Abstract
The Yersinia protein kinase A (YpkA) and outer protein J (YopJ) are co-expressed from a single transcript and are injected directly into eukaryotic cells by the plague bacterium Yersinia pestis. When overexpressed in vertebrate or yeast cells, YpkA disrupts the actin-based cytoskeletal system by an unknown mechanism, whereas YopJ obstructs inductive chemokine expression by inhibiting MAPK and NF-κB signaling. Previously, we showed that the fission yeast Schizosaccharomyces pombe was sensitive to the kinase activity of YpkA. Here, we screened yeast for cellular processes important for YpkA activity and found that the eIF2α kinases mollify the toxicity imparted by the kinase activity of YpkA. Specifically, strains lacking the eIF2α kinase Hri2 were particularly sensitive to YpkA. Unexpectedly, the activity of YopJ, which conferred a phenotype consistent with its inhibitory effect on MAPK signaling, was also found to be dependent on Hri2. When expressed in S. pombe, YopJ sensitized cells to osmotic and oxidative stresses through a Hri2-dependent mechanism. However, when co-expressed with YpkA, YopJ protected cells from YpkA-mediated toxicity, and this protection was entirely dependent on Hri2. In contrast, YopJ did not confer protection against the toxic effects of the Yersinia virulence factor YopE. These findings are the first to functionally link YpkA and YopJ and suggest that eIF2α kinases, which are critically important in antiviral defenses and protection against environmental stresses, also play a role in bacterial virulence.
The plague bacterium Yersinia pestis, as well as the closely related enteric pathogens Yersinia pseudotuberculosis and Yersinia enterocolitica, are relatively resistant to the antimicrobial killing systems of innate immune cells. This property is largely dependent on a membrane-bound type 3 secretion system (T3SS)2 that injects proteins (here referred to as “effectors”) directly into the eukaryotic cell (1, 2). This study is focused on two such effectors: YpkA and YopJ (YopO and YopP in Y. enterocolitica). Although they are encoded by a single transcription unit on the 70-kb extrachromosomal virulence plasmid of Yersinia, these two effectors shape the bacterial host relationship in very different ways. YpkA and its associated kinase activity is necessary for the immediate survival of Yersinia following attachment to host cells; this was suggested by animal infection experiments and confirmed at the cellular level in a function-based “infectivity” assay that measures both survival and growth of macrophage-associated Yersinia (3, 4). In contrast, there are no discernible differences in the infectivity assay between the wild-type and ΔyopJ mutant strains (5). However, YopJ very efficiently blocks MAPK- and NF-κB-mediated signaling pathways and the resulting inductive expression of proinflammatory chemokines; the consequences of YopJ activity during an actual infection is a reduction in the local inflammatory response (6–10). Therefore, unlike YpkA, which is important for the immediate survival of Yersinia during its encounter with macrophages, YopJ acts to modulate the immune response at the systems level.
These effectors also differ as to how they have been studied. The enzymatic activity of YpkA was immediately recognized based on its striking similarity (residues 136–408) to eukaryotic Ser/Thr protein kinases (11). YpkA also contains a GTPase-binding domain (residues ∼442–721) that was independently identified bioinformatically and by yeast two-hybrid screening (12, 13). Potential cellular substrates have been identified for YpkA using in vitro phosphorylation and transfection systems (14, 15), but it is currently unknown whether these cellular factors are targeted during infection at normal YpkA expression levels. YpkA has been widely characterized as a “cytoskeletal disruptor” based, again, on either transfection systems or infection studies in which YpkA is overexpressed in trans in the absence of the other 5 Yops; predictably this activity is primarily dependent on the GTPase-binding domain, and, to a much lesser degree, on its kinase activity (12, 13, 16). Similarly, there has been considerable uncertainty concerning YopJs mechanism despite the fact that it has very potent activities in cell culture models. Using transfection models, several different cellular activities have been attributed to YopJ, including deubiquination of IκBα and tumor necrosis factor receptor-associated factors, and most recently, acetylation of MKKs (17–21). Whether any of these activities (which are not mutually exclusive) occur when YopJ is introduced into cells via the T3SS following infection is unknown. Therefore, like YpkA, precisely how YopJ exerts its activity during infection is presently unclear.
We believe that progress in understanding the cellular activities of T3SS effectors expressed by animal pathogens has been limited by a lack of discovery-based models. Cell culture-based models have been successfully developed for the discovery of bacterial virulence genes. However, there has been little success in developing screens to identify critical host factors and/or processes in such cell culture-based infection systems. This is in sharp contrast to studies of T3SS effectors expressed by plant pathogens in which enormous strides have been made due to the fact that these studies are founded on genetic systems dating back to the 1950s (22). In the past several years, it has been shown that a number of T3SS effectors expressed by animal pathogens are active in yeast cells (4, 23–26); however, to the best of our knowledge, there has only been a single report of using yeast to identify previously unknown host factors important for T3SS effector activity (27). Here, we used the fission yeast Schizosaccharomyces pombe to screen for host factors important for YpkA activity and unexpectedly identified a cellular process in which YpkA and YopJ activities intersect.
EXPERIMENTAL PROCEDURES
Yeast Strains
Yeast strains are listed in the order of their presentation: 574W, h+ ade6-M216 leu1-32 ura4-D18 pJK148 nmt1-eGFP-ypkA pUR19; 589W, h+ mut-222 ade6-M216 leu1-32 ura4-D18 pJK148 nmt1-eGFP-ypkA pUR19; 598W, h+ mut-222 ade6-M216 leu1-32 ura4-D18 pJK148 nmt1-eGFP-ypkA pUR19 207; 1000W, h+ mut-222 ade6-M216 leu1-32 ura4-D18 pJK148 nmt1-eGFP-ypkA pUR19 207(−tRNA); 428W, h− ade6-M210 leu1-32 ura4-D18 pRep3X eGFP-ypkA; 781W, h− Δgcn2::ura4+ ura4-D18 leu1-32 pRep3x eGFP-ypkA; 35W, h− ade6–704 leu1-32 ura4-D18 pRep41X eGFP-ypkA; 702W, h− ade6-M210 leu1-32 ura4-D18 eIF2α(S52A):ura4+ pRep41X eGFP-ypkA; 710W, h− ade6-M210 leu1-32 ura4-D18 eIF2α(S52A):ura4+ pRep41X eGFP-ypkA(D270A); 707W, h− ade6-M210 leu1-32 ura4-D18 eIF2α(S52A):ura4+ pRep3X eGFP-ypkA; 739W, Δhri2::ura4+ ura4-D18 leu1-32 ade6–704 pRep3X eGFP-ypkA; 670W, h− ade6-M210 leu1-32 ura4-D18 pRep3X eGFP; 672W, h− ade6-M210 leu1-32 ura4-D18 pRep3X eGFP-yopJ; 639W, h+ leu1-32 ura4-D18 ade6-M210 his7-366 sty1–6HisHA:ura4+ pRep3X eGFP-yopJ; 674W, h− ade6-M210 leu1-32 ura4-D18 pRep3X eGFP-yopJ(C172A); 677W, h− ade6-M210 leu1-32 ura4-D18 pRep3X eGFP-yopJ(58S); 685W, h− ade6-M210 leu1-32 ura4-D18 pRep3X eGFP-yopJ(231S); 772W, h− Δsty1::ura4+ eEF2-3HA:kan ura4-D18 leu1-32; 745W, h− Δhri1:ura4+ ura4-D18 leu1-32 ade6-M210 pRep3X eGFP-yopJ; 753W, h− Δhri2:ura4+ ura4-D18 leu1-32 pRep3X eGFP-yopJ; 790W, h− Δgcn2:ura4+ ura4-D18 leu1-32 pRep3X eGFP-yopJ; 755W, h− ade6-704 ura4-D18 leu1-32 int.pRep6X eGFP-ypkA pRep3X eGFP-yopJ; 759W, h− ade6-704 ura4-D18 leu1-32 int.pRep6X eGFP-ypkA pRep3X eGFP-yopJ(C172A); 806W, h− Δhri2:ura4+ ura4-D18 leu1-32 ade6-704 int.pRep6X eGFP-ypkA pRep3X eGFP-yopJ; 883W, h− Δgcn2:ura4+ ura4-D18 leu1-32 ade6-704 int.pRep6X eGFP-ypkA pRep3X eGFP-yopJ; 891W, h− ura4-D18 leu1-32 ade6–704 int.pRep6X eGFP-yopE pRep3X eGFP-yopJ; and 893W, h− ura4-D18 leu1–32 ade6–704 int.pRep6X eGFP-yopE pRep3X eGFP-yopJ(C172A).
Plasmids
The pRep3X GFP, pRep3X GFP-ypkA, pRep3X GFP-ypkA(D270A), pRep41X GFP-ypkA, and pRep41X GFP-ypkA(D270A) yeast expression plasmids have been described previously (4). Strains possessing an integrated gfp-ypkA were constructed using pJK148 nmt1-GFP-ypkA, pRep6X GFP-ypkA, and pRep6X GFP-ypkA(D270A) (28). The genomic library pURSP1 was purchased from ATCC and used for YpkA hypersensitivity screening. The isolated pUR19 207 clone contained genomic sequences 3589023 to 3592371 of chromosome 1, and the pUR19 207 (-tRNA) derivative was generated by deleting the 474-bp SacI fragment from the right end of pUR19 207. To generate pRep3X GFP-yopJ, the yopJ gene was amplified from the Y. pseudotuberculosis YPIII pIB102 strain, and the resulting fragment was cloned into pRep3X as described previously for pRep3X GFP-ypkA (4). The pRep3X GFP-yopJ(C172A) derivative was generated using QuikChange mutagenesis (Stratagene) and verified by sequencing. Using a transposon-based mutagenesis system (Finnzymes MCS), the gfp-yopJ plasmid was mutated in vitro such that 5-codon nonpolar “scars” were inserted randomly into the plasmid; plasmid was then prepared from 150,000 independent pooled bacterial transformants and used for the YopJ activity screen. The YopJ-HA-encoding Yersinia expression plasmids (and derivatives) were constructed by first amplifying yopJ sequences from the GFP-YopJ-encoding yeast expression plasmids isolated from the plate-based elongation assay and then ligating the resulting fragments downstream of the yopE promoter as previously described (8).
Screening
To select for YpkA resistant (YpkAR) variants, S. pombe(GFP-YpkA) was first exposed to nitrosoguanidine sufficient to yield a ∼50% kill rate and then plated out on GFP-YpkA inducing media (i.e. lacking thiamine). Of a total of 8 × 106 viable cells, ∼1,500 healthy-looking putative YpkAR colonies were recovered as judged by colony size and the ability to exclude the vital dye Phloxine B (included in the media and serves as a general readout for metabolic activity). To eliminate clones in which YpkA had been inactivated during the mutagenesis (as well as those clones with a dominant phenotype), putative YpkAR clones were crossed with the parental wild-type strain, and the resulting diploids were selected and maintained by intra-allelic complementation. The diploids were then replica-plated to media inductive for GFP-YpkA expression. A total of five diploids displayed the wild-type-like YpkA sensitive (YpkAS) phenotype, of which, 511W, was further examined and appeared to have a single locus (i.e. in the haploid parent) conferring the YpkAR phenotype. 511W was characterized with a slow growth rate and was used as a recipient for a YpkA hypersensitivity screen. Accordingly, 511W was transformed with a S. pombe-derived genomic library pURSP1 and 18,500 transformants were screened for YpkAS by replica plating on media with and without thiamine. Two different plasmids were recovered multiple times, one of which was p207; p207 conferred YpkAS to 511W but did not restore the wild-type growth rate. To isolate inactive YopJ variants, we developed a plate-based screening method that involved adjusting the salt and YopJ expression levels such that both yopJ+ and yopJ − strains would form colonies that could then be examined by low power magnification to ascertain the relative activity of YopJ; transformants expressing an active YopJ would have cells along the outer edge of the colony that were elongated in contrast to transformants expressing an inactive YopJ, in which case the cells would be notably shorter in length. S. pombe transformed with the pooled transposon mutant library described above were plated on media containing 400 mm KCl and of the primary colonies (∼19,000) microscopically inspected, 298 displayed a phenotype suggestive that YopJ was inactive (shortened cells), and of these, 133 maintained this phenotype upon restreaking as well as scoring positive for GFP expression. By examining the recovered plasmids, ∼80% contained the scar within the YopJ-encoding sequence.
YpkA and YopJ Activity Assays
For the “ghost” assay measuring YpkA activity, cells were grown exponentially (A595 < 0.4) for at least eight generations. Cells were then washed to remove thiamine and allowed to grow under inducing GFP-YpkA expressing conditions (without thiamine) for 11.5 h, and then diluted to A595 of 0.1. At various times, a sample of the culture was removed and concentrated by centrifugation, and the cells were resuspended in 1% SDS. Greater than 500 cells were examined as to whether they were intact or had lysed (i.e. formed ghosts). Cultures were never allowed to exceed an A595 of 0.4 during either the preinduction or induction phases of the assay. For the elongation assay measuring YopJ activity, cells were propagated and induced as described for the ghost assay except that after 16 h of induction cultures were diluted to A595 of 0.1 in media containing 1 m KCl. Cultures were not allowed to exceed an A595 of 0.4, and at the indicated time points, 9 ml of culture was removed, and then the cells were fixed with formaldehyde and stained with calcofluor (4). Pictures of the stained cells were recorded with a fluorescent microscope, and the lengths of the septum-positive cells were quantified using ImageJ (available from NIH). For the H2O2 sensitivity assay, cells were propagated as described above, and after 16 h of induction for GFP-YopJ expression cells were diluted to an A595 of 0.1 in the presence of 1 mm H2O2 (0.003%). After either 30 or 60 mins of H2O2 exposure, cells were collected and analyzed for either protein content or by microscopy (Fig. 5, A and B, respectively). For intracellular staining, infected cells were prepared using the Cytofix/Cytoperm kit and stained with rat anti-mouse tumor necrosis factor α Alexa Fluor 647 (Pharmingen).
FIGURE 5.
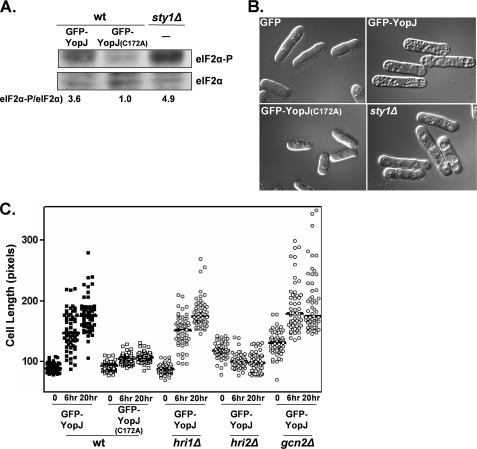
YopJ activity in S. pombe following oxidative and osmotic stress. A, actively growing cultures of the wild-type (wt) strain transformed with plasmids encoding either GFP-YopJ (672W), GFP-YopJ(C172A) (674W), or the untransformed sty1Δ strain (772W) were exposed to 1 mm H2O2 for 30 min. The resulting whole cell extracts were fractionated by SDS-PAGE, and the levels of total and phosphorylated eIF2α were determined by Western analysis. Signals were quantified and normalized to the eIF2α-P/eIF2α ratio observed in the extract derived from the GFP-YopJ(C172A) strain. B, live cells of the same strains were imagined following a 60-min exposure to 1 mm H2O2. C, cell elongation assay of the indicated strain (closed boxes, 672W; open boxes, 674W; open circles, 745W, 753W, and 790W) was performed as described in Fig. 3C.
Protein Methods
For detection of GFP hybrids and eIF2α, cells were lysed in H buffer (25 mm MOPS, pH 7.2, 80 mm β-glycerophosphate, 15 mm p-nitrophenylphosphate, 15 mm MgCl2, 15 mm EGTA, 1 mm dithiothreitol, 0.1 m sodium vanadate, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and additional protease inhibitors Sigma (P8215)) supplemented with 2% SDS using a FastPrep FP120 bead beater (MP Biomedicals), and the resulting extracts were boiled, clarified, and quantified before fractionation by SDS-PAGE and analysis by Western blotting using either anti-GFP (Chemicon), anti-eIF2α (Invitrogen), or phospho-specific anti-eIF2α (Invitrogen). For determination of phospho-eIF2α/eIF2α ratios, signals were quantified using ImageJ (rsb.info.nih.gov/ij). For Sty1-HA purification, cells were lysed in H buffer (without SDS) as described, and anti-HA antibody (Covance) was then added to the clarified lysate. After 1 h at 4 °C, protein G magnetic resin (New England Biolabs) was added, and following an additional 30 min of incubation, the resin was washed several times with H buffer, and the bound proteins were then eluted with SDS-loading buffer. The resulting Western blots were probed with anti-HA and anti-phosphotyrosine antibodies (Covance and Zymed Laboratories Inc., respectively).
RESULTS
YpkA Activity and eIF2α
Previously, we showed that S. pombe is extremely sensitive to YpkA (4). To identify host genes responsive to YpkA, we mutagenized S. pombe and selected for YpkA resistant (YpkAR) variants (see “Experimental Procedures” for details of the screen and analysis of the YpkAR isolates). One resulting YpkAR isolate, 511W, was subsequently transformed with a genomic library to identify sequences that restored the parent-like YpkA-sensitive (YpkAS) phenotype. Two unique YpkAS-conferring plasmids were recovered in several independent transformants; one of which, designated as p207, forms the basis of this report.
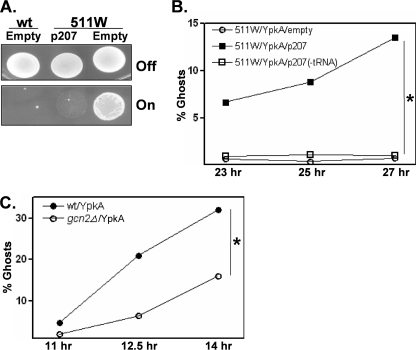
The 3.3-kb genomic insert in p207 conferred complete restoration of the YpkAS phenotype to 511W (Fig. 1A). The presence of p207 had no observable effect on the wild-type or 511W strains when propagated in non-YpkA-expressing conditions, indicating that this sequence was not generally toxic when present at multiple copies. By deletion analysis, it was found that the YpkAS-conferring activity of p207 was entirely dependent on sequences encoding a tRNALeu and its associated promoter (Fig. 1B). Because enhanced tRNA levels naturally occur following nutritional stress and directly activate the eIF2α kinase Gcn2 (29, 30), we tested whether the deletion of gcn2 affected YpkA sensitivity. Compared with wild-type gcn2+ cells, gcn2Δ cells were more resistant to YpkA (Fig. 1C). Collectively, these findings suggest that Gcn2 can modulate YpkA activity; relatively normal Gcn2 activity (wild-type and likely 511W/p207 cells) sensitizes cells to YpkA, whereas reduced Gcn2 activity (gcn2Δ and probably 511W cells) enables cells to better resist YpkA toxicity.
FIGURE 1.
YpkA sensitivity in S. pombe wild-type, 511W, and gcn2Δ cells. A, strain 511W was originally isolated in a mutagenesis screen for S. pombe variants possessing a YpkA-resistance (YpkAR) phenotype. Wild-type (wt) and 511W strains, each with an integrated gfp-ypkA gene, were transformed with either an “empty” control plasmid (strains 574W and 589W, see “Experimental Procedures”) or a plasmid, p207 (strain 598W), which contains a tRNALeu-encoding genomic fragment. Strains were plated on media that were either noninductive (Off) or inductive (On) for GFP-YpkA expression. B, actively growing cultures (A595 between 0.1–0.4) of 511W transformed with either the empty control plasmid (589W), p207 (598W), or a p207 derivative lacking the tRNALeu-encoding sequence (1000W) were shifted to GFP-YpkA-expressing conditions and at the indicated time points thereafter, cells were removed from the culture and mixed with SDS, and the number of lysed cells (ghosts) was determined microscopically. Shown is a representative experiment in which >500 cells were examined per sampling point. Parallel cultures growing in nonexpressing GFP-YpkA conditions contained <0.4% ghosts (not shown). C, the wild-type and gcn2Δ strains (428W and 781W, respectively), each transformed with GFP-YpkA-expressing plasmid driven by high-strength promoter, were assayed for YpkA toxicity as described in B. *, p < 0.05 using a two-sample test to compare proportions in two independent samples.
In addition to functioning as an eIF2α kinase, in S. pombe, Gcn2 also negatively regulates MAPK signaling (31). As has been shown by others (using MAPK mutant strains) as well as in our YopJ-related findings described below, abrogation of MAPK signaling results in enhanced levels of phosphorylated eIF2α (32), thus confounding the relationship between Gcn2 and eIF2α.
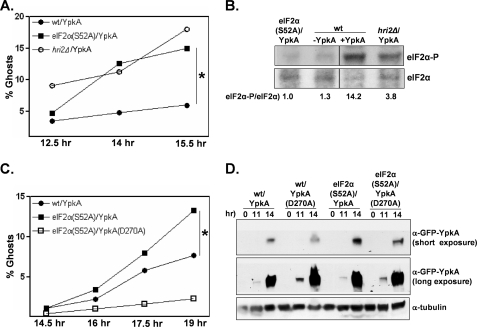
Therefore, to directly determine whether sensitivity to YpkA is modulated by eIF2α, we expressed YpkA in an S. pombe strain expressing a non-phosphorylatable eIF2α (eIF2α(S52A); equivalent to eIF2α(S51A) in vertebrates). We found that eIF2α(S52A) cells were particularly sensitive to YpkA (Fig. 2A). The relative sensitivity in wild-type and eIF2α(S52A) cells was observed at both medium and high GFP-YpkA expression levels (medium expression experiment shown in Fig. 2A). S. pombe possess, in addition to Gcn2, two other eIF2α kinases, Hri1 and Hri2 (33). We expressed GFP-YpkA in strains deleted for either of these kinases and found that YpkA sensitivity in hri1Δ cells was comparable with wild-type cells (data not shown), whereas hri2Δ cells were, like cells expressing eIF2α(S52A), especially sensitive to YpkA (Fig. 2A). The S. pombe Hri2 kinase has previously been shown to be specifically activated by oxidative, heavy metal, and heat-shock stresses (34). Similarly, we found that whereas YpkA expression is associated with heightened levels of phosphorylated eIF2α in extracts prepared from wild-type cultures, there was relatively less YpkA-induced phosphorylation of eIF2α in extracts prepared from the hri2Δ strain (Fig. 2B). In contrast to the hri2Δ strain, extracts prepared from the YpkA-expressing gcn2Δ strain had even higher levels of phosphorylated eIF2α as compared with the wild-type strain (data not shown), thus further strengthening the relationship between phosphorylated eIF2α levels and YpkA sensitivity at the cellular level. Taken together, our findings indicate that the detrimental effects of YpkA are counteracted by Hri2-mediated signaling through eIF2α.
FIGURE 2.
YpkA sensitivity in S. pombe wild-type, eIF2α(S52A), and hri2Δ cells. A, the wild-type (wt), eIF2α(S52A), and hri2Δ strains transformed with plasmids expressing GFP-YpkA driven by a relatively strong promoter (428W, 707W, and 739W, respectively) were subjected to the ghost assay as described in Fig. 1B. B, the indicated strains were propagated and, except where noted, induced for GFP-YpkA expression as described. Cells were collected at 13.5 h, and the resulting whole cell lysates were probed for total and phosphorylated eIF2α by Western analysis. For presentation purposes, the lanes were rearranged. Signals were quantified and normalized to the eIF2α-P/eIF2α ratio observed in the extract derived from the eIF2α(S52A) strain. C, the wild-type (35W) and eIF2α(S52A) (702W and 710W) strains transformed with plasmids expressing either GFP-YpkA or the kinase-inactive GFP-YpkA(D270A) driven by an intermediate-strength promoter were subjected to the ghost assay. D, cells of the transformed strains were collected at the indicated time points, and whole cell lysates were examined by Western analysis. *, p < 0.05 using a two-sample test to compare proportions in two independent samples.
The enhanced toxicity of YpkA in a eIF2α(S52A) background was almost entirely dependent on the kinase activity of YpkA (Fig. 2C), indicating that the increased sensitivity of eIF2α(S52A) cells is not simply due to trans gene overexpression. One explanation for these results is that eIF2α may affect the expression levels of YpkA such that increased “sensitivity” is due to enhanced YpkA expression levels. However, by both fluorescence microscopy and Western analysis, we found no detectable differences in either inductive or steady-state GFP-YpkA levels between wild-type and eIF2α(S52A) cells (Fig. 2D; fluorescence data not shown). These data indicate that the kinase activity of YpkA generates a Hri2-mediated stress-like condition in yeast cells; this is further supported by the experiments described below.
YopJ Activity and eIF2α
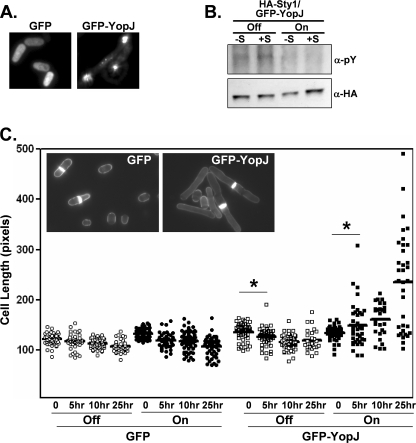
Because translational regulation mediated by eIF2α has been linked to MAPK signaling pathways in a number of experimental models including, as mentioned above, S. pombe (32), we examined whether YopJ, which is expressed from the same transcriptional unit as YpkA, is in any way dependent on eIF2α and its kinases. We therefore expressed GFP-YopJ in S. pombe and found that the hybrid protein was localized in very concentrated clusters in contrast to cells expressing GFP and GFP-YpkA, in which fluorescence is either evenly distributed throughout the cytoplasm or localized to the plasma membrane, respectively (Fig. 3A and Ref. 4).
FIGURE 3.
YopJ expression in S. pombe. A, fluorescence of live wild-type cells transformed with either GFP- or GFP-YopJ-encoding plasmids (strains 670W and 672W, respectively). B, the S. pombe (Sty1-HA) strain transformed with the GFP-YopJ-encoding plasmid (639W) was either left uninduced (Off) or induced (On) for GFP-YopJ expression for 12 h and then either left unexposed (-S) or exposed (+S) to osmotic stress (1 m KCl) for 20 min. Cells were then collected and lysed, and Sty1-HA was immunopurified. Immunoprecipitates were analyzed by Western analysis using the indicated antisera. pY, anti-phosphotyrosine. C, transformed strains (circles, 670W; boxes, 672W) were either left uninduced (Off, open symbols) or induced (On, closed symbols) for GFP or GFP-YopJ expression for 12 h and then exposed to osmotic stress (1 m KCl) for 0–25 h. At the indicated time points, cells were removed, stained with calcofluor, and examined by microscopy. Cells staining positive for a septum (as shown in the inset), indicative of the G2/M phase of the cell cycle when cells are at their maximal length, were measured using a molecular ruler (9.8 pixels/micron). Solid bars, population medium. *, p < 0.05.
We then tested whether YopJ affected signaling through the Sty1/Spc1 MAPK pathway, equivalent to the HOG- and p38-mediated kinase pathways of Saccharomyces cerevisiae and vertebrate cells, respectively. Exposure to osmotic stress normally results in a transitory tyrosine phosphorylation of Sty1; however, in YopJ-expressing S. pombe, phosphorylation of Sty1 was not observed (Fig. 3B). Additionally, exposure to osmotic stress causes a cell-shortening phenotype that is dependent on Sty1; sty1Δ cells become significantly elongated in response to osmotic stress due to a block in the G2/M transition (35). We found that in response to osmotic stress, YopJ-expressing S. pombe cells become elongated (Fig. 3C), resembling the phenotype of sty1Δ cells. This elongated phenotype was not observed in S. pombe cells expressing a YopJ variant (YopJ(C172A); Fig. 4B) that has previously been shown to be inactive in both S. cerevisiae and vertebrate cells (19, 26).
FIGURE 4.
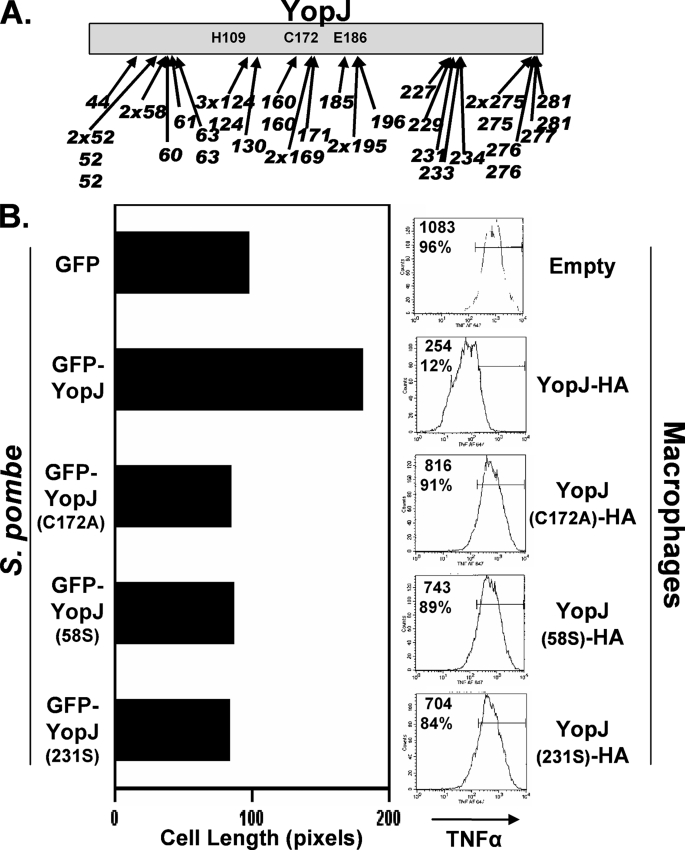
YopJ activity in S. pombe and infected macrophages. A, S. pombe transformed with a randomly generated library of GFP-YopJ variants containing 5-codon nonpolar inserts (scars) were plated on media that differentiated between transformants expressing active and inactive YopJ (elongated and short cells, respectively; see “Experimental Procedures” for details). Plasmids from transformants displaying the short cell phenotype were recovered, and the scar locations of 40 such YopJ variants are shown. (Indicated are the YopJ residues that are immediately followed by the scar sequence.) Both nonidentical and identical clones were recovered for three locations (residues 52, 124, and 275), whereas other locations were represented either singly (e.g. 44), by identical clones (e.g. 58 and 169), or by nonidentical clones (e.g. 63, 276). B, left panel, liquid-grown S. pombe transformed with plasmids encoding either GFP (670W), GFP-YopJ (672W), GFP-YopJ(C172A) (674W), or two variants recovered in the plate-based screen, GFP-YopJ(58S) and GFP-YopJ(231S) (677W and 685W, respectively), were evaluated following osmotic stress (1 m KCl for 20 h) in the cell elongation assay described in Fig. 3C. Right panel, sequences encoding the indicated YopJ variant were cloned into a Yersinia expression plasmid and transformed into the Y. pseudotuberculosis ΔyopJ strain (3). The transformed strains were added to cultured RAW 267 mouse macrophages, and 2 h later, cells were collected and intracellularly stained for tumor necrosis factor α and analyzed by flow cytometry. Shown within each histogram are the mean fluorescence intensity as well as the percentage of cells falling within the indicated gate.
To further verify that the cell elongation phenotype in S. pombe is an accurate indicator for YopJ activity in macrophages, we isolated a number of randomly generated YopJ variants that were inactive in S. pombe (see “Experimental Procedures” for details). These inactive YopJ variants contained mutations in one of four regions: region 1 between residues 44 and 63; region 2 encompassing the catalytic triad residues His109, Cys172, and Glu186; region 3 tightly clustered between residues 227 and 233; and region 4 being located at the carboxyl terminus (Fig. 4A). As expected, no inactive variants were recovered with mutations in the amino terminus, the site of the type 3 secretion signal sequence. All inactive YopJ variants were found to be stably expressed and cell length measurements of liquid grown cells showed that these variants were as inactive as YopJ(C172A) (Fig. 4B, left). To test whether the region 1 and 3 variants were active in macrophages, we expressed a representative variant from each of these regions in Yersinia (designated as YopJ(58S) and YopJ(231S), respectively) and tested the resulting bacterial strains in an infection assay. Neither the inactivating mutations nor the HA epitope tag used in the Yersinia expression plasmid affected the expression levels or export by the type 3 secretion system (data not shown). Macrophages infected with Yersinia expressing wild-type YopJ had notably lower levels of tumor necrosis factor α than macrophages infected with Yersinia expressing YopJ(C172A) (Fig. 4B, right). In this assay, the activity of YopJ(58S) and YopJ(231S) resembled that of YopJ(C172A), thus showing that the YopJ-mediated disruption of the osmotic stress response in S. pombe can serve as a convenient proxy for inductive chemokine expression in macrophages.
In S. pombe, genetic disruptions of the Sty1-mediated MAPK signaling pathway is associated with relatively higher levels of phosphorylated eIF2α following osmotic and oxidative stresses (32). As would be predicted, we found that S. pombe strains expressing active GFP-YopJ had notably higher levels of phosphorylated eIF2α following oxidative stress, similar to that observed in the sty1Δ strain, compared with strains expressing GFP-YopJ(C172A) (Fig. 5A). In the absence of oxidative stress, YopJ expression did not visibly affect eIF2α phosphorylation levels (data not shown).
At the cellular level, oxidative stress results in membrane pitting, creating an irregular surface; YopJ-expressing cells, as well as sty1Δ cells, display a greatly enhanced level of damage following oxidative stress (Fig. 5B). The YopJ-dependent cell elongation phenotype during osmotic stress was completely abolished in both eIF2α(S52A) cells (data not shown) as well as hri2Δ cells (Fig. 5C). In contrast, YopJ expression in wild-type, hri1Δ, and gcn2Δ cells were phenotypically indistinguishable in their osmotic and oxidative stress responses (Fig. 5C; data for oxidative stress not shown). Collectively, these data indicate that YopJ blocks the ability of S. pombe to appropriately respond to externally applied environmental stresses and that this phenotype is dependent on Hri2.
Co-expression of YpkA and YopJ in S. pombe
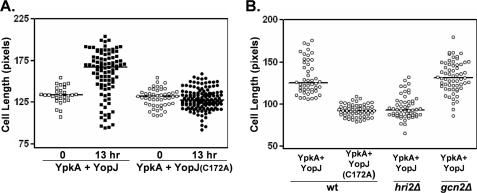
The findings described above indicate that when expressed individually, YpkA induces a stress-like response, whereas YopJ obstructs stress responses, and that both phenomena involve the eIF2α kinase Hri2. We therefore engineered S. pombe strains that co-express these two virulence factors to determine whether their respective activities are functionally intertwined. Co-expression affected neither the unique localization pattern of GFP-YopJ and GFP-YpkA nor their respective expression levels. Unexpectedly, in the absence of an externally applied environmental stress, YpkA/YopJ co-expressing cells elongated (Fig. 6A). Under these conditions, elongation was dependent on the enzymatic activity of YpkA (data not shown). In contrast, cells expressing YpkA and inactive YopJ (YopJ(C172A)) became shortened, reminiscent of the response to high salt conditions (compare Figs. 3C and 6A). These data indicate that YpkA activity mimics an externally applied stress in YopJ-expressing cells. Additionally, and as would be expected based on the single-expression studies, the elongation did not occur in hri2Δ cells co-expressing YpkA/YopJ (Fig. 6B). Interestingly, gcn2Δ cells co-expressing YpkA/YopJ not only displayed a wild-type-like elongation phenotype (Fig. 6B) but also exhibited much less visible signs of YpkA toxicity (compared with the wild-type strain), which would be expected from the single expression experiment shown in Fig. 1C.
FIGURE 6.
Assaying YopJ activity in S. pombe co-expressing YpkA and YopJ. A, actively growing cultures of strains possessing an integrated gfp-ypkA gene and plasmid-encoded gfp-yopJ (either wild-type (wt) or inactive: boxes, 755W; circles, 759W, respectively) were shifted to GFP-YpkA/GFP-YopJ expressing conditions and either immediately or 13 h later, cells (open and closed symbols, respectively) were removed from the culture and analyzed as described for Fig. 3C. Note that in this experiment no environmental stress was added to the cultures. Plotted are lengths of septum-positive cells. B, similar assay with strains used in A in addition to hri2Δ and gcn2Δ strains possessing an integrated gfp-ypkA gene and plasmid-encoded gfp-yopJ (806 and 883). Shown is the length measurements of septum-positive cells drawn from cultures that had been shifted to GFP-YpkA/GFP-YopJ expressing conditions 12 h previously (9.8 pixels/micron).
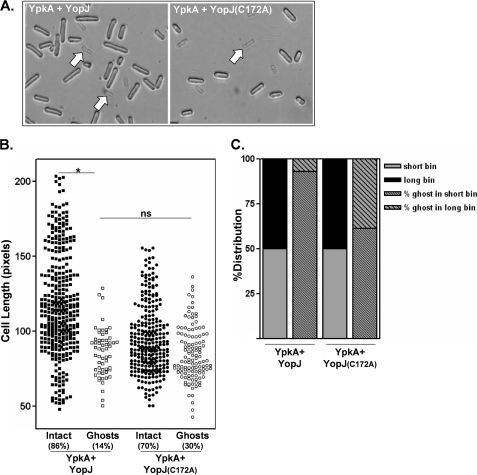
YopJ expression in S. pombe is associated with a heighten sensitivity to osmotic and oxidative stresses (Figs. 3 and 5). To determine whether YopJ-expressing S. pombe similarly affected the sensitivity to YpkA-induced stress (see Fig. 2A), we measured YpkA-mediated toxicity in cells co-expressing either YopJ or YopJ(C172A). Cells co-expressing YpkA/YopJ were relatively more resistant to lysis compared with cells co-expressing YpkA/YopJ(C172A) (data not shown), indicating that YopJ can at least partially reduce YpkA toxicity. Cultures of the YpkA/YopJ co-expressing strain contained a mixture of short and elongated cells; however, when these cells were treated with detergent, lysed cells (ghosts) were exclusively short, whereas the relatively longer cells remained intact (Fig. 7A; quantified in Fig. 7B). In contrast, even though cultures of the YpkA/YopJ(C172A) co-expressing strain contained a larger fraction of detergent-sensitive cells (∼30% compared with ∼14%), detergent-sensitive cells were more evenly distributed between the “short” and “long” cell length bins (Fig. 7C). These findings indicate that YopJ activity shields cells from the toxic effects of YpkA. This outcome is entirely consistent with what was found in the single-expression studies in which YopJ activity was associated with increased levels of phosphorylated eIF2α that, in turn, is associated with a mitigation of YpkA toxicity.
FIGURE 7.
Assaying YpkA activity in S. pombe co-expressing YpkA and YopJ. A, actively growing cultures of strains possessing an integrated gfp-ypkA gene and either plasmid-encoded gfp-yopJ or gfp-yopJ(C172A) (755W and 759W, respectively) were shifted to GFP-YpkA/GFP-YopJ-expressing conditions. Cells were then periodically removed from the culture, mixed with SDS, and examined microscopically. Shown are images of unfixed intact and detergent-lysed cells after 16 h in expressing conditions. Representative lysed cells (ghosts) are indicated by arrows. B, length measurements of unfixed cells (boxes, 755W; circles, 759W) were made and are plotted either as intact (filled symbols) or lysed (open symbols) cells (*, p < 0.001; not significant (ns), p = 0.70 using a two-sample test to compare proportions in two independent samples). C, drawn from the data shown in B, cells of the GFP-YpkA/GFP-YopJ and GFP-YpkA/GFP-YopJ(C172A) cultures were assigned to either a short or long bin depending on whether their length fell below or above the population median for each culture (solid gray or black bars, respectively). The stippled bars represent the distribution of the detergent-sensitive cells (ghosts) between the short and long cell length bins.
We were interested in examining whether the YopJ elongation phenotype triggered by either osmotic stress or YpkA could also be elicited by other Yersinia virulence proteins. The T3SS effector YopE, a GTPase activating protein of RhoA, is highly toxic in yeast (23, 24). Using conditions similar to what is shown for YpkA in Fig. 2A, YopE-expressing S. pombe cells display an extensive amount of internal damage yet, unlike YpkA-expressing cells, do not lyse when challenged with detergent. We therefore constructed S. pombe strains (891W and 893W) that possessed an integrated yopE gene and plasmids encoding either wild-type yopJ or yopJ(C172A). Following the induction of YopE and YopJ (or YopJ(C172A)) expression in an experiment similar to the one shown in Fig. 6A, we could detect no visible differences between these two strains in terms of YopE toxicity (data not shown). Significantly, the YopE/YopJ strain did not elongate indicating that YopE-mediated toxicity did not generate a YopJ-responsive stress. These data underscore the specificity of the YpkA-YopJ co-dependence on the eIF2α kinase Hri2.
DISCUSSION
We found by genetic screening in S. pombe that the eIF2α kinases play a modulatory role in the cellular response to the bacterial virulence factor YpkA. Specifically, the eIF2α kinase Hri2 was shown to serve a protective function against YpkA toxicity at the cellular level. At the biochemical level, YpkA expression resulted in enhanced eIF2α phosphorylation that was largely dependent on the presence of Hri2. Whereas Hri2 was found to be important for resistance to YpkA, reduced activity of Gcn2, an eIF2α kinase that is primarily responsive to nutritional-based stresses, actually enhanced the level of resistance to YpkA. Although at first glance, this latter result appears counterintuitive, in S. pombe reduced Gcn2 activity is associated with a reduction in MAPK activity that in turn is associated with enhanced levels of phosphorylated eIF2α following stress (31, 32). The proposition that heightened phosphorylated eIF2α levels are protective against YpkA toxicity was strengthened by our subsequent finding that expression of the bacterial virulence factor YopJ encoded on the same transcript as YpkA and in S. pombe confers a phenotype that is consistent with its previously described activity of abrogating MAPK signaling, results in higher levels of phosphorylated eIF2α, and counteracts YpkA-induced cell lysis. Strikingly, both the MAPK-blocking and YpkA-protecting activities of YopJ are absolutely dependent on the presence of Hri2. Interestingly, in contrast to playing a protective role against YpkA-induced stress, YopJ activity heightens the sensitivity of cells to oxidative stress suggesting that the pathogenic Yersinia shapes a cellular stress response for its own benefit. In summary, our findings support a model in which YopJ activity further amplifies a cellular process (in this case involving Hri2) that acts to mollify YpkA toxicity.
The findings described here raise a number of questions. To begin with, are YpkA and YopJ linked to Hri2 in the same way as other types of stresses are linked to this eIF2α kinase? The mammalian homolog, HRI, was originally discovered in erythroid cells in which it functions to match globin synthesis to the availability of heme (36) and was subsequently found to play a role in a variety of stress responses in nucleated cells (37–39). Most pertinent to this study were the findings that HRI is necessary for macrophage maturation and function and contributes ∼50% of the total eIF2α kinase activity in cultured macrophages following stimulation with lipopolysaccharide (40). HRI activity itself is controlled by multiple autophosphorylation sites that are responsive to heme levels (41–43). It will be of great interest to determine whether S. pombe YpkA and/or YopJ cause Hri2 to be phosphorylated at its normal sites or subjects it to novel modifications. Second, can the effect of YopJ on eIF2α phosphorylation levels (and the consequential protection from YpkA toxicity) be entirely explained by it blocking MAPK signaling? There have been a number of reports in a variety of experimental systems linking stress-activated protein kinase (SAPK) pathways and eIF2α-mediated translational control (31, 32, 44–46). And finally, it remains to be determined whether the functional readouts for YpkA and YopJ activities in macrophages (infectivity and the blockage of inductive chemokine expression, respectively) are dependent on eIF2α kinases.
Acknowledgments
We thank Andrea Echeverry, Fangfang Lai, Louis Gonzales, and Delia Gutman for technical assistance and Erik Boye, Ronald C. Wek, Jonathon Millar, Mohan K. Balasubramanian, Janni Petersen, Per Sunnerhagen, Fulvia Verde, and Larry Boise for strains, reagents, and wise counsel.
This work was supported by NIAID, National Institutes of Health Public Health Service Grant AI53459.
- T3SS
- type 3 secretion system
- MOPS
- 4-morpholinepropanesulfonic acid
- HA
- hemagglutinin
- eGFP
- enhanced green fluorescent protein.
REFERENCES
- 1.Galán J. E., Wolf-Watz H. (2006) Nature 444, 567–573 [DOI] [PubMed] [Google Scholar]
- 2.Viboud G. I., Bliska J. B. (2005) Annu. Rev. Microbiol. 59, 69–89 [DOI] [PubMed] [Google Scholar]
- 3.Galyov E. E., Håkansson S., Wolf-Watz H. (1994) J. Bacteriol. 176, 4543–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiley D. J., Nordfeldth R., Rosenzweig J., DaFonseca C. J., Gustin R., Wolf-Watz H., Schesser K. (2006) Microb. Pathog. 40, 234–243 [DOI] [PubMed] [Google Scholar]
- 5.Bartra S., Cherepanov P., Forsberg A., Schesser K. (2001) BMC Microbiol. 1, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boland A., Cornelis G. R. (1998) Infect. Immun. 66, 1878–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer L. E., Hobbie S., Galán J. E., Bliska J. B. (1998) Mol. Microbiol. 27, 953–965 [DOI] [PubMed] [Google Scholar]
- 8.Schesser K., Spiik A. K., Dukuzumuremyi J. M., Neurath M. F., Pettersson S., Wolf-Watz H. (1998) Mol. Microbiol. 28, 1067–1079 [DOI] [PubMed] [Google Scholar]
- 9.Monack D. M., Mecsas J., Bouley D., Falkow S. (1998) J. Exp. Med. 188, 2127–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meijer L. K., Schesser K., Wolf-Watz H., Sassone-Corsi P., Pettersson S. (2000) Cell. Microbiol. 2, 231–238 [DOI] [PubMed] [Google Scholar]
- 11.Galyov E. E., Håkansson S., Forsberg A., Wolf-Watz H. (1993) Nature 361, 730–732 [DOI] [PubMed] [Google Scholar]
- 12.Dukuzumuremyi J. M., Rosqvist R., Hallberg B., Akerström B., Wolf-Watz H., Schesser K. (2000) J. Biol. Chem. 275, 35281–35290 [DOI] [PubMed] [Google Scholar]
- 13.Barz C., Abahji T. N., Trülzsch K., Heesemann J. (2000) FEBS Lett. 482, 139–143 [DOI] [PubMed] [Google Scholar]
- 14.Juris S. J., Shah K., Shokat K., Dixon J. E., Vacratsis P. O. (2006) FEBS Lett. 580, 179–183 [DOI] [PubMed] [Google Scholar]
- 15.Navarro L., Koller A., Nordfelth R., Wolf-Watz H., Taylor S., Dixon J. E. (2007) Mol. Cell 26, 465–477 [DOI] [PubMed] [Google Scholar]
- 16.Juris S. J., Rudolph A. E., Huddler D., Orth K., Dixon J. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9431–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittal R., Peak-Chew S. Y., McMahon H. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18574–18579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S., Keitany G., Li Y., Wang Y., Ball H. L., Goldsmith E. J., Orth K. (2006) Science 312, 1211–1214 [DOI] [PubMed] [Google Scholar]
- 19.Orth K., Xu Z., Mudgett M. B., Bao Z. Q., Palmer L. E., Bliska J. B., Mangel W. F., Staskawicz B., Dixon J. E. (2000) Science 290, 1594–1597 [DOI] [PubMed] [Google Scholar]
- 20.Sweet C. R., Conlon J., Golenbock D. T., Goguen J., Silverman N. (2007) Cell. Microbiol. 9, 2700–2715 [DOI] [PubMed] [Google Scholar]
- 21.Zhou G., Golden T., Aragon I. V., Honkanen R. E. (2004) J. Biol. Chem. 279, 46595–46605 [DOI] [PubMed] [Google Scholar]
- 22.Mudgett M. B. (2005) Annu. Rev. Plant Biol. 56, 509–531 [DOI] [PubMed] [Google Scholar]
- 23.Lesser C. F., Miller S. I. (2001) EMBO J. 20, 1840–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Von Pawel-Rammingen U., Telepnev M. V., Schmidt G., Aktories K., Wolf-Watz H., Rosqvist R. (2000) Mol. Microbiol. 36, 737–748 [DOI] [PubMed] [Google Scholar]
- 25.Nejedlik L., Pierfelice T., Geiser J. R. (2004) Yeast 21, 759–768 [DOI] [PubMed] [Google Scholar]
- 26.Yoon S., Liu Z., Eyobo Y., Orth K. (2003) J. Biol. Chem. 278, 2131–2135 [DOI] [PubMed] [Google Scholar]
- 27.Kramer R. W., Slagowski N. L., Eze N. A., Giddings K. S., Morrison M. F., Siggers K. A., Starnbach M. N., Lesser C. F. (2007) PLoS Pathog. 3, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keeney J. B., Boeke J. D. (1994) Genetics 136, 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wek R. C., Jackson B. M., Hinnebusch A. G. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 4579–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A. G. (2000) Mol. Cell. 6, 269–279 [DOI] [PubMed] [Google Scholar]
- 31.Petersen J., Nurse P. (2007) Nat. Cell Biol. 9, 1263–1272 [DOI] [PubMed] [Google Scholar]
- 32.Dunand-Sauthier I., Walker C. A., Narasimhan J., Pearce A. K., Wek R. C., Humphrey T. C. (2005) Eukaryot. Cell. 4, 1785–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan K., Vattem K. M., Bauer B. N., Dever T. E., Chen J. J., Wek R. C. (2002) Mol. Cell. Biol. 22, 7134–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhan K., Narasimhan J., Wek R. C. (2004) Genetics 168, 1867–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiozaki K., Russell P. (1995) Nature 378, 739–743 [DOI] [PubMed] [Google Scholar]
- 36.Chen J. J., London I. M. (1995) Trends Biochem. Sci. 20, 105–108 [DOI] [PubMed] [Google Scholar]
- 37.Lu L., Han A. P., Chen J. J. (2001) Mol. Cell. Biol. 21, 7971–7980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uma S., Barret D. J., Matts R. L. (1998) Exp. Cell Res. 238, 273–282 [DOI] [PubMed] [Google Scholar]
- 39.McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J. J., Anderson P., Kaufman R. J. (2005) J. Biol. Chem. 280, 16925–16933 [DOI] [PubMed] [Google Scholar]
- 40.Liu S., Suragani R. N., Wang F., Han A., Zhao W., Andrews N. C., Chen J. J. (2007) J. Clin. Invest. 117, 3296–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagard R., London I. M. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer B. N., Rafie-Kolpin M., Lu L., Han A., Chen J. J. (2001) Biochemistry 40, 11543–11551 [DOI] [PubMed] [Google Scholar]
- 43.Chen J. J. (2007) Blood 109, 2693–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiaville M. M., Pan Y. X., Gjymishka A., Zhong C., Kaufman R. J., Kilberg M. S. (2008) J. Biol. Chem. 283, 10848–10857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goh K. C., deVeer M. J., Williams B. R. (2000) EMBO J. 19, 4292–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., Wek R. C. (2004) Mol. Cell. Biol. 24, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]