is little information on how NPY cleavage by these enzymes occurs in serum, in part because reliable techniques are lacking to distinguish different NPY immunoreactive (NPYir) forms and also because the factors affecting the expression of these enzymes have been poorly studied. Recently, Frerker et al. (21) reported by MALDI-TOF mass spectrometry that NPY1–36 is exclusively degraded by DPPIV into NPY3–36 in EDTA-plasma but they did not provide kinetics of NPY cleavage efficiency of DPPIV. Beck-Sickinger and co-workers (22) studied with the same technique the metabolic stability of fluorescent N-terminally labeled NPY analogues incubated in human plasma and found that the 36th, 35th, and 33rd residues of NPY analogues may also be removed by unknown carboxypeptidases.
We have set up a method using liquid chromatography coupled with tandem mass spectrometry (LC-MSn) to selectively quantify NPY and its C-terminal fragments NPY2–36 and NPY3–36 digested by human serum. The assays used the internal standard methodology with stable isotopes NPY1–36 (IDA) (23, 24) or porcine NPY1–36 as internal standard.
The goal of this work was: 1) to determine to which extent NPY1–36 is degraded by proteases present in human serum and whether an inhibition of DPPIV and AmP by vildagliptin and apstatin (two specific protease inhibitors), respectively, may affect the metabolism of NPY in serum; 2) to assign kinetic values to the proteases involved in the cleavage process toward NPY; and 3) to characterize new NPY-truncated forms and to check for their possible binding capacities on NPY receptors.
EXPERIMENTAL PROCEDURES
Peptides, Chemicals, and Protease Inhibitors
NPY1–36, NPY2–36, and NPY3–36 were purchased at American Peptide Company (Sunnyvale, CA). NPY3–35 was purchased at Altergen (Schiltigheim, France); Vildagliptin (NVP-LAF237) was custom-synthesized by GLSynthesis Inc. (Worcester, MA). Apstatin was synthesized by organic synthesis following the procedure described by Prechel et al. (25). EDTA was purchased at Merck, and p-choloromercuriphenylsulfonic acid and trifluoroacetic acid came from Sigma. Glycyl-l-proline-4-methylcoumaryl-7-amide (Gly-ProAMC) and l-lysyl (Nϵ-2-aminobenzoyl)-l-prolyl-l-prolyl-4-nitroanilide (Lys(Abz)-Pro-Pro-pNA) were purchased at Bachem (Bubendorf, Switzerland). Leupeptin, chymostatin, 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), aprotinin, N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide (E64), and gabexate mesylate were from Sigma. Purified human plasma kallikrein was purchased at Athens Research and Technology Inc. (Athens, GA). Prekallikrein-deficient plasma was obtained from Sigma. 125I-NPY was purchased at PerkinElmer Life Sciences.
Isotopic Peptides
Peptides were synthesized with an Applied Biosystems 433 apparatus (Foster City, CA), using Fmoc-protected amino acids of the highest purity commercially available from Novabiochem (Darmstadt, Germany) or Advanced Chemtech (Louisville, KY). (15%-13C, 95%-15N)-labeled amino acid were provided by Senn Chemicals (Dielsdorf, Switzerland) and used to label NPY at positions 3, 5, 8, 13, 19, 22, 24, 30, 33, and 35. The peptides were then purified by reversed-phase HPLC with a Vydac C18 column (Vydac, Deerfield, IL), (10 × 250 mm; flow rate: 3 ml × min−1, linear gradient: 10% B to 50% B in 5 min, then 50% to 58% B in 29 min, A: 0.1% trifluoroacetic acid, B: 60% CH3CN, 0.06% trifluoroacetic acid).
Determination of DPPIV and AmP Activities in Serum
DPPIV (26) and AmP activities were determined in the serum from 6 healthy individuals (4 women and 2 males, aged 27–43 yrs) who gave informed consent. DPPIV activity was determined as previously described (26). For the AmP assay, the serum was diluted 1:3.3, 1:5, and 1:10 in a buffer containing 100 mm Tris-HCl at pH 8.0 and 0.5 mm MnCl2 to a final volume of 50 μl with 5 mm of the AmP substrate Lys(Abz)-Pro-Pro-pNA. The protease inhibitor concentration was 50 μm for both vildagliptin (DPPIV inhibitor) and apstatin (AmP inhibitor). Samples containing protease inhibitors were prewarmed at 37 °C, 15 and 30 min for the DPPIV and AmP assays, respectively, and shaken at 1,000 rpm in a thermomixer prior to adding the substrates. The incubation time was 3 h for the AmP assay. Reactions were stopped by the addition of 5 μl of acetic acid 100%, and 3.5 ml of bidistilled water were added to the incubation mixtures for the AmP assay. AmP activity was expressed as arbitrary fluorescent units (AFU) at excitation and emission wavelengths of λex 310 nm, λem 410 nm on a PerkinElmer LS-5 fluorometer.
Quantitative LC-MSn Analysis of NPY Fragments
The LC-MSn system consisted of a Rheos Allegro UHPLC pump (Flux Instruments, Basel, Switzerland), an HTC PAL autosampler (CTC Analytics AG, Zwingen, Switzerland) and a linear ion trap mass spectrometer (LTQ-MS) from ThermoFisher performing with an electrospray ion source in positive mode (ESI+). The analytical column was an Hypersil Gold, 50-mm length, 1-mm inner diameter, 1.9 μm particle size (ThermoFisher). The mobile phase was composed of phase A: 10 mm ammonium formate (Sigma) with 0.1% formic acid (Merck) in deionized water and phase B: 0.1% formic acid in acetonitrile (JT Baker). The mobile phase was delivered at a flow rate of 0.15 ml/min with the following stepwise gradient: 10% of B at 0 min, 90% of B at 2 min, 100% of B at 3.5 min for 4.5 min, and back to 10% of B at 8 min for 4 min. The total run time was 12 min. The injection volume was 10 μl.
For the LTQ-MS ion source, the sheath and auxiliary gas (nitrogen) flow-rate was set at 40 psi and 10 arbitrary units, respectively, the capillary voltage was +4 kV, the heated capillary temperature was set at 250 °C, and the tube lens voltages ranged from 210 V to 250 V. LTQ-MS was operating in product ion scan mode acquisition. Each NPY and IS-NPY parent ions were isolated in the LTQ-MS with an isolation width of 5 arbitrary units before their collision-induced dissociation and the generation of MS2 product ions. In the trap, He2 gas was set at pressure at 275 kPa, and a collision energy of 25% was chosen and corresponded to the maximum abundance for the total ion current of the NPY product spectra. The maximum injection time was set at 10 ms. Pure standards of NPY forms were oxidized and tuned on the LTQ-MS to determine their parent and product ions. The following ion transitions were chosen with the isolation of [M+3H]3+ parent ions:
NPY1–36: m/z at 1430.2 → extracted product ion chromatogram (EIC) at m/z 1200.5/1343.6/1397.06/1402.9/1408.0/ 1409.0/1410.0/1412.2/1544.4/1545.4.
NPY2–36: m/z at 1375.9 → EIC at m/z 1201.3/1342.0/1343.0/ 1347.8/1348.8/1353.8/1354.8/1463.5/1545.4.
NPY3–36: m/z at 1343.5 → EIC at m/z 1200.5/1315.5/1316.5/ 1321.8/1322.65/1330.5/1535.9/1543.9.
NPY3–35: m/z at 1289.2 → EIC at m/z 1147.0/1261.5/ 1262.0/1268.0/1269.0/1463.0.
IS-NPY1–36: m/z at 1437.6 → EIC at m/z 1405.0/1410.6/ 1416.9/1415.8.
IS-Porcine NPY1–36: m/z at 1418.9 → EIC at m/z 1331.9/ 1400.8/1527.0/1783.0.
Qualitative LC-MSn Analysis of NPY Fragments
The HPLC analyses were performed with a Rheos 2200 HPLC pump (Flux Instruments) and a HTC PAL autosampler (CTC Analytics AG). Separation was carried out using the same columns and chromatographic conditions as the one already described. Mass spectrometry was performed on an LTQ Orbitrap mass spectrometer (ThermoFisher) with an electrospray ionization interface in positive mode. Source settings were identical to the one described above. MS spectra (from m/z 350–2000) were acquired in the orbitrap with resolution r = 60,000 at m/z 400 after accumulation to a target value of 500,000 charges in the linear ion trap (automatic gain control, 1 μscan, maximum injection time of 500 ms). For accurate mass measurements, the lock mass option was enabled and the bis(2-ethylhexyl)phthalate, a plastic contaminant present in HPLC solvents (protonated [C24H38O4]H+, m/z = 391.2843), was used for internal mass recalibration in real time. MS/MS experiments were performed targeting selectively the m/z values of the compounds of interest with an isolation width of 5 Da, a normalized collision energy of 25 and a resolution of r = 30,000 at m/z 400 after accumulation to a target value of 500,000 charges in the linear ion trap (automatic gain control, 1 μscan, maximum injection time of 500 ms). MSn data were collected on the triple- and fourth-charged precursor ions of NPY X1-X2 (X1 = 1, 2, 3 and X2 = 32, 34, 35).
Time Course: in Vitro Cleavage of NPY1–36
NPY1–36 (50 μm) was oxidized in H202 (10%) over 1 h at room temperature and diluted to a concentration of 10 μm. Human serum from volunteer 01 (2.5 μl in 12.5 μl of digestion buffer: 100 mm Tris-HCl, pH 8.0, 0.5 mm MnCl2) was preincubated with or without protease inhibitors (vildagliptin and apstatin at a concentration of 50 μm) for 15 min prior to the addition of 10 μl of 10 μm mono-oxygenated NPY. The samples were then incubated at 37 °C under agitation for 5, 10, 20, 30, 60, 120, and 240 min. Digested samples were diluted 2× in 1% trifluoroacetic acid containing Internal Standard (isotopic NPY1–36) at a final concentration of 0.5 μm. Samples were analyzed by LC-MSn, and the NPY signal was quantified using Excalibur software (ThermoFisher).
Kinetic Parameters of DPPIV, AmP, and Plasma Kallikrein on NPY Substrate
Mono-oxygenated NPY1–36 and NPY3–36 at concentrations of 5, 10, 20, 30, 40, 50, 60, 70 μm were incubated at 37 °C as described above. The incubation times were 10, 30, and 45 min for the determination of Km and Vmax constants of DPPIV, AmP, and plasma kallikrein, respectively. Samples were diluted 2× in 1% trifluoroacetic acid containing 5 μm (final concentration) of porcine NPY1–36 as internal standard. The calibration curve showed that the signal obtained for the three peptides (NPY2–36, NPY3–36, and NPY3–35) was linear for concentrations ranging from 50 to 1000 nm. The cleavage products obtained after partial digestion were quantified, and their concentrations were within the levels of calibration curves. The determined NPY fragment/internal standard ratios were transformed into pmol/min according to the calibration curve made for each NPY-truncated form. Km and Vmax constants were determined from initial velocity measurements plotted versus various substrate concentrations using hyperbolic representations (GraphPad Prism Software, San Diego, CA). Km and Vmax values were expressed as μm and pmol × min−1 × μl serum−1, respectively.
Radioreceptor Assay of NPY Fragments
NPY binding assays to Y1 and Y2 and Y5 receptors are described elsewhere (27, 28). Displacement curves were obtained by incubation of various concentrations of competitive peptides (0.0312 nm to 10 μm) together with a nonsaturating dose of radiolabeled NPY. At least three independent experiments were performed for each receptor assay, and each experiment was done in duplicate. Results were expressed as percent of 125I-NPY1–36 bound to receptors upon incubation with competitors (NPY1–36 or NPY3–35 at various concentrations).
Identification of the Enzyme Responsible for NPY3–35 Formation
The strategy used to identify the enzyme that exhibited carboxypeptidase and esterase activity was based on blockade of NPY3–35 formation using enzyme inhibitors. Serum was preincubated with various protease inhibitors for 45 min at 37 °C before adding NPY3–36 (4 μm final concentration in digestion buffer) for 2 h. The reaction was stopped by adding trifluoroacetic acid (1%) and diluted 2× with 5 μm or 2 μm (final concentration) of internal standard (porcine NPY1–36). Samples were analyzed by LC-MSn, and the NPY signal was quantified using the Excalibur software.
RESULTS
NPY1–36 Is Cleaved into NPY2–36 and NPY3–36
Because NPY is likely to be found in serum as an oxidized peptide, because of a methionine on residue 17, we decided to oxidize with H202 all NPY forms before testing their concentrations with LC-MSn. Once we were able to clearly separate NPY forms from a commercial source based on ion transitions (see “Experimental Procedures”), we conducted an initial assay that was aimed to confirm that the sera used in this study contained both DPPIV and AmP enzyme activity and that inhibitors for both enzymes were efficient in a classic enzyme substrate assay. The sera sampled from 6 volunteers exhibited a mean DPPIV activity of 35.2 ± 7.1 nmol/min/ml. In the absence of appropriate calibrators, AmP activity in these sera was estimated in arbitrary fluorescence unit (AFU) at 3.2 ± 2.1 AFU/ml/min. We arbitrarily chose the serum from volunteer 01 to perform the kinetic study. The serum chosen exhibited a DPPIV activity of 43.8 nmol/min/ml and an AmP activity of 4.4 AFU/ml/min. In addition, vildagliptin and apstatin inhibitory efficiencies were tested using the enzyme-substrate assay described above; vildagliptin almost entirely abolished (>95%) the cleavage of Gly-ProAMC in the fluorometric assay, whereas apstatin inhibited ∼80% of Lys(Abz)-Pro-Pro-pNA cleavage.
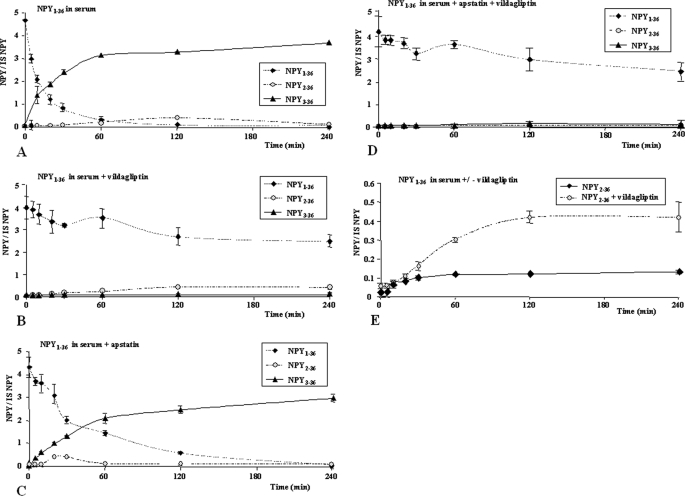
Considering that our material was relevant for the study, we incubated NPY1–36 in serum and observed that after 120 min of incubation, the peptide was almost entirely truncated at its N terminus, giving rise to NPY3–36 and NPY2–36 (Fig. 1, A and B). We then performed a time course assay at a fixed concentration of NPY1–36 (4 μm in serum) to assess the cleavage rate of NPY1–36 into NPY2–36 and NPY3–36. NPY3–36 concentration gradually increased, and reached a plateau between 1 and 2 h of incubation (Fig. 2A). After 4 h of incubation without inhibitors, NPY1–36 was predominantly cleaved into NPY3–36. To confirm that DPPIV and AmP were indeed responsible for, respectively, NPY3–36 and NPY2–36 production, 50 μm vildagliptin and/or apstatin were added in the incubation buffer. Vildagliptin was able to fully prevent NPY3–36 generation (inhibition rate of 96%, Fig. 2B). In the absence of inhibitors, around 50% of NPY1–36 was cleaved within the first 30 min mainly into NPY3–36, (Fig. 2A). Conversely, apstatin inhibition efficacy was far lower, which is not surprising, since NPY1–36 is only marginally degraded into NPY2–36 (Fig. 2C). Both inhibitors added in the same incubation sample only minimally reinforced the inhibition of NPY1–36 degradation compared with incubation with vildagliptin alone (Fig. 2D). A compensatory increase (around 4×) of NPY2–36 production was observed when NPY1–36 was incubated in serum complemented with vildagliptin compared with control incubation mixture (Fig. 2E). Even though the difference was significant, NPY2–36 did not reach the concentration of NPY3–36 observed in the absence of inhibitor.
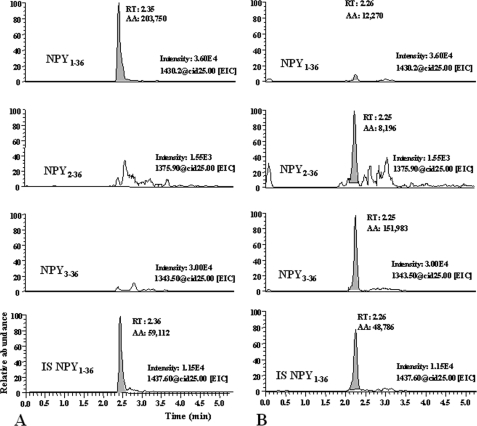
FIGURE 1.
Representative LC-MSn recording for NPY1–36, NPY2–36, and NPY3–36 in incubation mix. A, time 0, and B, time 120 min. Fixed scales intensity were chosen at both times to compare left and right chromatograms.
FIGURE 2.
Time course of NPY1–36 degradation in 10% human serum at 37 °C. The values on the y axis represent the ratio digestion products considered versus internal standard (isotopic NPY1–36). A, NPY1–36 degradation in serum without protease inhibitors. B, in the presence of 50 μm vildagliptin. C, in the presence of 50 μm apstatin. D, in the presence of both vildagliptin and apstatin at 50 μm each. E, NPY1–36 digested with or without vildagliptin.
Determination of the Kinetic Constants of DPPIV and AmP on NPY
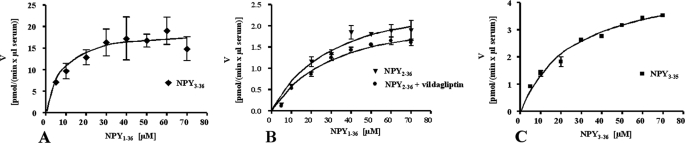
Several concentrations of NPY1–36 were digested as described under “Experimental Procedures,” and kinetic constants are reported in Table 1. The Km calculated for DPPIV for NPY3–36 production was 9.4 μm and the Vmax of 19.9 pmol × min−1 × μl serum−1 (Fig. 3A). We previously observed by ELISA that CD26 antigen concentration in human plasma reached ∼500 ng/ml and represents most of the DPPIV activity in blood (26); therefore, we were able to estimate a kcat (Vmax/mol of enzyme in 1 μl of serum) of 115.8 × s−1 and a resulting specificity constant kcat/Km of 12.3 liters × mol−1 × s−1. For the production of NPY2–36 by AmP, the Km was 35.4 μm and Vmax 3.0 pmol × min−1 × μl serum−1 in the absence of vildagliptin (Fig. 3B). It is noteworthy that Km and Vmax values for AmP toward NPY1–36 in the presence of vildagliptin showed comparable values (42.3 μm and 2.8 pmol × min−1 × μl serum−1, respectively), confirming that in both incubation mixtures, the production of NPY2–36 is linear and independent from the formation of NPY3–36.
TABLE 1.
Kinetic values measured for DPPIV, AmP, and plasma kallikrein toward NPY1–36 and NPY3–36
| Kma | Vmaxb | kcatc | kcat/Kmd | Intrinsic clearancee | |
|---|---|---|---|---|---|
| Dipeptidyl peptidase IV | 9.4 | 19.9 | 115.8 | 12.3 | 2.11 |
| Aminopeptidase P | 35.4 | 3 | NDf | ND | 0.08 |
| Plasma kallikrein | 28.1 | 4.9 | ND | ND | 0.17 |
aKm is expressed in μm.
b Vmax is expressed in pmol × min−1 × μl serum−1.
c kcat represents Vmax/mol of enzymes in 1 μl of serum and is expressed in s−1.
d kcat/Km is expressed in liter × mol−1 × s−1.
e The IC (Vmax/Km) is expressed in μl cleared × min−1 × μl serum−1.
f ND, not determined.
FIGURE 3.
Representation of enzyme kinetics: velocity (v) against concentration (s). Kinetic values are reported in Table 1. A, NPY1–36 was digested at concentrations ranging from 5 to 70 μm (x axis), NPY3–36 production was quantified using LC-MS analysis (porcine NPY1–36 as internal standard). The enzyme velocity is expressed as pmol generated per min and per μl of serum used in the incubation mixture (y axis). B, a similar approach was used for the determination of kinetic constants of AmP toward NPY1–36 without vildagliptin in the incubation mixture. C, determination of the kinetic values of plasma kallikrein toward NPY3–36.
Identification of New Truncated NPY Forms
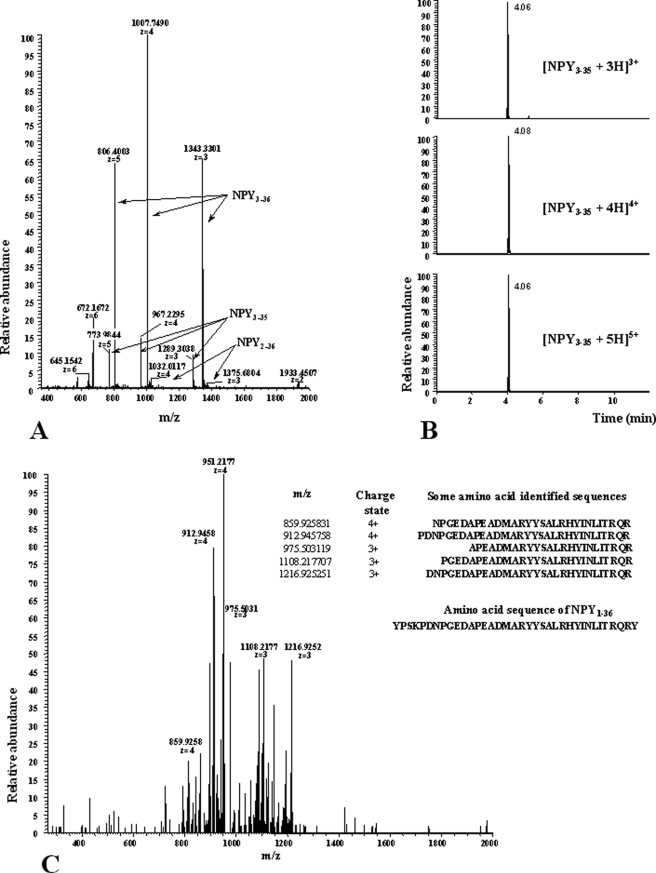
Because vildagliptin blunted almost completely NPY3–36 generation (Fig. 2B) without preventing (to a similar extent) NPY1–36 degradation, we hypothesized that alternative cleavages by unknown proteases might occur. To check this assumption, a full scan MS acquisition was performed on a digested sample of NPY1–36. Apart from the NPY forms already described above, ion series of m/z 1289.3, 967.2, and 773.9 were observed, corresponding to the truncated NPY3–35 form with z = 3, 4, and 5, respectively (Fig. 4, A and B). This result was confirmed by the accurate mass determination of these ion series and by the identification of their product ions (performed using a LTQ-Orbitrap MS) (Fig. 4C). With respect to the full-MS data measured with the LTQ-Orbitrap, the mass accuracy of all ions was better than 0.7 ppm using the internal lock-mass option. The signals and the mass accuracy in the MSn spectra of the fourth-charged ion (m/z 967.2) of NPY3–35 are in accordance with the theoretically calculated b- and y-ions of this peptide and confirm the presence of NPY3–35. Additional fragments such as NPY1–32, NPY1–34, NPY1–35, NPY2–32, NPY2–34, NPY2–35, NPY3–32, and NPY3–34 were also detected using the LTQ-Orbitrap mass spectrometer. The presence of these fragments as trace amount was confirmed by using the full-MS-data (mass-window 2 ppm) and the MSn data of the corresponding triple- and fourth-charged products (data not shown).
FIGURE 4.
Mass spectrometry characterization of NPY3–35. A, LTQ Orbitrap, full-MS spectra m/z 350–2000; signals of NPY3–35, NPY3–36, and NPY2–36 (charge states +3, +4, and +5 are in bold). B, LC-MS chromatograms for the most abundant isotopes of the triple- (top), fourth- (middle), and fifth-charged ions of NPY3–35 by using a mass window of 2 ppm ([C166H259O54N51S+×H]x+, × = 3, 4, 5). C, MSn spectra of NPY3–35 (charge state +4, m/z 967,2 CID-fragmentation) using a normalized collision energy of 25 and an isolation width of 5 Da; the amino acid sequences listed show the positive hits of the calculated Y-ion type of NPY3–35.
Binding of NPY3–35 to NPY Y1, Y2, and Y5 Receptors
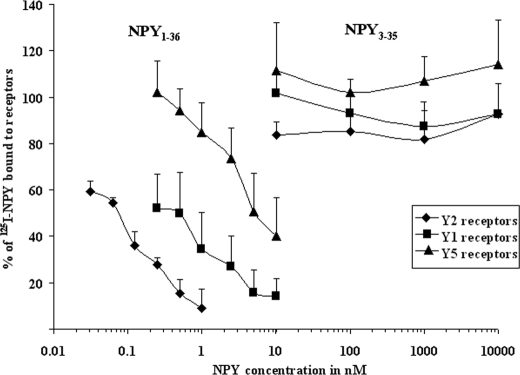
Considering that N-terminal NPY cleavages give rise to truncated peptides that retain bioactive activity and switch their affinity for different receptors, we investigated whether NPY3–35 was still able to bind to the known NPY Y1, Y2, and Y5 receptors despite the C-terminal removal of tyrosinamide. Receptor assays showed indeed that NPY3–35 was not able to bind significantly to any of the receptors studied at concentrations tested up to 10 μm. In contrast, NPY1–36 exhibited IC50 values at nanomolar affinities (27, 28) in LN319, SK-N-MC, and HEC-1B cell lines stably transfected or constitutively expressing, respectively, Y2, Y1, and Y5 receptors (Fig. 5).
FIGURE 5.
Concentration-response curves of the displacement of 125I-NPY by NPY1–36 and NPY3–35 for the Y1 (■), Y2(♦), and Y5(▴) receptors in SK-N-MC cells (Y1), LN319 cells (Y2), and HEC-1B Y5 cells. Three experiments were performed in duplicate. The percentage of 125I-NPY bound to the receptor, which is caused by the increasing concentrations of NPY1–36, is shown on the y axis. High affinity binding to all NPY receptors was found for NPY1–36. NPY3–35 did not bind to Y1, Y2, or Y5 receptors.
Identification of the Enzyme Responsible for NPY3–35 Formation
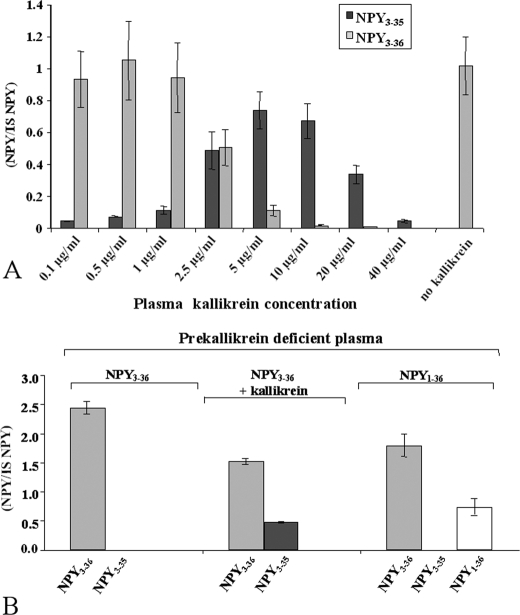
No inhibition of NPY3–35 production was observed with EDTA and EGTA at concentrations up to 10 mm, indicating that the enzyme is not a metalloprotease. A 34% inhibition of peptide degradation was observed with the cysteine protease inhibitor E64 that appeared to be nonspecific. Then, we found that the serine proteases inhibitors AEBSF, aprotinin, and leupeptin at 1 mm, 1 mg/ml, and 100 μm concentrations respectively, completely abolished NPY3–35 generation. Chymase, chymotrypsin-like, cathepsin A, B, D, and G involvement was excluded because 100 μm of the serine protease inhibitor chymostatin inhibited NPY3–36 degradation only by 32%. We then chose the serine protease inhibitor gabexate mesylate (29, 30), an antithrombotic agent able to inhibit several proteases involved in multiple proteolytic systems in plasma, including the classic complement pathway, plasmin, and kallikrein in the fibrinolytic pathway, and the activation of factor IX in the coagulation cascade (31, 32). A dose-dependent inhibition of NPY3–35 production was observed with gabexate mesylate from 1 mm up to a complete inhibition at 10 mm (data not shown). The fact that gabexate mesylate was active at millimolar concentrations allowed us to exclude thrombin, plasmin, and trypsin. This narrowed the choice to plasma kallikrein (EC 3.4.21.34) as the enzyme giving rise to NPY3–35. To check this hypothesis, 4 μm NPY3–36 was incubated with purified plasma kallikrein at concentrations ranging from 0.1 μg/ml to 40 μg/ml for 1 h in phosphate-buffered saline. The results showed that plasma kallikrein was sufficient to generate NPY3–35. The optimal kallikrein concentration tested was 5 μg/ml, corresponding to 10% of the physiological concentration of its proenzyme, prekallikrein observed in a healthy human population (33). Under these conditions, we found a cleavage of around 70% of NPY3–36 initially added in the incubation buffer (Fig. 6A). Higher concentrations of kallikrein conversely resulted in a decrease of NPY3–35 levels without an increase of NPY3–36 levels, suggesting that high concentrations of kallikrein enable additional and nonspecific NPY cleavages. To confirm that plasma kallikrein is responsible for NPY3–35 production, NPY3–36 was incubated with prekallikrein-deficient plasma; under these conditions, no cleavage was observed. In contrast, NPY3–35 production was restored when 5 μg/ml of purified kallikrein was added in the incubation mixture (Fig. 6B).
FIGURE 6.
Plasma kallikrein is the carboxypeptidase/esterase enzyme responsible for tyrosinamide removal from NPY. A, purified plasma kallikrein in phosphate-buffered saline is sufficient to cleave NPY3–36 into NPY3–35. NPY3–35 signal was expressed as the ratio versus porcine NPY1–36 used as internal standard; 1 mg of kallikrein corresponds to an activity of 15 units. One unit is defined as the amount of enzyme that hydrolyzes 1 μmol of d-Pro-Phe-Arg-pNA per min at 25 °C, pH 7.8. B, kallikrein-free plasma is unable to degrade NPY3–36 into NPY3–35. Left panel, NPY3–36 added as a substrate in 10% kallikrein-deficient plasma; middle, 0.5 μg/ml purified kallikrein added in the incubation mix; right, NPY1–36 added as a substrate.
Interindividual Variation of NPY1–36 Cleavages Resulting in NPY2–36, NPY3–36, and NPY3–35 Production
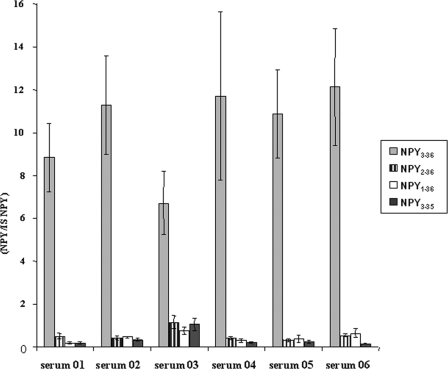
To ascertain that our study was relevant in a general population, 10 μm NPY1–36 was digested for 1 h, as described above in 6 samples of serum from healthy individuals. This incubation time was chosen to observe all NPY forms characterized in the present study. Longer digestion times resulted in a complete disappearance of NPY1–36, a decrease of NPY3–36, and an increase of NPY3–35 (data not shown). NPY3–36, NPY2–36, NPY3–35, and the remnant NPY1–36 signal intensities were normalized in terms of ratio versus an internal standard (porcine NPY1–36 at 0.5 μm). For each of the healthy individuals, NPY3–36 was by far the most abundant NPY form generated by in vitro incubation of NPY1–36 with serum (Fig. 7).
FIGURE 7.
Incubation of NPY1–36 in digestion buffer containing serum from 6 different individuals for 1 h. Results are expressed as the ratio of NPY form considered versus porcine standard; experiments were done in triplicate. Serum 01 was the material used to study kinetic parameters of NPY cleavage.
Determination of the Kinetic Constant of Plasma Kallikrein toward NPY3–36
The Vmax and the Km of plasma kallikrein for NPY3–36 were determined following the same approach as the one described for the determination of the kinetic constants of DPPIV and AmP. Because two different enzymes are involved in the production of NPY3–35 starting from NPY1–36 (DPPIV cuts at the N-terminal extremity of the peptide and plasma kallikrein at the C-terminal extremity), we digested NPY3–36 instead of NPY1–36. The Km obtained for the production of NPY3–35 by plasma kallikrein was 28.1 μm and Vmax was 4.9 pmol × min−1 × μl serum−1. It appears that plasma kallikrein exhibited an intermediate potency between AmP and DPPIV to hydrolyze NPY (Fig. 3C and Table 1).
DISCUSSION
We found that NPY1–36 degradation in human serum results from the combined action of amino- and carboxypeptidases. NPY3–36, which loses its affinity for the Y1 receptor and becomes a Y2/Y5 receptor agonist, represents the main NPY form in serum generated by the action of DPPIV, whereas AmP is poorly involved in the cleavage of NPY. The N-terminal NPY truncation by DPPIV observed in serum is in agreement with the recent findings by Frerker et al. (21) by MALDI-TOF mass spectrometry. However, in contrast with their data, we found that AmP is also present as a soluble protease in the serum and may also cleave NPY1–36 into NPY2–36. The discrepancies between the present study and the work by Frerker et al. may be due to the fact that EDTA as anticoagulant was used in this reports (AmP is a metalloprotease, Ref. 21). Besides minor cleavage products, we found an unexpected fragment in considerable amounts, identified as NPY3–35. This peptide is unable to bind to the NPY receptors, indicating that its formation is likely to represent a degradation process of the active NPY3–36 form. Although, NPY3–35 does not bind to any known NPY receptor, we cannot exclude that a putative unknown NPY receptor may be affected by this peptide or may compete with amidated NPY. We identified plasma kallikrein as the enzyme responsible for the removal of the C-terminal tyrosinamide.
It is noteworthy that the ability to generate NPY3–35 from NPY1–36 was successfully tested in plasma and serum from healthy individuals. Plasma kallikrein is derived from prekallikrein that undergoes proteolytic cleavage. Plasma kallikrein is part of the kallikrein-kinin system that activates the intrinsic pathway of coagulation by releasing bradykinin from kininogenes. It is also involved in fibrin degradation by catalyzing plasminogen cleavage into plasmin (34, 35). Moreover, plasma kallikrein has been reported to cleave a large panel of peptides both in vitro and in vivo (36–39). Based on kinetic values of the three enzymes responsible for NPY degradation, we were able to calculate the intrinsic clearances (IC) (Vmax/Km), which represented 2.11, 0.08, and 0.17 μl cleared × min−1 × μl serum−1 for DPPIV, AmP, and plasma kallikrein, respectively (Table 1). In agreement with our results, Mentlein et al. (17) reported a Km of 8 μm for DPPIV toward NPY. Intrinsic clearance values suggest that the metabolic pathway of NPY1–36 to NPY3–36 is 26 times more efficient than NPY1–36 to NPY2–36. In addition, NPY2–36 production from NPY1–36 is twice less efficient than NPY3–35 production from NPY3–36. The absolute amount of NPY3–35 produced remains to be assessed, but considering the Km value of plasma kallikrein (28.1 μm) and the fact that NPY3–35 is clearly observed for the charge state +3, +4, and +5 in the full scan LC-MS chromatogram of digested NPY1–36 (Fig. 4, A and B) suggests an intermediate amount between NPY2–36 and NPY3–36. We have also identified shorter C-terminal NPY-truncated forms in serum by using LTQ-Orbitrap (NPY1–35, NPY1–34, NPY1–32, NPY2–35, NPY2–34, NPY2–32, NPY3–34, and NPY3–32), but all of them were found in trace amounts. Our findings could explain in some cases poor correlation observed between concentrations of immunoreactive NPY and its pathophysiological action. Indeed, normal NPY concentrations in the human plasma have been reported to range from 0.25 to 129 pm (40–44). This variability in the measurement of circulating NPY concentrations likely reflects cross-reactivities of antisera directed against NPY toward immunoreactive fragments as well as to peptide YY and pancreatic polypeptide.
There are several lines of evidence suggesting that NPY action could be modulated by DPPIV and AmP. First, the activity of DPPIV is inversely correlated with inflammation observed in the nasal mucosa of patients with rhinitis (45), a condition known to involve NPY as a modulator of sympathetic and parasympathetic activities through Y1 and Y2 receptors, respectively (46). Second, in endothelial cells, cytokines increase the expression of 3 players in the system: NPY, its receptors, and DPPIV (47). Third, the correlation found between increased heart rate and mean arterial blood pressure, normally attributed to an increase in catecholamine release, may underestimate the role of NPY. Its elevation during stress or pheochromocytoma indeed suggests a counter-regulatory inactivation of the peptide (48). In this context, an enhanced sympathetic tone might be expected from direct postsynaptic effects of native NPY in the presence of a DPPIV inhibitor. This hypothesis has been assessed in a recent study reporting that male rats exhibit basal endogenous Y1-receptor modulation in their hindlimb vascular bed. On the other hand, the systemic inhibition of DPPIV and AmP elicited a Y1 receptor-dependent decrease in hindlimb vascular conductance in females. Thus, lack of basal endogenous Y1 receptor activation in female hindlimb vasculature is (at least partially) due to proteolytic processing of NPY (48).
The main physiological relevance of kallikrein action on NPY is highlighted by DPPIV inhibitors that have been recently marketed for the treatment of type 2 diabetes by preventing the degradation of glucagon-like peptide 1 (GLP-1), an incretin hormone (49). DPPIV inhibitors could interfere with the metabolism of NPY and increase sympathetic tonus by affecting its degradation. Consequently, kallikrein may be considered as the sole alternative enzyme pathway in blood to inactivate the vasoconstrictive Y1 activity of NPY1–36 in patients treated with a DPPIV inhibitor. Indeed, NPY1–36 incubation with vildagliptin resulted in an increase of NPY1–35 production (data not shown).
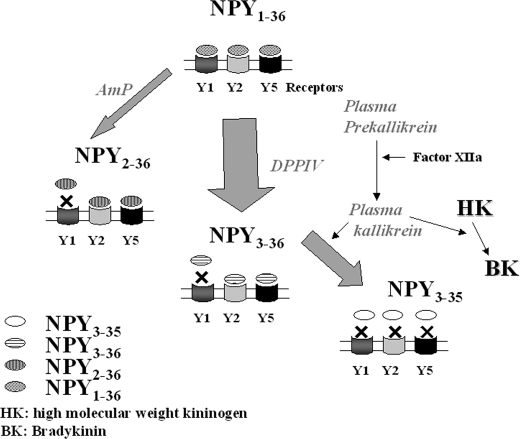
In light of our results, we propose a model regarding NPY1–36 processing in human serum (Fig. 8): NPY1–36 is predominantly cleaved by DPPIV into NPY3–36. Cleavage by AmP into NPY2–36 represents a minor pathway for the degradation of NPY1–36. A significant proportion of NPY3–36 is further transformed into the inactive NPY3–35 by plasma kallikrein. NPY3–35 is produced at all the concentrations of kallikrein tested between 0.2 and 100% of those of prekallikrein found in normal human plasma. It remains to be demonstrated whether in specific disease or metabolic states, active plasma kallikrein reaches concentration levels high enough to accelerate the degradation of NPY and possibly modulate its regular biological activities.
FIGURE 8.
Proposed kinetic model of NPY1–36 cleavage into truncated forms. NPY1–36 is predominantly cleaved into NPY3–36 by DPPIV, and through a slower process by AmP into NPY2–36, resulting for both peptides in a loss of affinity for NPY Y1 receptors. A fraction of NPY3–36 is further degraded by plasma kallikrein into NPY3–35, which do not retain the ability to bind to any NPY receptors. Unknown proteases are likely to be involved in the degradation of NPY1–36, NPY2–36, and NPY3–35 to yield shorter forms, found in trace amounts. See text for further details.
Acknowledgments
We thank the Foundation Michel Tossizza for helping with the acquisition of the LC-LTQ-MS system. We are grateful to Gilles Mourier and Roger Genet for helping with isotopic peptide synthesis and Drs. Christophe Kündig, David Deperthes, and Jair Chagas from Med Discovery, Geneva, Switzerland for advice regarding kallikrein. We thank Dr. T. Buclin for advice and criticism.
This work was supported by Swiss National Science Foundation Grant 3100 AO-101999 (to E. G.).
- NPY
- neuropeptide Y
- LC-MSn
- liquid chromatography coupled with tandem mass spectrometry
- DPPIV
- dipeptidyl peptidase IV
- AmP
- aminopeptidase P
- LTQ-MS
- linear ion trap mass spectrometer
- ESI
- electrospray ion source
- EIC
- extracted product ion chromatogram
- IC
- intrinsic clearance.
REFERENCES
- 1.Wahlestedt C., Edvinsson L., Ekblad E., Håkanson R. (1985) J. Pharmacol. Exp. Ther. 234, 735–741 [PubMed] [Google Scholar]
- 2.Walker P., Grouzmann E., Burnier M., Waeber B. (1991) Trends Pharmacol. Sci. 12, 111–115 [DOI] [PubMed] [Google Scholar]
- 3.Abdel-Samad D., Jacques D., Perreault C., Provost C. (2007) Peptides 28, 281–287 [DOI] [PubMed] [Google Scholar]
- 4.Cavadas C., Céfai D., Rosmaninho-Salgado J., Vieira-Coelho M. A., Moura E., Busso N., Pedrazzini T., Grand D., Rotman S., Waeber B., Aubert J. F., Grouzmann E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10497–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahu A., Kalra S. P. (1993) Trends Endocrinol. Metab. 4, 217–224 [DOI] [PubMed] [Google Scholar]
- 6.Stanley B. G., Leibowitz S. F. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3940–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arora S., Anubhuti (2006) Neuropeptides 40, 375–401 [DOI] [PubMed] [Google Scholar]
- 8.Naveilhan P., Hassani H., Canals J. M., Ekstrand A. J., Larefalk A., Chhajlani V., Arenas E., Gedda K., Svensson L., Thoren P., Ernfors P. (1999) Nat. Med. 5, 1188–1193 [DOI] [PubMed] [Google Scholar]
- 9.Kuo L. E., Kitlinska J. B., Tilan J. U., Li L., Baker S. B., Johnson M. D., Lee E. W., Burnett M. S., Fricke S. T., Kvetnansky R., Herzog H., Zukowska Z. (2007) Nat. Med. 13, 803–811 [DOI] [PubMed] [Google Scholar]
- 10.Michel M. C., Beck-Sickinger A., Cox H., Doods H. N., Herzog H., Larhammar D., Quirion R., Schwartz T., Westfall T. (1998) Pharmacol. Rev. 50, 143–150 [PubMed] [Google Scholar]
- 11.Pons J., Lee E. W., Li L., Kitlinska J. (2004) Curr. Opin. Investig. Drugs 5, 957–962 [PubMed] [Google Scholar]
- 12.Pedrazzini T., Pralong F., Grouzmann E. (2003) Cell Mol. Life Sci. 60, 350–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiest R., Jurzik L., Moleda L., Froh M., Schnabl B., von, Hörsten S., Schölmerich J., Straub R. H. (2006) J. Hepatol. 44, 512–519 [DOI] [PubMed] [Google Scholar]
- 14.Westfall T. C., Carpentier S., Chen X., Beinfeld M. C., Naes L., Meldrum M. J. (1987) J. Cardiovasc. Pharmacol. 10, 716–722 [DOI] [PubMed] [Google Scholar]
- 15.Bitran M., Tapia W., Eugenín E., Orio P., Boric M. P. (1999) Brain Res. 851, 87–93 [DOI] [PubMed] [Google Scholar]
- 16.de Meester I., Lambeir A. M., Proost P., Scharpé S. (2003) Adv. Exp. Med. Biol. 524, 3–17 [DOI] [PubMed] [Google Scholar]
- 17.Mentlein R., Dahms P., Grandt D., Krüger R. (1993) Regul. Pept. 49, 133–144 [DOI] [PubMed] [Google Scholar]
- 18.Mentlein R., Roos T. (1996) Peptides 17, 709–720 [DOI] [PubMed] [Google Scholar]
- 19.Yaron A., Mlynar D. (1968) Biochem. Biophys. Res. Commun. 32, 658–663 [DOI] [PubMed] [Google Scholar]
- 20.Medeiros M. D., Turner A. J. (1994) Endocrinology 134, 2088–2094 [DOI] [PubMed] [Google Scholar]
- 21.Frerker N., Wagner L., Wolf R., Heiser U., Hoffmann T., Rahfeld J. U., Schade J., Karl T., Naim H. Y., Alfalah M., Demuth H. U., von Hörsten S. (2007) Peptides 28, 257–268 [DOI] [PubMed] [Google Scholar]
- 22.Khan I. U., Reppich R., Beck-Sickinger A. G. (2007) Biopolymers 88, 182–189 [DOI] [PubMed] [Google Scholar]
- 23.Kippen A. D., Cerini F., Vadas L., Stöcklin R., Vu L., Offord R. E., Rose K. (1997) J. Biol. Chem. 272, 12513–12522 [DOI] [PubMed] [Google Scholar]
- 24.Stöcklin R., Vu L., Vadas L., Cerini F., Kippen A. D., Offord R. E., Rose K. (1997) Diabetes 46, 44–50 [DOI] [PubMed] [Google Scholar]
- 25.Prechel M. M., Orawski A. T., Maggiora L. L., Simmons W. H. (1995) J. Pharmacol. Exp. Ther. 275, 1136–1142 [PubMed] [Google Scholar]
- 26.Busso N., Wagtmann N., Herling C., Chobaz-Péclat V., Bischof- Delaloye A., So A., Grouzmann E. (2005) Am. J. Pathol. 166, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grouzmann E., Buclin T., Martire M., Cannizzaro C., Dörner B., Razaname A., Mutter M. (1997) J. Biol. Chem. 272, 7699–7706 [DOI] [PubMed] [Google Scholar]
- 28.Moser C., Bernhardt G., Michel J., Schwarz H., Buschauer A. (2000) Can. J. Physiol. Pharmacol. 78, 134–142 [PubMed] [Google Scholar]
- 29.Ohno H., Kosaki G., Kambayashi J., Imaoka S., Hirata F. (1980) Thromb. Res. 19, 579–588 [DOI] [PubMed] [Google Scholar]
- 30.Ohno H., Kambayashi J., Chang S. W., Kosaki G. (1981) Thromb. Res. 24, 445–452 [DOI] [PubMed] [Google Scholar]
- 31.Schmaier A. H. (2008) Int. Immunopharmacol. 8, 161–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sainz I. M., Pixley R. A., Colman R. W. (2007) Thromb. Haemost. 98, 77–83 [PubMed] [Google Scholar]
- 33.Veloso D. (2003) Scand. J. Clin. Lab. Invest. 63, 279–286 [PubMed] [Google Scholar]
- 34.Peek M., Moran P., Mendoza N., Wickramasinghe D., Kirchhofer D. (2002) J. Biol. Chem. 277, 47804–47809 [DOI] [PubMed] [Google Scholar]
- 35.Almeida P. C., Chagas J. R., Cezari M. H., Juliano M. A., Juliano L. (2000) Biochim. Biophys. Acta 1479, 83–90 [DOI] [PubMed] [Google Scholar]
- 36.Leduc R., Hendy G. N., Seidah N. G., Chrétien M., Lazure C. (1990) Life Sci. 46, 1427–1433 [DOI] [PubMed] [Google Scholar]
- 37.Metters K. M., Rossier J., Paquin J., Chrétien M., Seidah N. G. (1988) J. Biol. Chem. 263, 12543–12553 [PubMed] [Google Scholar]
- 38.Ichinose A., Fujikawa K., Suyama T. (1986) J. Biol. Chem. 261, 3486–3489 [PubMed] [Google Scholar]
- 39.Cardin A. D., Jackson R. L., Donaldson V. H., Chao J., Margolius H. S. (1986) Adv. Exp. Med. Biol. 198, 195–202 [DOI] [PubMed] [Google Scholar]
- 40.Allen J. M., Yeats J. C., Adrian T. E., Bloom S. R. (1984) Regul. Pept. 8, 61–70 [DOI] [PubMed] [Google Scholar]
- 41.Corder R., Lowry P. J. (1985) Peptides 6, 1195–1200 [DOI] [PubMed] [Google Scholar]
- 42.Lundberg J. M., Martinsson A., Hemsén A., Theodorsson-Norheim E., Svedenhag J., Ekblom B., Hjemdahl P. (1985) Biochem. Biophys. Res. Commun. 133, 30–36 [DOI] [PubMed] [Google Scholar]
- 43.Theodorsson-Norheim E., Hemsén A., Lundberg J. M. (1985) Scand. J. Clin. Lab. Invest. 45, 355–365 [DOI] [PubMed] [Google Scholar]
- 44.Grouzmann E., Comoy E., Bohuon C. (1989) J. Clin. Endocrinol. Metab. 68, 808–813 [DOI] [PubMed] [Google Scholar]
- 45.Grouzmann E., Monod M., Landis B., Wilk S., Brakch N., Nicoucar K., Giger R., Malis D., Szalay-Quinodoz I., Cavadas C., Morel D. R., Lacroix J. S. (2002) FASEB J. 16, 1132–1134 [DOI] [PubMed] [Google Scholar]
- 46.Malis D. D., Grouzmann E., Morel D. R., Mutter M., Lacroix J. S. (1999) Br. J. Pharmacol. 126, 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva A. P., Cavadas C., Baïsse-Agushi B., Spertini O., Brunner H. R., Grouzmann E. (2003) Regul. Pept. 116, 71–79 [DOI] [PubMed] [Google Scholar]
- 48.Jackson D. N., Milne K. J., Noble E. G., Shoemaker J. K. (2005) J. Physiol. 568, 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drucker D. J., Nauck M. A. (2006) Lancet 368, 1696–1705 [DOI] [PubMed] [Google Scholar]