Abstract
The Forkhead box A2 transcription factor (Foxa2/HNF-3β) has been shown to be a key regulator of genes involved in the maintenance of glucose and lipid homeostasis in the liver. It is constitutively inactivated in several hyperinsulinemic/obese mouse models, thereby enhancing their metabolic phenotypes. Foxa2 is activated under fasting conditions but is inhibited by insulin signaling via phosphatidylinositol 3-kinase/AKT in a phosphorylation-dependent manner, which results in its nuclear exclusion. However, the mechanism and relative importance of its nuclear export has not yet been elucidated. Here we show that Foxa2 contains a functional nuclear export signal and is excluded from the nucleus via a CRM1-dependent pathway in response to insulin signaling. Furthermore, direct evidence is provided that nuclear export-defective Foxa2 is phosphorylated and inactivated by insulin in vitro and in vivo. These data demonstrate for the first time that phosphorylation itself is the main event regulating the activity of Foxa2, suggesting that export-independent mechanisms have evolved to ensure inhibition of Foxa2 under conditions in which insulin signaling is present.
The hepatocyte nuclear factor 3 (HNF-3)2/Forkhead family of transcription factors in mammals includes three genes, designated Foxa1 (HNF-3α), Foxa2 (HNF-3β), and Foxa3 (HNF-3γ), which have overlapping patterns of tissue expression including gut, central nervous system, neuroendocrine cells, and lung (1, 2). FoxA proteins were first identified as a binding activity to an enhancer region of the transthyretin gene in liver nuclear extracts (3). They all have in common a highly conserved winged helix motif that is responsible for monomeric recognition of specific DNA target sites and is similar in structure to that of the linker histone H5 (4). However, in contrast to linker histones, which compact DNA in chromatin and repress gene expression, FoxA proteins are associated with transcriptionally active chromatin and may decompact DNA from the nucleosome (5, 6).
Although FoxA proteins share very high sequence homology within the DNA binding domain, outside of this region Foxa1 and Foxa2 are only 39% identical, with Foxa3 even more distinct (2). This sequence divergence allows for unique posttranslational modifications (e.g. AKT phosphorylation of Foxa2 at residue Thr-156 (7)) and differential DNA and protein interactions (e.g. interaction of the transcriptional coactivator PGC-1β with Foxa2 (8, 9)). Accordingly, FoxA proteins are not entirely redundant in function. Mice homozygous for a null mutation in Foxa2 exhibit an embryonic lethal phenotype, lack a notochord, and exhibit defects in foregut and neural tube development, whereas Foxa3-deficient mice develop normally (10–12). Mice lacking Foxa1 expression develop neonatal persistent hypoglycemia, hormonal insufficiencies, pancreatic alpha and beta cell dysfunction, and die between postnatal days 2 and 14 (13, 14).
Mouse genetic studies have also revealed important roles for Foxa genes in metabolism. In the livers of adult mice, Foxa2 activity has been shown to mediate fasting responses, including fatty acid oxidation, ketogenesis, and increased very low density lipoprotein and high density lipoprotein (VLDL and HDL) secretion by activating gene expression of key enzymes of these pathways (8, 15, 16). In the postprandial state, when insulin levels rise, Foxa2 is phosphorylated at threonine residue 156 through phosphatidylinositol 3-kinase/Akt signaling, which results in nuclear exclusion of Foxa2 and inhibition of its target genes (7, 17). In hyperinsulinemic/obese mice, Foxa2 is permanently excluded from the nucleus and its inactivation contributes to the development of hepatic steatosis and insulin resistance. This has been demonstrated by re-expression of constitutive active Foxa2 (Foxa2-T156A) in livers of obese mouse models which led to increased fatty acid oxidation, increased VLDL secretion, reduced hepatic triglyceride content, increased insulin sensitivity and normalization of blood glucose levels (15).
The negative regulation of Forkhead transcription factors by nutritional or stress signals is not unique to Foxa2. Phosphatidylinositol 3-kinase/Akt signaling in the nematode Caenorhabditis elegans suppresses the function of DAF-16, a transcription factor that also belongs to the Forkhead/winged-helix family (18). Mutations in the insulin/Igf-1 receptor homologue (daf-2) (19, 20), the catalytic subunit of phosphatidylinositol 3-kinase (age-1) (21), or Akt (akt1 and akt2) (22) result in increased longevity and constitutive dauer formation, a stage of developmental arrest and reduced metabolic activity that enhances survival during periods of food deprivation and other environmental stresses. In each case, mutation of daf-16 restores normal life span and prevents entry into the dauer stage.
In mammals, this regulation has also been described for Fkhr (Foxo1), Fkhrl1 (Foxo3), and AFX (Foxo4) (23–26). Foxo1 can be phosphorylated by Pkb/Akt at multiple sites causing repression of transcriptional activity of target genes such as insulin growth factor-binding protein 1, glucose-6-phosphatase, and phosphoenolpyruvate carboxykinase (27, 28). Similar to Foxa2, this regulation has been shown to occur, at least in part, by nuclear exclusion, although recent findings suggest that additional mechanisms are involved (29). Thus, at the moment it is unclear whether nuclear export is the key mechanism regulating FoxO transcription factors or whether other nucleus-specific regulatory pathways are involved in the regulation of this factor as well.
Although it is clear that phosphorylation is necessary for nuclear exclusion of Foxa2, the mechanism and importance of nuclear exclusion in the inactivation of Foxa2 by insulin has not yet been investigated. Here, we have explored the molecular mechanisms controlling nuclear exclusion of Foxa2 in response to insulin signaling and phosphorylation at residue Thr-156. We show that Foxa2 contains a functional, leptomycin B (LMB)-sensitive (CRM1-dependent), leucine-rich nuclear export sequence, which is necessary for nuclear exclusion of Foxa2. Furthermore, we demonstrate that phosphorylation of this factor, rather than nuclear exclusion, is the key regulatory event modulating Foxa2 activity in response to insulin signaling.
EXPERIMENTAL PROCEDURES
Materials
Human recombinant insulin (I9278), collagen (type I, C3867), and leptomycin B (L2913) were purchased from Sigma. The following antibodies were used: HA (Covance and Santa Cruz Biotechnology); rabbit polyclonal to Foxa2 (Abcam); anti-phosphorylated Foxa2 (Thr-156, Cell Signaling) (generated and described previously (15)); anti-LSD1 (Cell Signaling); anti- glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Abcam); anti-γ-tubulin (Sigma).
Plasmids and Adenovirus
HA-tagged rat Foxa2, Foxa2-T156A, and AKT2 were in pcDNA3 expression vectors as described previously (7). Emut (L110A,L113A) and TAE (L110A,L113A,T156A) constructs were generated by site-directed mutagenesis using overlap extension PCR. Adenoviruses were generated using the Rapid Adenovirus Production System (Viraquest). With the exception of Ad-GFP-C1Foxa2, GFP was coexpressed from an independent promoter in addition to HA-Foxa2 or Foxa2 variants. Ad-GFP, Ad-Foxa2, and Ad-T156A were described previously (15). For in vivo experiments, mice were injected with 8 × 108 plaque-forming units of adenovirus through the tail vein.
Cell Lines and Primary Hepatocytes
HepG2 cells and primary hepatocytes were maintained on collagen-coated plates in Dulbecco's modified Eagle's medium (Invitrogen; containing 4.5 g/liter glucose, 110 mg/liter sodium pyruvate, 4 mm l-glutamine) supplemented with 10% fetal bovine serum and 100 units/ml penicillin/streptomycin in a humidified incubator at 37 °C and 5% CO2. Serum starvation was carried out for 18 h in Dulbecco's modified Eagle's medium without fetal bovine serum or penicillin/streptomycin. Stable cell lines were generated by transfection of 4.5 × 106 HepG2 cells with 6 μg of plasmid DNA using FuGENE 6 transfection reagent (Roche Applied Science) and selection with 1 mg/ml G418 (Calbiochem) over a period of 2 to 3 weeks. Clonal populations were isolated and analyzed for expression of Foxa2 constructs by Western blotting using an anti-HA antibody (MMS-101P, Covance). Primary hepatocytes were isolated as described previously (16).
Animal Models
C57Bl/6 and ob/ob mice were purchased from Charles River and maintained on a normal chow diet and a 12-h light/dark cycle.
Nuclear/Cytosolic Extracts
HepG2 cells were grown in 10-cm plates to 80% confluency and serum-starved for 18 h followed by the indicated treatments (LMB: 2 h with 2.5 ng/ml leptomycin B; insulin: 500 nm human recombinant insulin for 15 min at 37 °C). Nuclear and cytoplasmic extracts were prepared as described previously (16). Briefly, cells were swollen on ice in hypotonic lysis buffer (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl) containing 1 mm dithiothreitol and protease inhibitors (Complete mixture, Roche Applied Science) and permeabilized by the addition of Nonidet P-40 to 0.6%. After centrifugation at 10,400 × g for 30 s at 4 °C, the supernatants (cytoplasmic extracts) were collected, and nuclear pellets were resuspended in nuclear lysis buffer (10 mm HEPES, pH 7.9, 100 mm KCl, 3 mm MgCl2, 0.1 mm EDTA) containing 1 mm dithiothreitol and protease inhibitors. Nuclei were lysed by the gradual addition of a one-tenth volume of 4 m (NH4)2SO4 over 30 min. For liver experiments, ∼50 mg of liver was Dounce-homogenized directly in hypotonic lysis buffer on ice 10 times with a tight piston.
Transfection and Transactivation Assays
HepG2 cells were plated at 70,000 cells/well in 24-well plates and transfected on the following day with 25 ng of p6xCdx-TkLuc reporter gene (7), 10 ng of pRL-Tk, and 25 ng of Foxa2 expression vectors alone or in combination with 5 ng of a human AKT2 expression vector using Lipofectamine 2000 (Invitrogen). Cells were harvested 40 h post-transfection, and luciferase was measured using the Dual Luciferase System (Promega).
Immunoprecipitation and Chromatin Immunoprecipitation
Primary hepatocytes from three mice were pooled and plated onto ten 10-cm collagen-treated plates. Cells were washed extensively and allowed to recover over 48 h after which they were infected with 2.5 × 107 plaque-forming units of the indicated virus. After 24 h, cells were serum-starved overnight followed by a 15-min stimulation in the absence or presence of 100 nm insulin. For chromatin immunoprecipitation, 11% formaldehyde was added to the cells to a final concentration of 1% and incubated for 10 min at 37 °C. Cells were put on ice, washed twice with ice-cold PBS, and scraped in 1 ml of chromatin immunoprecipitation cell lysis buffer (10 mm Tris-Cl, pH 8.0, 10 mm NaCl, 1.5 mm MgCl2, 0.2% Nonidet P-40, with protease inhibitors). Lysates were centrifuged for 5 min at 5400 × g at 4 °C. Pellets were resuspended in 450 μl of SDS lysis buffer (Upstate Biotechnology) and sonicated for 4 × 20 s on/30 s off at amplitude 16 on a Misonix Sonicator 4000. Lysates were further processed according to the manufacturer's protocol (chromatin immunoprecipitation assay kit, Upstate Biotechnology). DNA complexes were immunoprecipitated overnight using 2.6 μg of HA antibody (Santa Cruz Biotechnology, sc-805) or rabbit IgG (ChromPure, Jackson ImmunoResearch). Primer sequences are available on request.
Immunofluorescence Microscopy
Liver pieces were frozen directly in OCT (optimal cutting temperature) compound at −80 °C. 9 μm cryosections were fixed in 4% paraformaldehyde in PBS at 4 °C for 30 min, permeabilized in 0.2% Nonidet P-40 in PBS, blocked in 5% normal donkey serum, 1% bovine serum albumin, 0.1% Nonidet P-40 in PBS, and incubated with anti-HA antibody (1:25, Santa Cruz Biotechnology, sc-805) overnight at 4 °C in a humidified chamber. Donkey anti-rabbit IgG Alexa Fluor 488 (Molecular Probes) was used as a secondary antibody and mounted with VECTASHIELD mounting media with 4,6-diamidino-2-phenylindole (Vector Laboratories) to visualize nuclei. Immunofluorescent staining was visualized with a Leica confocal microscope at ×40 magnification.
Gene Expression
Extraction of mRNA and cDNA synthesis were performed from 50 mg of liver tissue with the μMACS One-step cDNA Kit (Miltenyi Biotec) following the manufacturer's protocol. PCR products were measured as a function of SYBR Green incorporation using LightCycler 480 SYBR Green I Master Mix (Roche Applied Science) and the Mx3005P Real-Time QPCR Detection System (Stratagene). Values shown are given in arbitrary units based on a standard curve and normalized to glyceraldehyde-3-phosphate dehydrogenase. Primer sequences are available on request.
Physiological Measurements
Retro-orbital blood samples were taken into nonheparinized capillary tubes. Blood glucose was measured using a standard glucometer (Ascensia Contour, Bayer). Plasma insulin was measured with the sensitive rat insulin RIA kit (Linco Research). Liver triglycerides were extracted by the Folch method and quantitated by colorimetric assay (Roche Applied Science).
Mitochondrial β-Oxidation/Ketone Body Formation
Mitochondria were isolated from 200 mg of PBS-perfused mouse livers by differential centrifugation as described previously (30). We assessed the β-oxidation of [1-14C]palmitic acid by liver mitochondria in samples normalized to mitochondrial protein as described (31). CO2 trapped on the filter papers was counted for 14C activity by scintillation counter. To measure ketone body formation, the incubation mixture was centrifuged at 4000 × g for 10 min, and 14C-labeled acid-soluble products of mitochondrial palmitate metabolism were counted from an aliquot of the supernatant (31).
Statistical Analysis
Results are given as mean ± S.E. if not otherwise indicated. Statistical analyses were carried out by using a two-tailed Student's unpaired t test, and the null hypothesis was rejected at the 0.05 level (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
Foxa2 Contains a LMB-sensitive Nuclear Export Sequence
To identify possible mechanisms responsible for the nuclear export of Foxa2, we first established a robust assay to allow us to semiquantitatively determine the nuclear and cytoplasmic localization of both endogenous and exogenous Foxa2. Whereas GFP fusion to Foxa2 abolished its insulin-induced nuclear export, HA-FLAG double-tagged Foxa2 was found to display normal export dynamics after insulin treatment when stably expressed in HepG2 cells (supplemental Fig. 1). We therefore used this assay to determine whether nuclear export of Foxa2 could be inhibited by the most common nuclear export factor, CRM1 (also called exportin1/Xpo1).
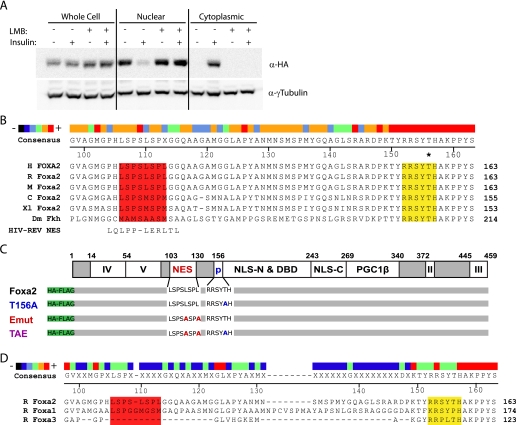
Because CRM1-mediated nuclear export can be potently inhibited by the pharmacological agent LMB (32, 33) we pretreated cells with 2.5 ng/ml LMB for 2 h prior to insulin stimulation and cellular fractionation. Analysis of nuclear and cytoplasmic extracts showed that LMB prevents the export of Foxa2 from the nucleus after stimulation with insulin, whereas untreated control cells display normal export dynamics (Fig. 1A). This finding demonstrates that nuclear export of Foxa2 is mediated by CRM1.
FIGURE 1.
Foxa2 contains an LMB-sensitive nuclear export signal. A, cellular fractionation of HepG2 cells stably expressing HA-tagged Foxa2. Cells were serum-starved overnight and stimulated with 500 nm insulin with or without pretreatment with 2.5 ng/ml LMB. Foxa2 localization was determined by Western blotting. B, partial sequence alignment of Foxa2 from six different species and the HIV-REV NES (32, 33). The putative NES (red) and AKT phosphorylation site (yellow) are highlighted, with an asterisk indicating the phosphorylated Thr-156. The heat map illustrates the degree of conservation, and numbering is based on the rat Foxa2 amino acid sequence. C, schematic depiction of Foxa2 showing the putative NES in relation to other known domains along with mutant constructs. D, partial sequence alignment of FoxA family members with regions corresponding to the putative NES (red) and AKT phosphorylation site (yellow) highlighted. H, human; R, rat; M, mouse; C, chicken; Xl, Xenopus laevis; Dm, Drosophila melanogaster; II–V, transactivation domains; p, phosphorylation site; NLS, nuclear localization signal (N or C terminus of the DNA binding domain); DBD, DNA binding domain; PGC1β, PGC1β interaction domain; Foxa2, rat wild-type Foxa2; T156A, mutated at residue T156A; Emut, mutated at residues L110A and L113A; TAE, mutated at residues T156A, L110A, and L113A.
To identify potential CRM1 regulatory sites in Foxa2, we performed a comparative analysis of the primary amino acid sequence using the generally accepted CRM1-mediated nuclear export signal (NES) consensus sequence LX2,3(L/I/V/F/M)X2,3LX(L/I) (32, 34). This analysis revealed that Foxa2 contains one leucine-rich region at amino acid position 106–113 that closely resembles this consensus sequence (shown in Fig. 1B aligned to the prototypical HIV-REV NES). The region lies in the N-terminal half of Foxa2, about 40 amino acids upstream of the AKT phosphorylation site (Thr-156) between the transactivation and DNA binding domains, and is evolutionarily well conserved (Fig. 1, B and C). Consistent with the constitutively nuclear localization of Foxa1 and Foxa3, there was little to no sequence conservation observed in the region of these proteins corresponding to the Foxa2 NES (Fig. 1D).
Foxa2 NES Is Necessary for Nuclear Export
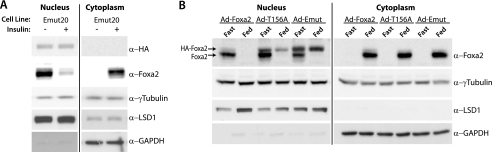
To elucidate whether the putative CRM1 NES is responsible for CRM1-mediated nuclear export of Foxa2 and thus important for its regulation in different physiological states, we performed mutational analyses. Because the C-terminal hydrophobic residues of the CRM1 consensus sequence have been shown to be the most critical for nuclear export (35), we mutated both terminal leucines (Leu-110 and Leu-113) of the putative Foxa2 NES to determine whether this sequence is necessary for nuclear exclusion in response to insulin. The L110A,L113A Foxa2 mutant (Fig. 1C, Emut) was stably expressed in HepG2 cells and analyzed for its ability to shuttle from the nucleus in response to insulin stimulation. Endogenous Foxa2 was used as an internal control. Whereas endogenous Foxa2 still shuttles in these cells in response to insulin stimulation, the export mutant remains nuclear, demonstrating that the NES is functional and necessary for export in vitro (Fig. 2A).
FIGURE 2.
Nuclear/cytoplasmic shuttling of Foxa2 requires a functional NES. A, immunoblots showing cellular fractionation of HepG2 cells stably expressing HA-Emut Foxa2, serum-starved overnight, and stimulated with 500 nm insulin. B, immunoblots showing cellular fractionation of livers from fasted or fed C57Bl/6 mice infected with adenoviruses expressing GFP, Foxa2, T156A, or Emut. Livers of mice that were either fasted for 18 h or fed ad libitum were harvested 5 days post-infection. γ-Tubulin was used as a loading control, and LSD1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression levels were used as the nuclear and cytosolic markers, respectively.
To test whether this sequence is also necessary for export in vivo and thus might play an important role in the hormonal regulation of Foxa2 activity we generated a recombinant adenovirus-containing export mutant (Ad-Emut) Foxa2 and injected it into C57Bl/6 mice. As controls we treated mice with recombinant adenoviruses expressing wild-type (Ad-Foxa2) or phosphorylation-deficient constitutively active Foxa2 (Ad-T156A), which have been characterized previously (15). All constructs were HA- and FLAG-tagged, and a GFP-expressing adenovirus was used as an additional control. As shown in Fig. 2B, both endogenous and exogenous Foxa2 were nuclear in all fasted animals (plasma insulin 0.3 ± 0.05 ng/ml). However, although endogenous and Ad-Foxa2 were excluded from the nucleus in ad libitum fed animals (which exhibit 5–6- fold increased plasma insulin levels compared with the fasting state), the export mutant, as well as the T156A control, remained nuclear. Thus, the newly identified NES in Foxa2 is responsible for the active nuclear export of Foxa2 in hepatocytes both in vitro and in vivo.
Emut Foxa2 Is Inhibited by Insulin Signaling
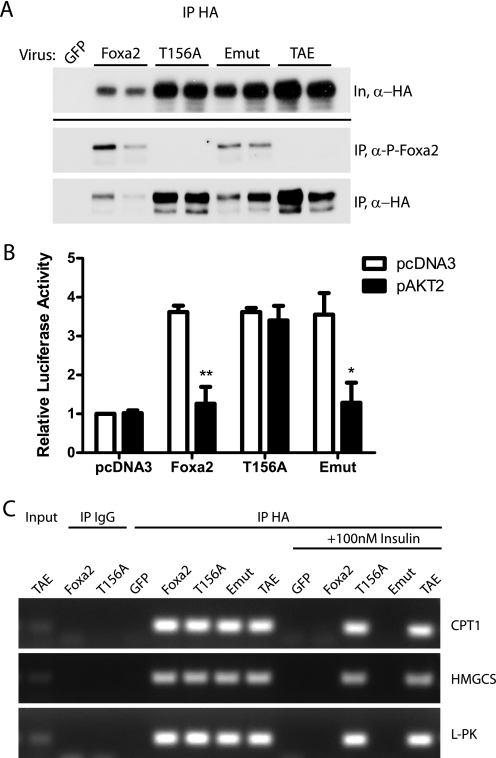
The ability to study the effect of phosphorylation uncoupled from subcellular localization of Foxa2 led us to ask whether regulation of Foxa2 activity by insulin is controlled mainly by nuclear export or by phosphorylation at Thr-156. To explore this possibility, we first sought to determine whether Emut Foxa2 could be efficiently phosphorylated through insulin signaling. Primary hepatocytes isolated from C57Bl/6 mice were infected with the corresponding adenoviruses and serum-starved overnight, followed by a 15-min incubation with insulin. Exogenous Foxa2 was immunoprecipitated and subjected to Western blot analysis. Western blots probed with an anti-Foxa2Thr-156-specific phosphopeptide antibody show that Emut Foxa2 is phosphorylated to a similar extent as wild-type Foxa2, suggesting a segregation of posttranslational modification and cellular localization. To further clarify this point, we generated recombinant adenovirus expressing a double Foxa2 mutant protein (Fig. 1C, TAE) containing both the Emut and T156A mutations. The T156A and TAE mutants, both of which lack the phosphorylation site and thus serve as negative controls, were not detected by the phospho-specific antibody (Fig. 3A).
FIGURE 3.
Emut Foxa2 is phosphorylated and inactivated by insulin. A, primary hepatocytes isolated from C57Bl/6 mice were infected with recombinant adenovirus expressing the indicated HA-tagged Foxa2 prior to overnight serum starvation and stimulation with 100 nm insulin. HA-Foxa2 and variants were immunoprecipitated using a mouse anti-HA antibody; immunoblots of whole cell lysates (In) and precipitates (IP) were probed with rabbit antibodies against HA- and phospho-Foxa2 (P-Foxa2). B, HepG2 cells were transfected with expression vectors containing Foxa2 constructs alone or in combination with AKT2. Vector p6xCdx-TkLuc was used as a reporter gene. Luciferase activity, normalized to Renilla luciferase, is shown relative to vector only controls. All experiments were performed in triplicate, and values shown represent the mean of three independent experiments ± S.E. *, p < 0.05; **, p < 0.01 by unpaired t test. C, chromatin immunoprecipitations from serum-starved or insulin-stimulated primary hepatocytes that were infected with the indicated adenoviruses. Chromatin was immunoprecipitated with HA or IgG (control) antibodies. Binding of Foxa2 and mutants to promoter sites in target genes was assayed by PCR. CPT1, carnitine palmitoyltransferase 1; HMGCS, HMG-CoA synthase; l-PK, liver pyruvate kinase.
Because the export mutant is phosphorylated in response to insulin signaling, we hypothesized that, despite its nuclear localization, Foxa2 could still be inactivated by Akt-dependent phosphorylation. To test this hypothesis, we first expressed wild-type Foxa2, T156A, or Emut plasmids together with a reporter plasmid containing six Foxa binding sites of the murine Cdx-2 gene upstream of firefly luciferase (6xCdx), with or without AKT2, in HepG2 cells. Reporter assays showed that although the export mutant retained transcriptional activity under basal conditions, cotransfection with AKT ablated this activity to a similar extent as the wild type (Fig. 3B). As a control, the Foxa2-T156A remained fully active with or without AKT coexpression. These data demonstrate that, on a functional level, Foxa2 transcriptional activity is primarily regulated by posttranslational modification rather than by subcellular localization.
To test whether these functional changes correlate with binding of Foxa2 to its target promoter regions, we performed chromatin immunoprecipitations from C57Bl/6-derived primary hepatocytes infected with Ad-GFP, Ad-Foxa2, Ad-T156A, Ad-Emut, and Ad-TAEmut. In a serum-starved state, all Foxa2 constructs were able to bind to known Foxa2 interaction sites in CPT1, HMG-CoA synthase, and L-PK genes. However, after insulin stimulation, the wild type and export mutant constructs were no longer found to bind the target sites, whereas binding of the T156A and TAE mutant Foxa2 proteins was unaffected by insulin (Fig. 3C). These data confirm the results obtained from the transactivation assays and show that Thr-156 phosphorylation determines binding of Foxa2 to its responsive promoter elements and that cellular localization is a secondary effect.
Emut Is Constitutively Nuclear but Inactive in Hyperinsulinemic ob/ob Mice
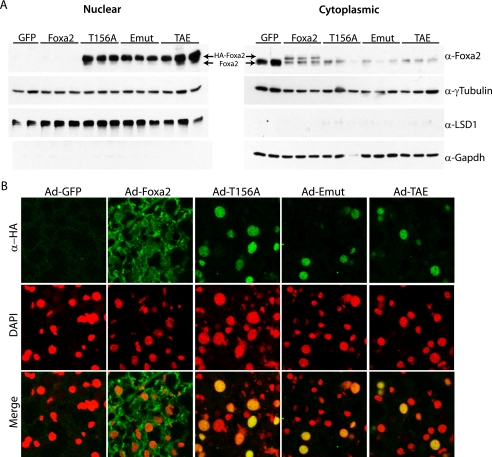
To assess the importance of nuclear export on the physiological activity of Foxa2 in the context of metabolic disorders, and to elucidate whether the biological activities of Foxa2 mutants show the same regulation in vivo, we expressed wild-type and mutant T156A, Emut, and TAE Foxa2 proteins in the livers of ob/ob mice using recombinant adenoviruses. We demonstrated previously that endogenous and Ad-Foxa2 are constitutively cytoplasmic and inactive in hyperinsulinemic mouse models of type 2 diabetes, whereas Ad-T156A remains in the nucleus and is able to restore Foxa2 activity (15). Nuclear and cytoplasmic extracts from the livers of these mice confirmed that endogenous and Ad-Foxa2 are constitutively cytoplasmic, whereas the Ad-T156A, Ad-Emut, and Ad-TAE Foxa2 constructs remain in the nucleus (Fig. 4A). Immunofluorescence microscopy analysis was also used to confirm these results (Fig. 4B).
FIGURE 4.
Emut Foxa2 is nuclear in hyperinsulinemic mice. Ob/ob mice were injected with the indicated adenovirus, and livers were analyzed 10 days post-injection by cellular fractionation and immunoblotting (A) or immunofluorescence microscopy (B). γ-Tubulin was used as a general loading control, and LSD1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as the nuclear and cytoplasmic extraction controls, respectively. Recombinant Foxa2 is shown in green, and red staining indicates the nuclear dye 4,6-diamidino-2-phenylindole (DAPI).
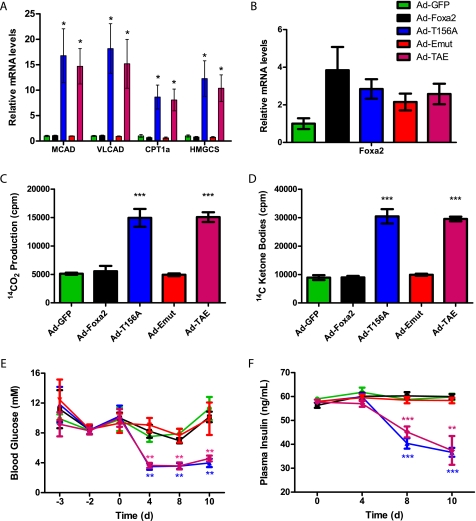
To analyze the activity of the export mutant in these mice, which is the most important functional readout, we measured target gene activation by real-time PCR and assayed various metabolic parameters over a span of 10 days. As expected, mRNA levels of Foxa2 target genes involved in mitochondrial fatty acid oxidation and ketone body formation (MCAD, VLCAD, CPT1α, and HMG-CoA synthase, respectively) were up-regulated in the livers of T156A-injected mice, but no significant change was observed in mice injected with wild-type Foxa2. Consistent with our previous findings, Emut Foxa2 was unable to activate transcription of target genes in these hyperinsulinemic mice, whereas the double mutant TAE Foxa2 restored constitutive activation (Fig. 5A). As a control, we checked the Foxa2 mRNA levels, which were up-regulated roughly 2-fold in all mice relative to Ad-GFP controls (Fig. 5B).
FIGURE 5.
Emut Foxa2 is inactive in livers of ob/ob mice. Transcript levels of Foxa2 and target genes and metabolic parameters were measured in ob/ob mice injected with Ad-GFP (green), Ad-Foxa2 (black), Ad-T156A (blue), Ad-Emut (red), or Ad-TAE (magenta) after a 6-h fast. A, mean mRNA levels of Foxa2 target genes from livers, n ≥ 3. MCAD, medium chain acyl-CoA dehydrogenase; VLCAD, very long chain acyl-CoA dehydrogenase; CPT1α, carnitine palmitoyltransferase 1α; HMGCS, HMG-CoA synthase. B, mean mRNA levels of Foxa2. Primers were designed to recognize both rat and mouse isoforms. β-Oxidation (C) and ketone body generation (D) by liver mitochondria was measured by the formation of 14CO2 and 14C-labeled acid-soluble products, respectively, from 14C-palmitic acid. E, blood glucose levels. Time point zero indicates the time of adenovirus injection. F, plasma insulin levels. Values shown represent the mean ± S.E. *, p < 0.05; **; p < 0.01; ***, p < 0.001 by unpaired t test compared with Ad-GFP.
As a physiological readout, we measured hepatic mitochondrial fatty acid β-oxidation and ketone body production in the livers of hyperinsulinemic ob/ob mice that were infected with Ad-GFP, Ad-Foxa2, Ad-T156A, Ad-Emut, or Ad-TAE. Consistent with the gene expression data, fatty acid oxidation and ketone body production remained unchanged in the Ad-Foxa2 and Ad-Emut injected mice, indicating that both are functionally inactive in hyperinsulinemic conditions. On the contrary, the phosphorylation-deficient Ad-T156A and Ad-TAE groups were immune to inactivation by insulin and showed significant increases in fatty acid metabolism (Fig. 5, C and D). Likewise, blood glucose and plasma insulin levels decreased over 10 days in the T156A- and TAE-injected mice, whereas no decrease was seen in the Ad-Foxa2 or Ad-Emut groups (Fig. 5, E and F). Hepatic triglyceride content also showed a similar trend (data not shown). In contrast to previous assumptions, these data demonstrate that nuclear exclusion is not necessary for transcriptional inactivation of Foxa2. We show here that Akt-dependent phosphorylation is a more direct determinant of Foxa2 activity and thus of its physiological functions in maintaining hepatic lipid metabolism.
DISCUSSION
In this study we investigated the molecular mechanisms that govern the regulation of Foxa2 activity in response to insulin signaling. First, we show that Foxa2 contains a functional CRM1-dependent (LMB-sensitive) nuclear export site, which is necessary for its nuclear exclusion in response to insulin stimulation. Interestingly, alignment of the Foxa2 NES shows that it is well conserved throughout vertebrate homologues of Foxa2. Minor variations within the sequence occur from Xenopus to human (methionines instead of leucines at amino acids 110 and 113), but these changes conserve the overall character of the export sequence and are not likely to hinder nuclear export capability. Indeed, it has been shown that hydrophobic residues other than leucine (including isoleucine, valine, methionine, and phenylalanine) may constitute a functional CRM1 NES (32, 36).
In contrast to this relatively high degree of NES conservation from Xenopus laevis to Homo sapiens, we observed little if any sequence homology in the NES region of Foxa1, Foxa3, and Fork head, the Drosophila homologue of Foxa2. This was expected for Foxa1 and Foxa3, neither of which has been shown to be regulated via nuclear export or to contain the Foxa2 AKT phosphorylation site or shuttle in response to insulin (7). Likewise, the lack of NES conservation in the Drosophila orthologue suggests that Fork head may not be capable of shuttling in response to insulin, and that the nuclear export capability may have arisen later in evolution. Unlike Foxa1 or Foxa3, however, the AKT phosphorylation site is conserved in Fork head. It will therefore be interesting to see whether the same mechanism of insulin-induced phosphorylation exists in flies and whether this also results in Fork head inactivation.
The second major finding we report here is that nuclear export of Foxa2 is not necessary for inhibition of its transcriptional activity by insulin. Previous data from our laboratory provide evidence for a strong correlation among insulin signaling, Thr-156 phosphorylation, nuclear exclusion, and inactivation of Foxa2 (7, 15). Although nuclear exclusion certainly leads to inhibition of transcriptional activity, there was previously no means of differentiating between these correlative observations. The novel identification of a functional NES in Foxa2 has now allowed us to uncouple these events. Our new data clearly demonstrate that ablation of the nuclear export site, both in vitro and in vivo, results in constitutively nuclear Foxa2, which is still rendered transcriptionally inactive upon insulin stimulation. In addition, we have shown that phosphorylation at Thr-156 is necessary for both nuclear export and transcriptional inactivation, as export mutants that are constitutively located in the nucleus are still phosphorylated by insulin signaling, and T156A mutation results in constitutive Foxa2 activity.
Mechanisms to this effect have also been reported for the FoxO family of transcription factors. Phosphorylation of three critical residues in Foxo1 have been shown to contribute to nuclear export and subsequent loss of transcriptional activity in response to insulin/insulin growth factor (IGF) signaling (25, 26, 37). In vitro data suggest that Ser-256 acts as a “gatekeeper” to subsequent regulatory events, with phosphorylation at this residue resulting in transcriptional inhibition of nuclear Foxo1 (29, 38). However, additional posttranscriptional modifications have also been shown to contribute to the regulation of FoxO family members, including acetylation, ubiquitination, and methylation (reviewed in Refs. 39 and 40). The relative contribution and exact mechanisms of these modifications are still being worked out.
Our data support a model whereby phosphorylation of Foxa2 acts as the dominant signal for transcriptional inactivation; however, we cannot exclude the possibility that additional post-translational modifications exist. Whether this single phosphorylation event is sufficient for inactivation remains to be determined. One plausible hypothesis is that Thr-156 phosphorylation, which occurs N-terminally to the DNA binding domain, directly decreases the DNA binding affinity of Foxa2. Although electrophoretic mobility shift assay data suggest that this is not the case, it was not possible to monitor the phosphorylation state of Foxa2 in these assays (7). Alternatively, or additionally, phosphorylation (or other modifications) could affect coactivator or histone binding. Our data thus provide a novel perspective on the regulation of Foxa2 by insulin, which could potentially be applied to the hormonal regulation of other transcription factors. Additionally, the discovery of an essential NES controlling insulin-induced nuclear export of Foxa2 lays a strong foundation for the identification of additional signaling pathways and/or post-translational modifications involved in the regulation of Foxa2 activity.
The constitutive inactivation of Foxa2 by insulin, in addition to the beneficial effects of constitutively active Foxa2 in mouse models of obesity, make understanding the molecular mechanisms of its regulation of great scientific and potentially therapeutic interest. Taken together, our data show that Foxa2 is subject to active nuclear export in response to insulin signaling, that this export is not a prerequisite for transcriptional inactivation, and that Foxa2 phosphorylation at Thr-156 is currently the most direct readout of Foxa2 activity.
Supplementary Material
Acknowledgments
We thank Christian Wolfrum for advice and insightful discussions and Manuela Hitz for technical assistance.
This work was supported in part by a LiverX Grant from SystemsX.ch (Swiss Initiative in Systems Biology).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- HNF
- hepatocyte nuclear factor
- LMB
- leptomycin B
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFP
- green fluorescent protein
- PBS
- phosphate-buffered saline
- NES
- nuclear export signal
- Emut
- export mutant
- HA
- hemagglutinin.
REFERENCES
- 1.Besnard V., Wert S. E., Hull W. M., Whitsett J. A. (2004) Gene Expr. Patterns 5, 193–208 [DOI] [PubMed] [Google Scholar]
- 2.Lai E., Prezioso V. R., Tao W. F., Chen W. S., Darnell J. E., Jr. (1991) Genes Dev. 5, 416–427 [DOI] [PubMed] [Google Scholar]
- 3.Costa R. H., Grayson D. R., Darnell J. E., Jr. (1989) Mol. Cell. Biol. 9, 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark K. L., Halay E. D., Lai E., Burley S. K. (1993) Nature 364, 412–420 [DOI] [PubMed] [Google Scholar]
- 5.Cirillo L. A., Lin F. R., Cuesta I., Friedman D., Jarnik M., Zaret K. S. (2002) Mol. Cell 9, 279–289 [DOI] [PubMed] [Google Scholar]
- 6.Cirillo L. A., McPherson C. E., Bossard P., Stevens K., Cherian S., Shim E. Y., Clark K. L., Burley S. K., Zaret K. S. (1998) EMBO J. 17, 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfrum C., Besser D., Luca E., Stoffel M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11624–11629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfrum C., Stoffel M. (2006) Cell Metabolism 3, 99–110 [DOI] [PubMed] [Google Scholar]
- 9.Oberkofler H., Hafner M., Felder T., Krempler F., Patsch W. (2009) J. Mol. Med. 87, 299–306 [DOI] [PubMed] [Google Scholar]
- 10.Kaestner K. H., Hiemisch H., Schütz G. (1998) Mol. Cell. Biol. 18, 4245–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinstein D. C., Ruiz i Altaba A., Chen W. S., Hoodless P., Prezioso V. R., Jessell T. M., Darnell J. E., Jr. (1994) Cell 78, 575–588 [DOI] [PubMed] [Google Scholar]
- 12.Ang S. L., Rossant J. (1994) Cell 78, 561–574 [DOI] [PubMed] [Google Scholar]
- 13.Shih D. Q., Navas M. A., Kuwajima S., Duncan S. A., Stoffel M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10152–10157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaestner K. H., Katz J., Liu Y., Drucker D. J., Schütz G. (1999) Genes Dev. 13, 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfrum C., Asilmaz E., Luca E., Friedman J. M., Stoffel M. (2004) Nature 432, 1027–1032 [DOI] [PubMed] [Google Scholar]
- 16.Wolfrum C., Howell J. J., Ndungo E., Stoffel M. (2008) J. Biol. Chem. 283, 16940–16949 [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaka T., Shimano H., Yahagi N., Kato T., Atsumi A., Yamamoto T., Inoue N., Ishikawa M., Okada S., Ishigaki N., Iwasaki H., Iwasaki Y., Karasawa T., Kumadaki S., Matsui T., Sekiya M., Ohashi K., Hasty A. H., Nakagawa Y., Takahashi A., Suzuki H., Yatoh S., Sone H., Toyoshima H., Osuga J., Yamada N. (2007) Nat. Med. 13, 1193–1202 [DOI] [PubMed] [Google Scholar]
- 18.Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., Ruvkun G. (1997) Nature 389, 994–999 [DOI] [PubMed] [Google Scholar]
- 19.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. (1993) Nature 366, 461–464 [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb S., Ruvkun G. (1994) Genetics 137, 107–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris J. Z., Tissenbaum H. A., Ruvkun G. (1996) Nature 382, 536–539 [DOI] [PubMed] [Google Scholar]
- 22.Paradis S., Ruvkun G. (1998) GenesDev. 12, 2488–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kops G. J., de Ruiter N. D., De Vries-Smits A. M., Powell D. R., Bos J. L., Burgering B. M. (1999) Nature 398, 630–634 [DOI] [PubMed] [Google Scholar]
- 24.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 25.Tang E. D., Nuñez G., Barr F. G., Guan K. L. (1999) J. Biol. Chem. 274, 16741–16746 [DOI] [PubMed] [Google Scholar]
- 26.Rena G., Guo S., Cichy S. C., Unterman T. G., Cohen P. (1999) J. Biol. Chem. 274, 17179–17183 [DOI] [PubMed] [Google Scholar]
- 27.Durham S. K., Suwanichkul A., Scheimann A. O., Yee D., Jackson J. G., Barr F. G., Powell D. R. (1999) Endocrinology 140, 3140–3146 [DOI] [PubMed] [Google Scholar]
- 28.Ayala J. E., Streeper R. S., Desgrosellier J. S., Durham S. K., Suwanichkul A., Svitek C. A., Goldman J. K., Barr F. G., Powell D. R., O'Brien R. M. (1999) Diabetes 48, 1885–1889 [DOI] [PubMed] [Google Scholar]
- 29.Tsai W. C., Bhattacharyya N., Han L. Y., Hanover J. A., Rechler M. M. (2003) Endocrinology 144, 5615–5622 [DOI] [PubMed] [Google Scholar]
- 30.Hoppel C., DiMarco J. P., Tandler B. (1979) J. Biol. Chem. 254, 4164–4170 [PubMed] [Google Scholar]
- 31.Fréneaux E., Labbe G., Letteron P., Dinh T. L., Degott C., Genève J., Larrey D., Pessayre D. (1988) Hepatology 8, 1056–1062 [DOI] [PubMed] [Google Scholar]
- 32.la Cour T., Gupta R., Rapacki K., Skriver K., Poulsen F. M., Brunak S. (2003) J. Nucleic Acids Res. 31, 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. (1997) Nature 390, 308–311 [DOI] [PubMed] [Google Scholar]
- 34.Kudo N., Wolff B., Sekimoto T., Schreiner E. P., Yoneda Y., Yanagida M., Horinouchi S., Yoshida M. (1998) Exp. Cell Res. 242, 540–547 [DOI] [PubMed] [Google Scholar]
- 35.Wen W., Meinkoth J. L., Tsien R. Y., Taylor S. S. (1995) Cell 82, 463–473 [DOI] [PubMed] [Google Scholar]
- 36.Yanai H., Kobayashi T., Hayashi Y., Watanabe Y., Ohtaki N., Zhang G., de la Torre J. C., Ikuta K., Tomonaga K. (2006) J. Virol. 80, 1121–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biggs W. H., 3rd, Meisenhelder J., Hunter T., Cavenee W. K., Arden K. C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Gan L., Pan H., Guo S., He X., Olson S. T., Mesecar A., Adam S., Unterman T. G. (2002) J. Biol. Chem. 277, 45276–45284 [DOI] [PubMed] [Google Scholar]
- 39.Vogt P. K., Jiang H., Aoki M. (2005) Cell Cycle 4, 908–913 [DOI] [PubMed] [Google Scholar]
- 40.Calnan D. R., Brunet A.Oncogene 27, 2276–2288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.