Abstract
Regulation by the NK and T cell surface receptor CD244 in mice and humans depends both on engagement at the cell surface by CD48 and intracellular interactions with SAP and EAT-2. Relevance to human disease by manipulating CD244 in mouse models is complicated by rodent CD2 also binding CD48. We distinguish between contributions of mouse CD244 and CD2 on engagement of CD48 in a mouse T cell hybridoma. CD2 and CD244 both contribute positively to the immune response as mutation of proline-rich motifs or tyrosine motifs in the tails of CD2 and CD244, respectively, result in a decrease in antigen-specific interleukin-2 production. Inhibitory effects of mouse CD244 are accounted for by competition with CD2 at the cell surface for CD48. In humans CD2 and CD244 are engaged separately at the cell surface but biochemical data suggest a potential conserved intracellular link between the two receptors through FYN kinase. We identify a novel signaling mechanism for CD244 through its potential to recruit phospholipase C-γ1 via the conserved phosphorylated tyrosine motif in the tail of the adaptor protein EAT-2, which we show is important for function.
The CD2 family of cell surface receptors is differentially expressed on immune cells (1, 2) and is involved in regulating both innate and adaptive immunity (3). These receptors have related extracellular immunoglobulin superfamily domains and interact either homophilically or heterophilically within the CD2 family (1, 2). The CD2 family contains a subgroup of receptors termed the SLAM family that have a conserved tyrosine signaling motif in their cytoplasmic region TXYXX(I/V) referred to as an immunoreceptor tyrosine-based switch motif (ITSM).2 The SLAM family of receptors include CD244 (2B4), NTB-A (Ly-108), CD319 (CRACC, CS-1), CD150 (SLAM), CD84, and CD229 (Ly-9). Defects in signaling and aberrant expression of these receptors have been implicated in several immunodeficiency and autoimmune disorders in humans and mice (4–8). Within the SLAM family, CD244 is unusual in that it shares its ligand CD48 with the receptor CD2 in rodents, whereas in humans CD2 has evolved to interact with CD58 (9). The affinity of CD244 for CD48 in rodents is 6–9-fold higher than the still functionally relevant CD2/CD48 interaction (10). CD244 and CD2 have different cytoplasmic regions comprised of tyrosine motifs or proline-rich motifs, respectively.
CD244 is predominantly found on NK cells and cytotoxic T cells and primarily characterized as an activating receptor (11–15). CD2 is found on the same cells as CD244 but is also expressed on all T cells, both activated and resting, and has an activating or costimulatory function upon engagement of ligand (9). The tyrosine motifs found in the cytoplasmic tail of CD244 have been shown to bind the SH2 domains of cytoplasmic adaptor proteins SAP and EAT-2 and FYN kinase (16–18) and are important to its function (5, 19–21). In contrast to SH2 interactions of CD244, several SH3 domain-mediated interactions have been reported for the cytoplasmic region of CD2 including CD2AP/CMS, CIN85, FYN, and LCK (22–26).
The activating function of CD244 was called into question when a study using cells from a CD244 knock-out mouse showed that CD244 had an inhibitory effect as loss of CD244 resulted in enhanced NK killing of target cells (27). This suggested that previous results in mice where positive effects were seen may have been due to blocking CD244 ligand engagement as opposed to cross-linking with antibodies against CD244 (27). This has led to proposals that there are differences in function between mouse and human CD244 as there is more evidence to suggest that human CD244 is a positive regulator enhancing cytotoxicity and cytokine production (13, 15, 28). However, other more recent studies have shown the mouse CD244/CD48 interaction to be important for cytokine production and effector functions such as cytotoxicity against tumor targets in CD244-deficient mice (29). Long and short forms of CD244 have been cloned from mice with the short form being described as activating and the long form inhibitory (27, 30). Only the long form of CD244 is present in humans and is regarded as activating (14).
Positive signaling by CD244 has been attributed to the recruitment of SAP (18), which is a signaling adaptor molecule comprised of a single SH2 domain encoded by the SH2D1A gene and has the ability to recruit the kinase FYN by binding its SH3 domain (31, 32). Loss of the SAP/FYN interaction can lead to X-linked lymphoproliferative disease in humans (17). The molecular basis of in vitro inhibitory effects observed with CD244 in mice on ligation with mAb or ligand remains elusive (33). Protein tyrosine and inositol phosphatases have been reported to associate with CD244 (18, 19, 34) but our studies using surface plasmon resonance found them to be very weak and unlikely to bind competitively compared with the SAP family of adaptors or FYN (16). The SAP-related adaptor EAT-2 has been reported to have an active inhibitory effect that is dependent on tyrosine motifs in the tail of EAT-2 (35) but its mechanism is not understood. The only interaction reported for the tail of EAT-2 is with FYN kinase and studies overexpressing EAT-2 in a T cell hybridoma resulted in increased IL-2 production upon antigen stimulation (16).
The conservation between mouse and human CD244 cytoplasmic regions and associated adaptors suggests that both function in a similar way. We have explored the main difference between mouse and human CD244, which is the extracellular interaction through CD48 ligation in the mouse. This has revealed that inhibitory effects of CD244 ligation in mice can be due to competition between CD244 and CD2 for CD48. We have also found that the adaptor protein EAT-2 binds PLCγ1 providing a molecular basis for the important role CD244 plays in regulating cellular cytotoxicity (13, 36). We demonstrate that there is a potentially shared signaling mechanism through the FYN kinase that links CD2 and CD244 intracellularly even though in humans CD2 and CD244 no longer share a cell surface ligand.
EXPERIMENTAL PROCEDURES
Constructs
Mutated forms of mouse CD2 and mouse CD244 (the long form (10)) were constructed by overlapping PCR with mutations made to CD2 at amino acids (aa) 330 and 331 (PR to AA) and aa 288 and 289 (HH to DE). Mutations to mouse CD244 consisted of single aa substitutions of Tyr to Phe in the 4 ITSM motifs, TXYXX(I/V). CD2 was cloned into pBabe puro vector (16) as a BamHI-SalI fragment. Mutant CD2 was made by replacement of an EcoRI-SalI fragment. EAT-2 and mutated EAT-2 were constructed by PCR and cloned into BglII-SalI cut pLEGFP-C1 (Clontech) as a BamHI-SalI fragment. CD244 was cloned into pBabe puro and pFBNEO (Stratagene) as an EcoRI-SalI fragment and mutant CD244 into pBabe puro as EcoRI-SalI fragments. Mouse CD2 and CD2 mutants were expressed in 2B4 Reay T hybridoma cells by transfection into the retroviral packaging line EcoPhoenix by FuGENETM and subsequent transduction (16). Wild type CD244 in pFBNEO was transduced into these cells. 2B4 Saito T hybridoma cells expressing mouse CD2 or a tailless CD2 (37) were transduced with wild type or mutant mouse CD244 in pBabe puro vector. Cells were selected in puromycin (1 μg/ml) and G418 (1 mg/ml) in RPMI 1640 medium containing 5–10% fetal calf serum. Mouse CD48+ve major histocompatibility complex class II+ IEK Chinese hamster ovary cells (CHO IEk) were previously described (37) and maintained in Dulbecco's modified Eagle's medium + 5% fetal calf serum, 20 μm methylsulfoximine, and 0.4 mg/ml G418. Expression levels were checked by standard flow cytometry techniques.
BIACoreTM Analyses
BIACore analyses using a BIACoreTM 3000 were carried out essentially as described previously (16). All experiments were carried out at 37 °C. Streptavidin (∼3000 response units) was directly coupled to CM5 chips by amine coupling for immobilization of biotinylated phosphorylated peptides (Table 1) (Peptide Protein Research Ltd. (Fareham, UK). Full-length human SAP-glutathione S-transferase (GST) was immobilized by anti-GST mAb amine coupled to the surface of the chip (∼7000–8000 response units). In measurements of binding to the four ITSM peptides, one flow cell was used as a negative control and then coated with the fourth peptide, and equilibrium binding was repeated. At the level of immobilization of peptide (∼25 response unit) there was no difference between negative controls of streptavidin or an irrelevant phosphorylated peptide. For the SAP-GST a control flow cell of immobilized anti-GST mAb was used as a control. Measurements of FYN-SH3-SH2 binding to CD2 peptides were carried out as above but with ∼100 response units of CD2 peptide (Table 1) bound to the surface of the chip. In FYN-SH3-SH2 complex binding experiments the concentration of FYN-SH3-SH2 was constant at 5 μm, whereas nonbiotinylated phosphorylated or nonphosphorylated peptides in solution were titrated.
TABLE 1.
Synthetic peptides of human receptors
| Name | Sequence |
|---|---|
| EAT-2 127 | Biotin-NSNSD(pY)VDVLP-COOH |
| CTLA-4 | Biotin-KQFQP(pY)FIPIN-COOH |
| CD2-322 PR mutant | Biotin-QKGPPLPRAAVQPKPPHG-COOH |
| CD2-281-305 | Biotin-HPPPPPGHRSQAPSHRPPPPGHRVQ-COOH |
| CD2-322 | Biotin-QKGPPLPRPRVQPKPPHG-COOH |
| CD244 ITSM 1 | Biotin-EFLTI(pY)EDVKD-COOH |
| LAT 132 | Biotin-HNPG(pY)LVVLP-COOH |
Cellular Assays
Assays for antigen-specific IL-2 production by T hybridoma cells were performed in a 96-well round bottom plate with 105 cells/well of the T cell hybridomas and CHO IEK mouse CD48+ve cells in the presence of 1 μm moth cytochrome c peptide (MCC) (16) and relevant mAbs at 5 μg/ml or Fab fragments at 10 μg/ml. Assays were performed in RPMI + 5% fetal bovine serum for 16 h at 37 °C in a total volume of 200 μl. Supernatants were harvested and assayed for IL-2 by enzyme-linked immunosorbent assay (BD Biosciences).
Antibodies
mAbs used were as follows: anti-mouse CD3 (KT3) rat IgG2a, anti-mouse CD2 (RM2.1) rat IgG2a, anti-mouse CD48 (OX78) rat IgG2a, anti-rat κ-chain (OX11) rat IgG2a, anti-phosphotyrosine (clone PT66; Sigma), anti-PLCγ1 (Cell Signaling), anti-EAT-2 (Santa Cruz), phycoerythrin-coupled secondary antibodies (Sigma), fluorescein isothiocyanate-coupled secondary antibodies (Serotec), and horseradish peroxidase-coupled secondary reagents (Sigma and Bio-Rad). Fab fragments were produced by papain digestion using standard techniques and subjected to gel filtration on an S200 Superdex column (GE Healthcare).
Recombinant Proteins
Recombinant soluble proteins representing SH3 and/or SH2 domains of human signaling proteins were provided by Louise Bird (Oxford Module Consortium). Proteins were expressed as N-terminal His-tagged fusion proteins and purified using nickel-agarose affinity chromatography. Proteins were checked by mass spectrometry and subjected to gel filtration prior to BIACore analysis. Concentration was determined using absorbance at 280 nm using theoretical extinction coefficients (Vector NTI): FYN-SH3-SH2 (aa 82 to 248), LCK-SH3-SH2 (aa 58 to 230), PLCγ1 SH2 N+C (aa 545 to 749), SAP-GST (full-length), and CMS SH3 d1 (aa 2 to 60).
Peptide Pulldowns
Peptide pulldowns were performed essentially as described (25, 38). Human T blasts (108) cultured with 5 μg/ml phytohemagglutinin and 100 IU human IL-2/ml in RPMI + 5% fetal calf serum were pelleted and washed twice in phosphate-buffered saline before being lysed in 500 μl of lysis buffer containing 1% Nonidet P-40 + mammalian protease inhibitor mixture (Sigma). Cell lysates were spun at 13,000 rpm in a microcentrifuge for 20 min at 4 °C before being pre-cleared with 25 μl of streptavidin-coated Dynabeads (Invitrogen). Biotinylated synthetic peptides were coupled to streptavidin Dynal beads at saturating concentrations (8 μl of peptide (2 mg/ml (1 mm)) added to 25 μl beads) at 4 °C for 30 min with agitation. Beads were washed 3 times in 1 ml of phosphate-buffered saline before being added to cleared lysates. Beads and lysates were incubated at 4 °C with rotation for 1 h. Beads were washed prior to magnetic separation with a tube change between washes. Beads were resuspended in 30 μl of reducing SDS-PAGE sample buffer and subjected to SDS-PAGE. A sample was excised from the resolving gel immediately after passing through the stacking gel so as to avoid separation of proteins and potential loss of sample.
Immunoprecipitation and Western Blotting
Cell lysates were prepared at 108 cells/ml using Triton X-100 detergent as described above (31). Immunoprecipitates were prepared using protein G-Sepharose (Amersham Biosciences). Western blot analysis was carried out using 4–12% SDS-polyacrylamide gels under reducing conditions and ECL detection methods (Amersham Biosciences) (29).
Mass Spectrometry
Gel pieces were digested with trypsin and desalted on a C18 packed pipette tip before being injected onto an Ultimate 3000 nano-HPLC (Dionex) system coupled to an Orbitrap mass spectrometer (Thermo Electron). Samples were resolved on a 5-cm × 100-μm inner diameter picotip column (New Objective) that was packed in-house with Reprosil-Pur C18-AQ phase (Dr. Maisch). The mass spectrometer was operated in a data-dependent acquisition mode. Peak lists were generated using DTAsupercharge and searched using Mascot (Matrixscience). Data were searched against the IPI human data base.
RESULTS
Positive Signaling by Mouse CD2 and CD244
To understand signaling by CD244 and the differences between mouse and human it is necessary to understand the contribution of both CD244 and CD2 as they share CD48 as a ligand in rodents. First, we needed a system in which we could manipulate the extracellular and intracellular interactions of CD2 and then ask how these are influenced by CD244. The aim is to understand how manipulation of a particular molecular interaction affects the functional outcome to interpret data from more complex biological systems. We chose to use a model that involved ligand engagement with antigen-specific stimulation of a CD4+ mouse T cell hybridoma and measuring IL-2 production. 2B4 Reay cells expressed very low levels of endogenous CD2, which facilitated detection of the effects of transduced single point mutants. Functional effects of extracellular and intracellular engagement of CD2 have been demonstrated in this hybridoma (37). As CD244 is not primarily expressed on CD4+ cells and not detectable on CD4+ hybridoma cells interpretation would not be complicated by the endogenous receptor as it is in experiments with CD8+ hybridoma cells.3 We established conditions where we could detect the effect of manipulation of extracellular interactions at the cell surface by blocking with well characterized mAbs and intracellular interactions using single point mutants of sites that have been shown to bind intracellular ligands.
Positive Signaling by Mouse CD2
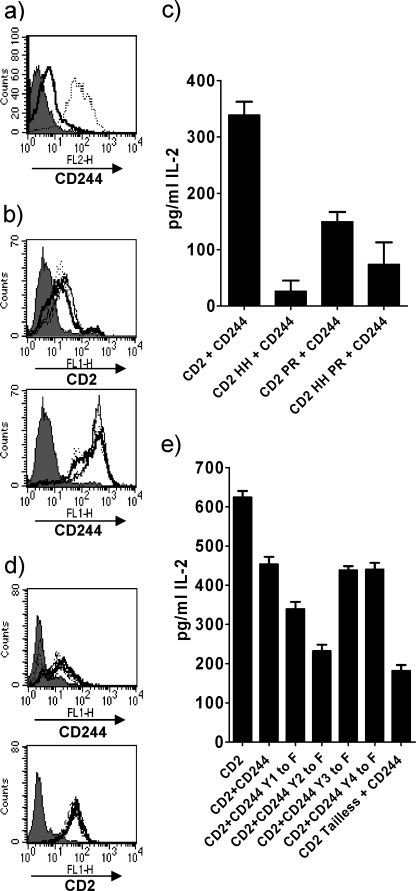
Biochemical data have shown that specific sites in the cytoplasmic region of CD2 are important for interactions with intracellular ligands (23, 25, 39). The functional importance of these sites has been implicated in studies using antibody cross-linking of receptors (40, 41) but it is not clear how they contribute to an immune response upon ligand engagement. We transduced 2B4 Reay hybridoma cells with mouse CD2 alone or mutated forms of mouse CD2 with amino acid substitutions at position 288–289 (HH to DE) or 335–336 (PR to AA), which have previously been shown to be functionally important in humans (23, 40), or a double mutant (HH/DE and PR/AA). However, this did not provide a robust assay in which we could detect the effects of ligand engagement at the cell surface or within the cell. Interestingly cotransduction of CD2 and CD244 into these cells resulted in increased expression of CD244, by more than an order of magnitude (Fig. 1a). Additionally, we found that cotransduction of CD2 and CD244 into these cells elicited responses that we could show were altered by blocking mAbs and mutation of the cytoplasmic region of CD2.
FIGURE 1.
Positive signaling by mouse CD2 and CD244. a, flow cytometric analysis of 2B4 Reay T hybridoma cells transduced with CD244 (thick black line) or CD244 and CD2 (dotted line). b, flow cytometric analysis of 2B4 Reay T hybridoma cells transduced with CD244 and either CD2 or mutated CD2 showing levels of CD244 and CD2 (thick black lines) or CD2 mutated at aa 288–289 (HH to DE) (dotted line), 335–336 (PR to AA) (dashed line), or a double mutant of 288–289 and 335–336 (HH/PR to DE/AA) (thin line) compared with untransduced cells (gray shading). c, IL-2 production by cells in b in response to 1 μm MCC peptide presented by CHO IEK mouse CD48+ve cells. d, flow cytometric analysis of 2B4 Saito T hybridoma cells transfected with CD2 or tailless CD2 and transduced with either CD244 or mutated CD244 showing levels of CD2 and CD244 (thick black lines), tailless CD2 and CD244 (dash dash dot), CD2 and mutated CD244 with a Tyr to Phe substitution in the indicated ITSM: Y1F (dotted line), Y2F (dashed line), Y3F (dot dash line), Y4F (thin line). e, IL-2 production by cells in d in response to 1 μm MCC peptide presented by CHO IEK mouse CD48+ve cells. The experiments shown are representative of at least three.
Cells expressing CD244 and either CD2 or mutated forms of CD2 were matched for surface expression of CD2, CD244 (Fig. 1b), and CD3 (data not shown). The cells were stimulated with peptide and CHO IEK cells or CHO IEK cells expressing mouse CD48. Antigen-specific IL-2 production by these cells was markedly reduced with the CD2 mutant (HH to DE) and the CD2 mutant (PR to AA) relative to wild type CD2 in response to both sets of CHO IEK cells (Fig. 1c and data not shown). As ligand-dependent effects of blocking mAbs (see below) could only be detected with CHO IEK cells expressing CD48 we continued with these as antigen presenting cells. Reduced responses by the CD2 cytoplasmic mutants are consistent with these sites being important for binding intracellular ligands under conditions where they have a positive effect on an immune response (23, 25, 42).
Positive Signaling by Mouse CD244
We next needed to clarify the effect of interactions of the cytoplasmic region of CD244 on an immune response under conditions where there is productive ligand engagement at the cell surface. The correlation between CD2 and CD244 expression suggested that there may be functional dependence of CD244 on CD2 so we transduced CD244 and mutants into the Saito 2B4 hybridoma cells, which express higher levels of endogenous CD2, and were also transfected with CD2 (43). This resulted in stable expression of CD244 (Fig. 1d). IL-2 production is dependent on CD2 in these cells as shown by cells expressing a tailless form of CD2 (Fig. 1e) (37, 43).
2B4 Saito cells transfected with CD2 (43) and transduced with CD244 or mutated forms of CD244 with a tyrosine substitution in 1 of the 4 ITSM motifs were tested for antigen-specific IL-2 production. Cells had comparable levels of CD2, CD244 (Fig. 1d), and CD3 (data not shown) by flow cytometry. As with the 2B4 Reay cells (Fig. 1c and data not shown) IL-2 production in CD2+ve CD244+ve 2B4 Saito cells was reduced compared with CD2+ve cells (Fig. 1e). However, mutations to ITSM 1 and 2 of CD244 by changing the tyrosine to phenylalanine resulted in a decrease in IL-2 production. Point mutations to ITSM motifs 3 and 4 of CD244 did not result in an increase in IL-2 production. These results show that like human (13, 15, 28), mouse CD244 does not give an inhibitory signal and establishes that both CD2 and CD244 are signaling in a positive manner.
Inhibition by Mouse CD244 Is Due to Competition for CD48
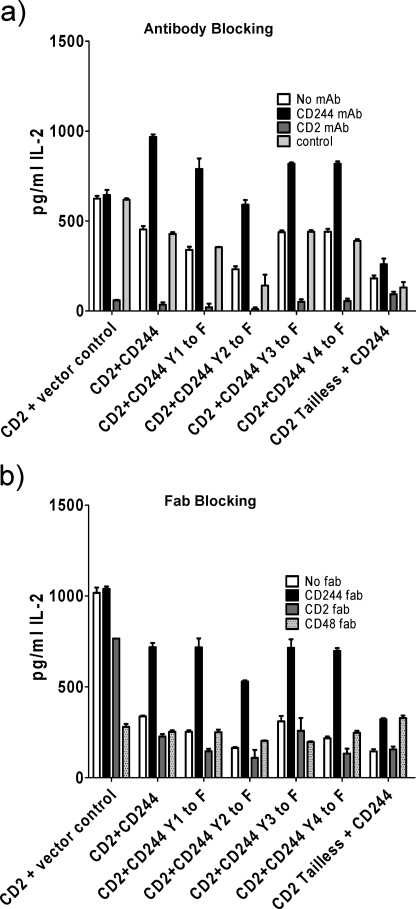
In cells cotransduced with CD2 and CD244 the overall level of IL-2 production in response to antigen stimulation is reduced (Fig. 1). A possible model for CD244 inhibition is competition with CD2 for its ligand CD48. An increase in the level of occupancy of CD2 by CD48 would be predicted to increase IL-2 production. To test this hypothesis we used blocking antibodies against CD2 and CD244. Cells were assayed for antigen-specific IL-2 production in response to CHO IEk cells expressing CD48. Blocking CD2 on 2B4 Saito cells transfected with CD2 resulted in a decrease in IL-2, consistent with positive signaling, whereas a CD244 antibody had no effect (Fig. 2a). Blocking CD244 on cells expressing CD2 and CD244 resulted in an increase in IL-2 (Fig. 2a). This was also true for cells expressing mutated forms of CD244 (Fig. 2a). This is consistent with the inhibitory effects of CD244 engagement being independent of signaling by CD244. This indicates that a mechanism for the inhibitory effects of CD244 is competition with CD2 for engagement of CD48 thereby changing the occupancy of CD2.
FIGURE 2.
Inhibition by mouse CD244. IL-2 production by 2B4 Saito T cell hybridoma cells transfected with CD2 or tailless CD2 and transduced with either CD244 or mutated CD244 with a Tyr to Phe substitution in the indicated ITSM motif in response to 1 μm MCC presented by CHO IEK mouse CD48+ve cells in the presence of the indicated blocking (a) antibodies or (b) Fab fragments from antibodies against mouse CD244, CD2, CD48, or isotype control. The experiments shown are representative of at least three.
Fab fragments of CD2, CD244, and CD48 mAbs were used as above in an antigen-specific IL-2 response to confirm that the antibodies were blocking ligand engagement and not cross-linking (Fig. 2b). IL-2 production was enhanced upon blocking CD244 in both the wild type and mutant forms and reduced when CD2 was blocked. CD2 Fab fragments were relatively ineffectual due to low affinity of the monomeric reagent (data not shown) but together with results of the whole mAbs (Fig. 2a) inhibition and activation by CD48 and CD244 Fab fragments, respectively, shows that the majority of positive signaling is due to CD2. The pattern changes in the absence of CD2 signaling (CD2 tailless) where CD48 and CD244 Fab fragments both enhance the response (Fig. 2b). In these cells CD244 signaling is marginal as competent signaling is dependent on SAP and EAT-2 (5, 19, 20). Nevertheless, dissection of the relative contributions of extracellular and intracellular contributions of CD2 and CD244 to the functional outcome shows that inhibition observed by CD244 in rodents can be accounted for by competition for CD48.
CD2 and CD244 Intracellular Regions Show Higher Affinity for FYN Than LCK
Biochemical and functional data have shown that CD2 and CD244 perform essentially the same function in the mouse and human. On ligand engagement they can mediate positive signaling and competition in rodents at the cell surface for CD48 can account for differences in the functional outcome between mouse and human. The link between rodent CD2 and CD244 in engagement at the cell surface has been lost in humans but as the cytoplasmic tails of CD2 and CD244 show a high degree of conservation between mouse and human (11, 44), potential signaling mechanisms common to the two molecules may be maintained. CD2 is present on resting cells and is unlikely to undergo post-translational modification of its cytoplasmic region for interactions through proline-rich motifs. CD244 function depends on phosphorylation of its cytoplasmic region (45). The SRC kinases FYN and LCK are potential ligands common to CD2 and CD244 (31, 39). LCK has been reported to bind directly to CD2 and be important for its function (22, 46). FYN has been implicated in both CD244 and CD2 function and has been reported to interact directly through SH2 and SH3 interactions, respectively (16, 22, 39, 47). FYN has also been shown to be activated in vitro by peptides based on the cytoplasmic tail of CD2 (48) suggesting a potential mechanism for the initiation of phosphorylation. As FYN has been reported to bind to many substrates, one way of determining the functionally relevant interactions is to measure the relative affinities for different substrates and compare them with previously characterized interactions.
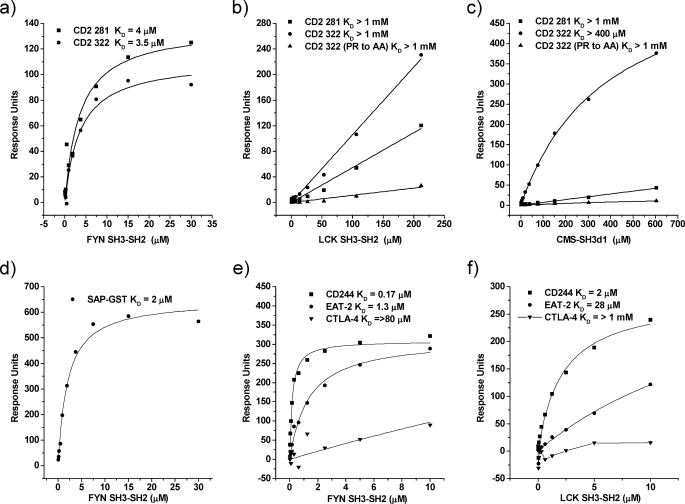
We tested recombinant human FYN SH3-SH2 and LCK SH3-SH2 for binding to synthetic peptides based on the signaling motifs found in the cytoplasmic regions of human CD2 and CD244 (Table 1). The relative affinities of SH3 binding to peptides based on CD2 proline-rich cytoplasmic regions 281–305 and 322–339 favors FYN over LCK (Fig. 3), which is in agreement with previous biochemical (47) and functional data (49). The affinities for FYN binding were KD ∼ 4 μm for both CD2 peptides (Fig. 3a). Very weak binding was detected for LCK, KD > 1 mm for both peptides (Fig. 3b). The related adaptor proteins CMS and CIN85 have also been shown to bind to CD2 at region 322–339 (25). Mutation of a proline and arginine at positions 335 and 336, respectively, abrogated binding of both FYN and CMS showing that the binding sites are overlapping for the two proteins (Fig. 3c and data not shown). Interaction of the SH3 domain of FYN with CD2 is of a similar affinity to the FYN/SAP interaction, KD ∼ 2 μm (Fig. 3d).
FIGURE 3.
CD2 and CD244 intracellular regions show higher affinity for FYN than LCK. Equilibrium binding data from which dissociation constants were calculated were obtained by passing various concentrations of soluble recombinant protein over immobilized peptides. a and b, FYN-SH3-SH2 and LCK-SH3-SH2 binding to proline-rich peptides based on the 2 regions of the human CD2 cytoplasmic region (Table 1). c, the interaction of CMS and FYN-SH3-SH2 (data not shown) with CD2 322 peptide is dependent on aa PR at positions 335–336, which are substituted to AA in the CD2 mutant peptide. d, FYN-SH3-SH2 binds SAP. e and f, FYN SH3-SH2 binds with a higher affinity than LCK-SH3-SH2 to phosphorylated synthetic peptides based on CD244 ITSM-1 and EAT-2 127 (Table 1). Lines show best fits except for CTLA-4 in f.
We had previously shown that the FYN SH2 domain can bind directly to the first ITSM of CD244 with an affinity that would compete with other interactions of this motif (16). We compared binding of SRC kinases FYN and LCK to the first ITSM motif of human CD244. Phosphorylated peptide based on the first ITSM motif of CD244 was tested as above with CD2 peptides with soluble FYN SH3-SH2 and LCK SH3-SH2 (Fig. 3, e–f). FYN SH3-SH2 had a 10-fold higher affinity for CD244, KD ∼ 0.2 μm, than LCK SH3-SH2, KD ∼ 2 μm. The same ratio was observed for FYN binding human EAT-2 with LCK being 10-fold weaker. LCK binding was consistently an order of magnitude weaker to all peptides that may be physiologically significant or a measure of protein activity. Binding of FYN to a phosphorylated peptide representing the tail of human EAT-2 was ∼5-fold weaker than to the CD244 ITSM. Negligible binding was observed to a peptide based on CTLA-4, which shows FYN has specificity in binding.
Interactions of the Tail of EAT-2
The 5-fold greater affinity of FYN binding to CD244 compared with binding to EAT-2 brings into question whether FYN is the dominant physiological ligand for EAT-2. Recruitment of EAT-2 by CD244 requires phosphorylation of CD244 and the tail of EAT-2 requires phosphorylation for its functional effects (35). This would suggest that a more downstream effector may play a role in its function. To identify any novel interactions of the tail of human EAT-2, a phosphorylated peptide based on the single tyrosine in the tail of human EAT-2 at position 127, which is conserved in mouse, was anchored to streptavidin beads and used to isolate interacting partners from human T blast lysates. Associated proteins were analyzed by mass spectrometry. The results showed that the protein with the highest score for percent coverage was human phospholipase C-γ (PLCγ1) with a score of 135. The only other protein with a significant score for coverage was LCK with a score of 70, which we have previously shown to be unlikely to interact directly with EAT-2 (Fig. 3f) and was probably pulled down indirectly via PLCγ1.
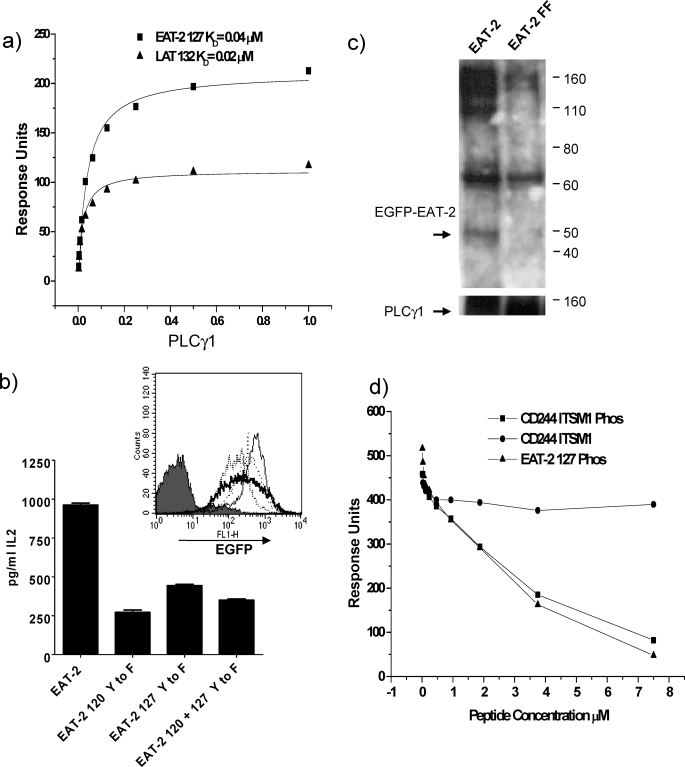
To determine whether this interaction was direct we tested recombinant human PLCγ1 SH2 N+C (a recombinant protein containing both SH2 domains of the protein), for binding to the phosphorylated peptide based on human EAT-2 (EAT-2 127). PLCγ1 bound EAT-2 127 with an affinity of KD ∼ 0.04 μm (Fig. 4a) comparable with binding to a LAT peptide, KD ∼ 0.02 μm (Fig. 4a),4 which has previously been shown to interact with PLCγ1 (50). This suggests that the interaction is physiological and is a ∼30-fold increase in affinity compared with FYN binding EAT-2 127 (Fig. 3e).
FIGURE 4.
Interactions of the tail of EAT-2. a, equilibrium dissociation constants were determined as described in the legend to Fig. 3 for PLCγ1 SH2 N+C binding to immobilized biotinylated synthetic peptides based on the phosphorylated tail of human EAT-2 and LAT (see Table 1). Lines show best fits. b, effects of EAT-2 on IL-2 production by transduced hybridoma cells in response to 1 μm MCC peptide presented by CHO IEK cells. 2B4 Reay T hybridoma cells transduced with mouse EAT-2 EGFP or mutated EAT-2 EGFP with tyrosine substitution of Tyr to Phe at either amino acid 120 or 127 or 120 and 127. Flow cytometric analysis (inset) levels of EGFP expression as an indication of EAT-2 levels of wild type EAT-2 (bold line), 120 (Tyr to Phe) mutant (thin line), 127 (Tyr to Phe) mutant (dotted line), 120 + 127 (Tyr to Phe) double mutant (dashed line), and untransduced (gray shading). The experiment shown is representative of two. Production of IL-2 in the absence of EAT-2 in the system has been shown in Ref. 16. c, immunoprecipitation of PLCγ1 from 2B4 Reay cells transduced with EGFP EAT-2 or EGFP EAT-2 120 + 127 (Tyr to Phe) double mutant (FF). Immunoprecipitates were blotted for EAT-2 and PLCγ1. d, FYN-SH3-SH2 (5 μm) was passed over immobilized proline-rich peptides based on CD2 aa 281–305 and 322–339 (see Table 1) in the presence of various concentrations of phosphorylated peptide based on EAT-2 127 or CD244 ITSM1 or a non-phosphorylated CD244 ITSM1 peptide as a control. FYN-SH3-SH2 binding to CD2 is inhibited by occupancy of its SH2 domain by soluble phosphorylated peptide.
The Tyrosine Motifs in the Tail of Mouse EAT-2 Are Important for Function
Recruitment with relatively high affinity of the effector enzyme, PLCγ1, to EAT-2 could explain the dramatic enhancement of antigen-specific IL-2 production that we observed with expression of mouse EAT-2 in the T cell hybridoma model (16). We had previously compared the effects of transduced enhanced green fluorescent protein (EGFP) and fusion proteins with mouse SAP and mouse EAT-2 on antigen-specific IL-2 production in the hybridoma model and shown that EAT-2 specifically had a significant effect in this model. As the effect of EAT-2 was observed independently of CD244 binding CD48 on antigen-presenting cells, we tested the effect of EAT-2 mutants in 2B4 Reay T cell hybridoma cells without further manipulation and relied on endogenous receptors for recruitment of EAT-2. Of the CD244-related receptors, these cells express at least CD150 (SLAM) (16). To see if enhancement by EAT-2 was dependent on tyrosine motifs in the tail of EAT-2 we produced mutants with a tyrosine substitution (Tyr to Phe) at positions 120 or 127 and a double mutant 120 + 127 (Tyr to Phe) fused with EGFP and compared them with responses by cells expressing wild type EAT-2 EGFP fusion protein. These were transduced into 2B4 Reay T hybridoma cells and tested for expression by fluorescence of EGFP by flow cytometry (Fig. 4b, inset). Antigen-specific IL-2 was measured in response to CHO IEk cells and the MCC peptide. IL-2 production was markedly decreased in cells expressing mutant forms of EAT-2 as compared with wild type EAT-2 (Fig. 4b). This shows that the enhancement is mediated through tyrosine motifs in the tail of EAT-2 and is consistent with recruitment of a positive effector enzyme such as PLCγ1.
To provide evidence that the native proteins interact in cells, we isolated complexes of PLCγ1 and phosphorylated EAT-2 from these cells (Fig. 4c). Immunoprecipitation of PLCγ1 from pervanadate-treated 2B4 Reay cells transduced with EGFP-EAT-2 showed that co-precipitation of EAT with PLCγ1 was dependent on tyrosine residues in the tail of EAT-2 (Fig. 4c). PLCγ1 did not co-precipitate a mutant form of EAT-2 with tyrosine substitutions to phenylalanine at positions 120 and 127 (Fig. 4c).
EAT-2 Tail Inhibits the FYN SH3 Domain Binding to Proline-rich Motifs
EAT-2 has been shown to have an active inhibitory effect when overexpressed in mice, which is dependent on tyrosine residues in its tail (35). As we have that shown positive regulators bind to these tyrosine residues we sought an explanation for these effects given the interactions determined. The structural conformation of the FYN SH3 domain has been shown to be affected by occupancy of its SH2 domain by phosphorylated peptides (51, 52). We used a novel technique employing surface plasmon resonance to determine whether binding of a phosphorylated human EAT-2 127 peptide or human CD244 ITSM1 peptide to the SH2 domain of FYN-SH3-SH2 affected binding of the SH3 domain to proline-rich peptides from CD2. Increasing the concentration of phosphorylated EAT-2 127 or CD244 ITSM1 dramatically decreased binding of FYN-SH3-SH2 to the CD2 peptides (Fig. 4d). The decrease in binding correlated with the increase in peptide concentration and was specifically due to phosphorylation of the peptide as no decrease in binding was seen with the unphosphorylated control peptide. This demonstrates that binding of a ligand to the SH2 domain of FYN can affect binding of its SH3 domain.
DISCUSSION
Cell Surface Connections
Dissection of how the molecular interactions of CD2 and CD244 contribute to the functional outcome has allowed us to resolve differences between mouse and human CD244. We have demonstrated that both CD2 and CD244 signal in a positive way in rodents using a hybridoma model. This approach has also allowed the dissection of the key difference between mouse and human CD244 and CD2, which is engagement at the cell surface by the same and different ligands, respectively.
Competition at the cell surface between mouse CD2 and CD244 for their ligand CD48 can account for the inhibitory effects seen in mouse models for studying the function of CD244. From initial studies in CD244-deficient mice showing enhanced NK killing of target cells when CD244 is not engaged by CD48, conclusions were made that CD244 was mediating inhibitory signaling (27). These did not correlate with later studies using SAP-deficient mice that highlighted the importance of CD244 and SAP in clearance of CD48+ve tumors (21).
As we have demonstrated using antibody blocking experiments in the mouse, if CD244 does not engage CD48 it favors CD2 engagement resulting in increased responses to antigen. This model can be applied to data from cytotoxic assays and blocking antibodies to dissect the function of mouse CD244. The increase in cytotoxicity could be a result of increased CD2 engagement, which would be even more evident if CD244 was not signaling fully as we have observed in the hybridoma model. The same would be true in mice lacking CD244 as there would be no competition for CD48, which would favor CD2 ligation. The positive and negative effects of murine CD244 have been attributed to long and short forms of the molecule with the long form being inhibitory and the short form being activating (30). In experiments where the long and short forms were transfected into cells, expression of the long form restored inhibition in 2B4−ve LAK cells (27). However, the long form expression was higher than the short form, which would fit with competitive binding of CD48 with CD2. This is a mechanism as to why CD244 in rodents can appear inhibitory, whereas in humans it is activating. A similar paradigm has also been demonstrated for the inhibitory role of CTLA-4 as it competes with CD28 for ligand at the cell surface (53). Humans only have the long form of CD244 that is stimulatory, have the same number of ITSM motifs as the mouse long form, and have conserved signaling interactions (16). The major difference between mouse and human is that CD2 binds CD58 meaning there is no competition with CD244. Experiments using mAbs in which engagement of human CD244 was claimed to be inhibitory did not examine how the mAbs were working (16, 54). Fab fragments of blocking mAbs revealed that the conclusions drawn with whole IgG were erroneous (16),5 and the data were consistent with human CD244 being an activating receptor (13, 15, 28). Re-examination of data from X-linked proliferative patients also reveals that the mechanism of action claimed to be blocking by mAbs is not rigorously determined (54). Fc effects were not conclusively excluded. Also, CD244 does not correctly localize to cell contact sites in X-linked proliferative patients (55). These points lead to the conclusion that CD244 is simply ineffective rather than actively inhibitory.
Intracellular Connections
The connection between CD244 and CD2 has always been a puzzle as a shared ligand in rodents implies a functional connection yet their cytoplasmic regions differ greatly. The mechanism by which CD244 aids in revealing positive effects of CD2 in the hybridoma model may include extracellular and/or intracellular interactions. In humans the extracellular link has been lost although a recent study has shown that CD244 and CD2 colocalize at synapses (56) implying a functional link is still important. The interaction of the SRC kinases FYN and LCK with CD2 have been widely reported (22, 24, 39). FYN has also been shown to be important for the function of CD244 as shown by FYN knock-out mice (21). As FYN SH3-SH2 bound CD2 with an affinity over 100-fold greater than LCK, it is more likely to be the dominant physiological binding partner unless there were very large differences in local concentrations of the two proteins. A dominance of FYN over LCK in interacting with CD2 physically (47) and functionally (49) has been reported by others. These results suggest that the two closely related receptors still share a common intracellular signaling molecule, with both being able to interact in a distinct way, CD2 by an SH3 interaction and CD244 by an SH2. A model in which there is direct recruitment of FYN to CD2 may be important for activation of FYN resulting in subsequent phosphorylation of CD244 and recruitment of SAP and EAT-2. This is supported by the fact that peptides from CD2 can activate FYN (48) and that CD2 does not need post-translational modifications to be able to function making it the first step in activation. Increased signaling by CD2 may enhance CD244 expression in the hybridoma, which may reflect the sequence of events on activation of normal cells. Any model of FYN binding CD2 must take into account other interactions mediated by CD2 and their physiological importance. CD2AP in rodents or CMS and CIN85 in humans also bind at the membrane distal proline-rich region 322–339 (23, 25) with a 10-fold weaker affinity than FYN. As CMS and CIN85 are dimeric the functional avidity would be increased considerably making it likely that competition with FYN would occur, particularly as we found that mutating the same 2 residues in the CD2 peptide abrogated binding of both FYN and CMS. A requirement for a link to the cytoskeleton (25) is lost on activation (57) possibly allowing FYN to bind. The CD244 binding adaptor SAP also binds FYN and this binding site on the SH3 domain of FYN is overlapped with the autoinhibitory binding site within FYN with which CD2 has also been shown to compete (31, 48). This raises the possibility that an exchange of interactions may occur on both CD2 and CD244 as EAT-2 and SAP have the potential to competitively bind CD244 (16). In keeping with the possibility of an exchange of interactions, the SAP/FYN interaction has been well characterized (31) but there are other contenders for binding to SAP (58, 59).
Our finding that PLCγ1 is a candidate partner for EAT-2 provides a molecular basis for the role of CD244 in NK cells. CD244 is important for NK killing as are the isoforms of phospholipase C-γ (60–63). In humans, EAT-2 has been shown to be critical for CRACC and NTB-A-mediated cytotoxicity and for their phosphorylation (20, 64, 65). Our data showing that PLCγ1 binds the tail of EAT-2 is consistent with these findings. Tyrosine residues in the tail of EAT-2 are crucial for the positive effect of EAT-2 on IL-2 production consistent with PLCγ1 being recruited. PLCγ1 is likely to compete with FYN for binding to EAT-2, but we cannot rule out some interaction of EAT-2 with FYN. In humans EAT-2 is expressed at the protein level at a steady state before and after activation, whereas SAP expression peaks after activation (28). One possibility is that SAP displaces EAT-2 and thus controls signaling during an immune response.
An active inhibitory role has been proposed for EAT-2 based on experiments using EAT-2-deficient and transgenic mice (35). Lack of EAT-2 may result in excessive SAP recruitment giving the impression that EAT-2 was having an active inhibitory effect. Overexpression of EAT-2 could lead to a high level of phosphorylated ligand for the FYN SH2 domain. This may result in a decrease in the binding of its SH3 domain as we demonstrated by surface plasmon resonance (Fig. 4d). As the binding site on the SH3 domain of FYN overlaps with the autoinhibitory site within FYN (31) for which CD2 competes (48) it is reasonable to assume that overexpression of EAT-2 could result in a loss of binding to SAP and CD2 by FYN. This mechanism is consistent with the lack of phosphorylation seen in mice overexpressing EAT-2 (35).
In conclusion the functional outcome of CD244 can be resolved as it acts solely in an activating manner once confusion over competition for ligand is ruled out. PLCγ1 is likely to be a key contributor to activation by CD244.
Acknowledgments
We thank Neil Barclay for scientific support and critical reading of the manuscript, Erica Lacey and Steve Simmonds for technical assistance, Dhaval Sangani for the LAT peptide, Ben Thomas for mass spectrometry, and Anton van der Merwe for helpful discussion.
This work was supported by Medical Research Council Grant G0400808.
N. G. Clarkson, unpublished data.
D. Sangani, unpublished data.
S. Simmonds, unpublished data.
- ITSM
- immunoreceptor tyrosine-based switch motif
- CD2AP
- CD2-associated protein
- CHO
- Chinese hamster ovary
- CIN85
- Cbl interating protein of 85 kDa
- CMS
- p130Cas ligand with multiple SH3 domains
- CTLA-4
- cytotoxic T-lymphocyte antigen 4
- EAT-2
- Ewing's sarcoma-activated transcript 2
- FYN
- FYN oncogene related to SRC
- GST
- glutathione S-transferase
- LAT
- linker for activation of T cells
- LCK
- lymphocyte-specific protein-tyrosine kinase
- MCC
- moth cytochrome c
- PLCγ1
- phospholipase C-γ1
- SLAM
- signaling lymphocytic activation molecule
- SAP
- SLAM-associated protein
- SH
- Src homology
- EGFP
- enhanced green fluorescent protein
- mAb
- monoclonal antibody
- IL
- interleukin
- aa
- amino acid(s).
REFERENCES
- 1.Nichols K. E., Ma C. S., Cannons J. L., Schwartzberg P. L., Tangye S. G. (2005) Immunol. Rev. 203, 180–199 [DOI] [PubMed] [Google Scholar]
- 2.Engel P., Eck M. J., Terhorst C. (2003) Nat. Rev. Immunol. 3, 813–821 [DOI] [PubMed] [Google Scholar]
- 3.Schwartzberg P. L., Mueller K. L., Qi H., Cannons J. L. (2009) Nat. Rev. Immunol. 9, 39–46 [DOI] [PubMed] [Google Scholar]
- 4.Cunninghame Graham D. S., Vyse T. J., Fortin P. R., Montpetit A., Cai Y. C., Lim S., McKenzie T., Farwell L., Rhodes B., Chad L., Hudson T. J., Sharpe A., Terhorst C., Greenwood C. M., Wither J., Rioux J. D. (2008) Genes Immun. 9, 93–102 [DOI] [PubMed] [Google Scholar]
- 5.Tangye S. G., Phillips J. H., Lanier L. L., Nichols K. E. (2000) J. Immunol. 165, 2932–2936 [DOI] [PubMed] [Google Scholar]
- 6.Suzuki A., Yamada R., Kochi Y., Sawada T., Okada Y., Matsuda K., Kamatani Y., Mori M., Shimane K., Hirabayashi Y., Takahashi A., Tsunoda T., Miyatake A., Kubo M., Kamatani N., Nakamura Y., Yamamoto K. (2008) Nat. Genet. 40, 1224–1229 [DOI] [PubMed] [Google Scholar]
- 7.Howie D., Laroux F. S., Morra M., Satoskar A. R., Rosas L. E., Faubion W. A., Julien A., Rietdijk S., Coyle A. J., Fraser C., Terhorst C. (2005) J. Immunol. 174, 5931–5935 [DOI] [PubMed] [Google Scholar]
- 8.Veillette A. (2006) Nat. Rev. Immunol. 6, 56–66 [DOI] [PubMed] [Google Scholar]
- 9.Davis S. J., Ikemizu S., Wild M. K., van der Merwe P. A. (1998) Immunol. Rev. 163, 217–236 [DOI] [PubMed] [Google Scholar]
- 10.Brown M. H., Boles K., van der Merwe P. A., Kumar V., Mathew P. A., Barclay A. N. (1998) J. Exp. Med. 188, 2083–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boles K. S., Nakajima H., Colonna M., Chuang S. S., Stepp S. E., Bennett M., Kumar V., Mathew P. A. (1999) Tissue Antigens 54, 27–34 [DOI] [PubMed] [Google Scholar]
- 12.Garni-Wagner B. A., Purohit A., Mathew P. A., Bennett M., Kumar V. (1993) J. Immunol. 151, 60–70 [PubMed] [Google Scholar]
- 13.Tangye S. G., Cherwinski H., Lanier L. L., Phillips J. H. (2000) Mol. Immunol. 37, 493–501 [DOI] [PubMed] [Google Scholar]
- 14.Boles K. S., Stepp S. E., Bennett M., Kumar V., Mathew P. A. (2001) Immunol. Rev. 181, 234–249 [DOI] [PubMed] [Google Scholar]
- 15.Valiante N. M., Trinchieri G. (1993) J. Exp. Med. 178, 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarkson N. G., Simmonds S. J., Puklavec M. J., Brown M. H. (2007) J. Biol. Chem. 282, 25385–25394 [DOI] [PubMed] [Google Scholar]
- 17.Morra M., Lu J., Poy F., Martin M., Sayos J., Calpe S., Gullo C., Howie D., Rietdijk S., Thompson A., Coyle A. J., Denny C., Yaffe M. B., Engel P., Eck M. J., Terhorst C. (2001) EMBO J. 20, 5840–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tangye S. G., Lazetic S., Woollatt E., Sutherland G. R., Lanier L. L., Phillips J. H. (1999) J. Immunol. 162, 6981–6985 [PubMed] [Google Scholar]
- 19.Chen R., Relouzat F., Roncagalli R., Aoukaty A., Tan R., Latour S., Veillette A. (2004) Mol. Cell. Biol. 24, 5144–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tassi I., Colonna M. (2005) J. Immunol. 175, 7996–8002 [DOI] [PubMed] [Google Scholar]
- 21.Bloch-Queyrat C., Fondanèche M. C., Chen R., Yin L., Relouzat F., Veillette A., Fischer A., Latour S. (2005) J. Exp. Med. 202, 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell G. M., Fargnoli J., Bolen J. B., Kish L., Imboden J. B. (1996) J. Exp. Med. 183, 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dustin M. L., Olszowy M. W., Holdorf A. D., Li J., Bromley S., Desai N., Widder P., Rosenberger F., van der Merwe P. A., Allen P. M., Shaw A. S. (1998) Cell 94, 667–677 [DOI] [PubMed] [Google Scholar]
- 24.Freund C., Kühne R., Yang H., Park S., Reinherz E. L., Wagner G. (2002) EMBO J. 21, 5985–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchings N. J., Clarkson N., Chalkley R., Barclay A. N., Brown M. H. (2003) J. Biol. Chem. 278, 22396–22403 [DOI] [PubMed] [Google Scholar]
- 26.Li J., Nishizawa K., An W., Hussey R. E., Lialios F. E., Salgia R., Sunder-Plassmann R., Reinherz E. L. (1998) EMBO J. 17, 7320–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K. M., McNerney M. E., Stepp S. E., Mathew P. A., Schatzle J. D., Bennett M., Kumar V. (2004) J. Exp. Med. 199, 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Endt J., Eissmann P., Hoffmann S. C., Meinke S., Giese T., Watzl C. (2007) Eur. J. Immunol. 37, 193–198 [DOI] [PubMed] [Google Scholar]
- 29.Lee K. M., Forman J. P., McNerney M. E., Stepp S., Kuppireddi S., Guzior D., Latchman Y. E., Sayegh M. H., Yagita H., Park C. K., Oh S. B., Wülfing C., Schatzle J., Mathew P. A., Sharpe A. H., Kumar V. (2006) Blood 107, 3181–3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stepp S. E., Schatzle J. D., Bennett M., Kumar V., Mathew P. A. (1999) Eur. J. Immunol. 29, 2392–2399 [DOI] [PubMed] [Google Scholar]
- 31.Chan B., Lanyi A., Song H. K., Griesbach J., Simarro-Grande M., Poy F., Howie D., Sumegi J., Terhorst C., Eck M. J. (2003) Nat. Cell Biol. 5, 155–160 [DOI] [PubMed] [Google Scholar]
- 32.Latour S., Roncagalli R., Chen R., Bakinowski M., Shi X., Schwartzberg P. L., Davidson D., Veillette A. (2003) Nat. Cell Biol. 5, 149–154 [DOI] [PubMed] [Google Scholar]
- 33.Chlewicki L. K., Velikovsky C. A., Balakrishnan V., Mariuzza R. A., Kumar V. (2008) J. Immunol. 180, 8159–816718523281 [Google Scholar]
- 34.Eissmann P., Beauchamp L., Wooters J., Tilton J. C., Long E. O., Watzl C. (2005) Blood 105, 4722–4729 [DOI] [PubMed] [Google Scholar]
- 35.Roncagalli R., Taylor J. E., Zhang S., Shi X., Chen R., Cruz-Munoz M. E., Yin L., Latour S., Veillette A. (2005) Nat. Immunol. 6, 1002–1010 [DOI] [PubMed] [Google Scholar]
- 36.Nakajima H., Cella M., Langen H., Friedlein A., Colonna M. (1999) Eur. J. Immunol. 29, 1676–1683 [DOI] [PubMed] [Google Scholar]
- 37.Wild M. K., Cambiaggi A., Brown M. H., Davies E. A., Ohno H., Saito T., van der Merwe P. A. (1999) J. Exp. Med. 190, 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan N. J., Simmonds S. J., Clarkson N. G., Hanrahan S., Puklavec M. J., Bomb M., Barclay A. N., Brown M. H. (2006) Mol. Cell. Biol. 26, 6727–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell G. M., Bolen J. B., Imboden J. B. (1992) Mol. Cell. Biol. 12, 5548–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang H. C., Moingeon P., Pedersen R., Lucich J., Stebbins C., Reinherz E. L. (1990) J. Exp. Med. 172, 351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Q., Beyers A. D., Barclay A. N., Williams A. F. (1988) Cell 54, 979–984 [DOI] [PubMed] [Google Scholar]
- 42.Nishizawa K., Freund C., Li J., Wagner G., Reinherz E. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14897–14902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohno H., Nakamura T., Yagita H., Okumura K., Taniguchi M., Saito T. (1991) J. Immunol. 147, 2100–2106 [PubMed] [Google Scholar]
- 44.Brown M. H., Sewell W. A., Mason D. Y., Rothbard J. B., Crumpton M. J. (1988) Eur. J. Immunol. 18, 1223–1227 [DOI] [PubMed] [Google Scholar]
- 45.Watzl C., Stebbins C. C., Long E. O. (2000) J. Immunol. 165, 3545–3548 [DOI] [PubMed] [Google Scholar]
- 46.Carmo A. M., Mason D. W., Beyers A. D. (1993) Eur. J. Immunol. 23, 2196–2201 [DOI] [PubMed] [Google Scholar]
- 47.Lin H., Hutchcroft J. E., Andoniou C. E., Kamoun M., Band H., Bierer B. E. (1998) J. Biol. Chem. 273, 19914–19921 [DOI] [PubMed] [Google Scholar]
- 48.Holdorf A. D., Green J. M., Levin S. D., Denny M. F., Straus D. B., Link V., Changelian P. S., Allen P. M., Shaw A. S. (1999) J. Exp. Med. 190, 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukai I., Hussey R. E., Sunder-Plassmann R., Reinherz E. L. (2000) Eur. J. Immunol. 30, 3507–3515 [DOI] [PubMed] [Google Scholar]
- 50.Zhang W., Trible R. P., Zhu M., Liu S. K., McGlade C. J., Samelson L. E. (2000) J. Biol. Chem. 275, 23355–23361 [DOI] [PubMed] [Google Scholar]
- 51.Panchamoorthy G., Fukazawa T., Stolz L., Payne G., Reedquist K., Shoelson S., Songyang Z., Cantley L., Walsh C., Band H. (1994) Mol. Cell. Biol. 14, 6372–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hofmann G., Schweimer K., Kiessling A., Hofinger E., Bauer F., Hoffmann S., Rösch P., Campbell I. D., Werner J. M., Sticht H. (2005) Biochemistry 44, 13043–13050 [DOI] [PubMed] [Google Scholar]
- 53.Carreno B. M., Bennett F., Chau T. A., Ling V., Luxenberg D., Jussif J., Baroja M. L., Madrenas J. (2000) J. Immunol. 165, 1352–1356 [DOI] [PubMed] [Google Scholar]
- 54.Parolini S., Bottino C., Falco M., Augugliaro R., Giliani S., Franceschini R., Ochs H. D., Wolf H., Bonnefoy J. Y., Biassoni R., Moretta L., Notarangelo L. D., Moretta A. (2000) J. Exp. Med. 192, 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dupré L., Andolfi G., Tangye S. G., Clementi R., Locatelli F., Aricò M., Aiuti A., Roncarolo M. G. (2005) Blood 105, 4383–4389 [DOI] [PubMed] [Google Scholar]
- 56.Schleinitz N., March M. E., Long E. O. (2008) PLoS ONE 3, e3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Douglass A. D., Vale R. D. (2005) Cell 121, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li C., Schibli D., Li S. S. (2009) Cell. Signal. 21, 111–119 [DOI] [PubMed] [Google Scholar]
- 59.Gu C., Tangye S. G., Sun X., Luo Y., Lin Z., Wu J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caraux A., Kim N., Bell S. E., Zompi S., Ranson T., Lesjean-Pottier S., Garcia-Ojeda M. E., Turner M., Colucci F. (2006) Blood 107, 994–1002 [DOI] [PubMed] [Google Scholar]
- 61.Chuang S. S., Kumaresan P. R., Mathew P. A. (2001) J. Immunol. 167, 6210–6216 [DOI] [PubMed] [Google Scholar]
- 62.Upshaw J. L., Schoon R. A., Dick C. J., Billadeau D. D., Leibson P. J. (2005) J. Immunol. 175, 213–218 [DOI] [PubMed] [Google Scholar]
- 63.Tassi I., Presti R., Kim S., Yokoyama W. M., Gilfillan S., Colonna M. (2005) J. Immunol. 175, 749–754 [DOI] [PubMed] [Google Scholar]
- 64.Eissmann P., Watzl C. (2006) J. Immunol. 177, 3170–3177 [DOI] [PubMed] [Google Scholar]
- 65.Cruz-Munoz M. E., Dong Z., Shi X., Zhang S., Veillette A. (2009) Nat. Immunol. 10, 297–305 [DOI] [PubMed] [Google Scholar]