Abstract
Interferon-γ (IFNγ) has an antiproliferative effect on a variety of tumor cells. However, many tumor cells resist treatment with IFNs. Here, we show that IFNγ fails to inhibit the growth of some types of oral squamous cell carcinoma (OSCC) cells that possess a fully functional IFNγ/STAT1 (signal transducer and activator of transcription-1) signaling pathway. IFNγ inhibited the growth of the HSC-2, HSC-3, and HSC-4 OSCC cell lines. However, Ca9–22 cells were resistant to IFNγ despite having intact STAT1-dependent signaling, such as normal tyrosine phosphorylation, DNA binding activity, and transcriptional activity of STAT1. The growth inhibition of HSC-2 cells resulted from S-phase arrest of the cell cycle. IFNγ inhibited cyclin A2 (CcnA2)-associated kinase activity, which correlated with the IFNγ-mediated down-regulation of CcnA2 and Cdk2 expression at both the transcriptional and post-transcriptional level in HSC-2 cells but not in Ca9–22 cells. RNAi-mediated knockdown of CcnA2 and Cdk2 resulted in growth inhibition in both cell lines. These results indicate that the resistance of OSCC to IFNγ is not due simply to the deficiency in STAT1-dependent signaling but results from a defect in the signaling component that mediates this IFNγ-induced down-regulation of CcnA2 and Cdk2 expression at the transcriptional and post-transcriptional levels.
Interferon-γ (IFNγ)2 is a cytokine produced by activated T cells and natural killer cells. It exhibits a number of biological activities in host-defense systems and immunoregulation, including anti-viral and anti-tumor responses (1, 2). The antiproliferative activity of IFNs has been well documented in a variety of tumor cell types. Multiple studies have shown that both type I (IFNα/β) and type II (IFNγ) IFNs induce cell cycle arrest at G0/G1, which is mediated by the up-regulation of the cyclin-dependent kinase inhibitors p21WAF1/Cip1 and p27Kip1 after IFN treatment (3–7). The p21 protein has been shown to inhibit cyclin/ Cdk activity, which phosphorylates the retinoblastoma (Rb) tumor suppressor and then activates members of the E2F transcription factor family (8, 9). IFNγ-induced signal transducer and activator of transcription 1 (STAT1) has been shown to induce transactivation of the p21WAF1/Cip1 gene (3). In some tumor cells, however, the arrest of IFNγ-mediated, cyclin-dependent kinase inhibitor-independent cell growth has been reported (10–12). Thus, IFNγ-mediated growth inhibition appears to be mediated by multiple pathways, depending on the cell type, and the molecular mechanisms by which IFNγ inhibits tumor cell growth remain to be fully elucidated.
Although IFN exhibits a potent antiproliferative and proapoptotic effects on many tumor cells, some types of tumor cells resist IFN treatment (13–17). Several studies have demonstrated the molecular mechanisms underlying this resistance to IFN. Defects in components of the IFN signaling pathway, such as the expression of the IFNγ receptor, Janus kinase (JAK), STAT1, STAT2, and interferon regulatory factor-9 (IRF-9/p48), have been identified in resistant cells (13–17). Furthermore, reduced expression of ISGF-3 (a tetramer complex with STAT1, STAT2, and IRF-9) has been detected in skin squamous carcinoma cells from surgical specimens (18). However, some types of tumor cells have been reported to resist IFNs despite having a normal JAK-STAT pathway (7, 12, 19, 20). Thus, both JAK-STAT-dependent and-independent mechanisms appear to explain IFN resistance. However, the mechanism of JAK-STAT-independent IFN resistance remains poorly understood.
To gain insight into the molecular mechanisms responsible for the antiproliferative effect of IFNγ and the resistance to the IFNγ-mediated effect in human oral squamous cell carcinomas (OSCC) cells, we examined the effect of IFNγ on the growth of human OSCC cell lines. We also explored the mechanisms underlying the antiproliferative effect of IFNγ and the unresponsiveness of cells to this molecule. We demonstrated that IFNγ inhibits the growth of the HSC-2, HSC-3, and HSC-4 human OSCC cell lines, whereas Ca9–22 cells are resistant to IFNγ despite the presence of intact STAT1-dependent signaling. IFNγ inhibited the expression of cyclin A (CcnA2) and cyclin-dependent kinase 2 (Cdk2) in HSC-2 cells, but not in Ca9–22 cells, and knockdown of either CcnA2 or Cdk2 by siRNA inhibited cell growth in both cell types. Furthermore, IFNγ suppressed the promoter activity of the CcnA2 and Cdk2 genes and destabilized CcnA2 and Cdk2 mRNAs in HSC-2 cells but not in Ca9–22 cells. These results suggest that the resistance of OSCC cells to the antiproliferative effect of IFNγ is not because of a deficiency in STAT1-dependent signaling but, instead, results from a defective signaling component that mediates the IFNγ-induced down-regulation of CcnA2 and Cdk2 expression.
EXPERIMENTAL PROCEDURES
Reagents
Recombinant human IFNγ was purchased from BioSource International Inc. (Camarillo, CA). A cell counting kit was obtained from Dojin Laboratories (Tokyo, Japan). Antibodies against CcnA2 (H432), Cdk2 (M-2), Cdk6 (C-21), p21 (C-19), STAT1 (E23), STAT3 (C-20), histone H1 (AE-4), and β-actin (I-19) were obtained from Santa Cruz Biotechnology (Hercules, CA). Antibodies against cyclin D1 (#2926), cyclin D2 (#2924), cyclin D3 (#2936), cyclin E (#4129), Cdk4 (#2906), phospho-STAT1 Tyr-701 (#9171S), phospho-STAT3 Tyr-705 (#9131S), phospho-Akt Thr-308 (#2965), phospho-Akt Ser-473 (#4060), pan-Akt (#4691), and phospho-PDK1 Ser-241 (#3061) were obtained from Cell Signaling Technology (Danvers, MA). Anti-α-tubulin antibody was obtained from Sigma. FuGENE transfection reagent and histone H1 were purchased from Roche Diagnostics. StealthTM Select RNAi oligonucleotides for CcnA2, Cdk2, and green fluorescent protein were obtained from Invitrogen.
Cell Culture and Proliferation Assay
The HSC-2, HSC-3, HSC-4, and Ca9–22 human OSCC cell lines were described previously (21–23). These cells were originally isolated from metastatic OSCC cells derived from the oral cavity (21). For the cell proliferation assays, cells were seeded in 96-well plates (3 × 103 cells/well) and grown for 20 h before being treated with IFNγ. After treatment with IFNγ, the viable cell number was determined using the Cell Counting kit (Dojin) according to the manufacturer's protocol. After incubation with the reagent, the optical density at 450 nm was measured using a microplate reader. The cell number after treatment with IFNγ was also counted using a hemocytometer after trypsinization.
Preparation of Nuclear Extracts and Electrophoretic Mobility Shift Assay
Nuclear extracts were prepared as described previously (24) using a modification of the method described by Dignam et al. (25). Nuclear extracts (5 μg of total protein) were incubated in 12.5 μl of 20 mm HEPES, pH 7.9, containing 50 mm KCl, 0.1 mm EDTA, 1 mm dithiothreitol, 5% glycerol, 200 μg/ml bovine serum albumin, and 1.25 μg of poly(dI-dC). A 32P-labeled, double-stranded oligonucleotide from the IRF-1 gene (5′-tcgaGCCTGATTTCCCCGAAATGAGGC-3′) (26) was then added to the reaction mixture. The reaction products were analyzed by electrophoresis in a 5% polyacrylamide gel.
Western Blotting
Total cell lysates were resolved in SDS-PAGE sample buffer (62.5 mm Tris, pH 6.8, containing 2% SDS, 20% glycerol, 5% β-mercaptoethanol, and 0.2% bromphenol blue) and separated by SDS-PAGE in a 7.5% polyacrylamide gel, as described previously (23).
siRNA Transfection
siRNA transfection was performed with Lipofectamine RNAiMaxTM according to the manufacturer's instructions (Invitrogen). Briefly, 5 × 104 cells were seeded in a 6-well plate. One day after plating, the siRNA (final concentration, 10 nmol) was suspended in 1 ml of RNAiMAX and added to each well. Cells were harvested 48 h after transfection and either used to determine the efficiency of knockdown or reseeded into a 96-well plate for the cell proliferation assays.
CcnA2, Cdk2, and c-Myc Promoter Constructs
Sequences encoding the 5′-flanking promoter/enhancer region of the human CcnA2 and Cdk2 genes were cloned from human genomic DNA using PCR with high fidelity Platinum PCR SuperMix (Invitrogen) and a set of primers corresponding to the human CcnA2 (from −881 to +216) (GenBankTM accession number X68303) (27) and the human Cdk2 (from −767 to +19) genomic sequences (GenBankTM accession number U50730) (28). The amplified PCR fragments were individually subcloned into a luciferase reporter construct (pGL3-Basic, Promega), and the nucleotide sequence was confirmed. The resulting plasmids were designated as pGL-CcnA2–881 (CcnA2) and pGL-Cdk2–767 (Cdk2). The luciferase reporter plasmid pDel-1, which contains a 2.5-kb sequence from the 5′-flanking region of the human c-myc gene (29), was obtained from Addgene (Cambridge, MA). The luciferase reporter construct pGL-IRF-1, which contains a 1.3-kb sequence from the 5′-flanking region of the IRF-1 gene (26, 30), and pTK GASluc, which contains four copies of the IFNγ activation site (GAS) motif from the IRF-1 gene upstream of the herpes simplex virus-thymidine kinase (TK) promoter, were described previously (31).
Luciferase Reporter Assay
The transfection procedure and luciferase reporter assay were described previously (23). To standardize the transfection efficiencies, the firefly luciferase activity from pGL luciferase reporter plasmids was normalized to the Renilla luciferase activity from the pRL-TK plasmid (Promega), and the relative luciferase activities in the different OSCC cells were normalized to the activity of the pCMVluc or the pGL3-Control plasmid.
Cell Cycle Analysis
Cells were cultured in 6-cm dishes 20 h before stimulation and were then treated with IFNγ for the indicated periods. After trypsinization, the cells were collected by centrifugation, washed in phosphate-buffered saline and fixed with cold 70% ethanol. Cells were then washed with phosphate-buffered saline and treated with 250 μg/ml of RNase at 37 °C for 40 min. Cellular DNA was stained with 50 μg/ml of propidium iodide, and 5 × 104 cells were analyzed on a FACScan flow cytometer (EPICS ALTRA, Beckman Coulter, Fullerton, CA). The proportions of cells in different stages of the cell cycle were determined using WinCycle software (Beckman Coulter).
DNA Synthesis
Cells were cultured in 24-well plates for 20 h in complete medium before treatment with IFNγ. After treatment with or without IFNγ for varying periods, cells were pulse-labeled with 1 μCi of [3H]thymidine (PerkinElmer Life Sciences) for the last hour of the cultivation. The cells were then washed with phosphate-buffered saline, fixed in cold 5% trichloroacetic acid, and washed with 5% trichloroacetic acid. Incorporated [3H]thymidine was extracted with 0.2% SDS and 0.5 n NaOH and measured in a liquid scintillation counter (Aloka, Tokyo, Japan).
Cdk Kinase Assay
CcnA2-dependent kinase activity was assessed in vitro by the phosphorylation of histone H1. Cells were lysed in an ice-cold kinase lysis buffer (50 mm Hepes, pH 7.4, 150 mm NaCl, 0.1% Nonidet P-40, 0.1% Triton X-100, 1 mm EDTA, 2.5 mm EGTA, 10 mm glycerophosphate, 50 mm NaF, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and proteinase inhibitor mixture (Sigma)). After centrifugation, 1 μg of anti-CcnA2 antibody was added, and the sample was agitated for 1 h at 4 °C. After incubation, protein A-agarose beads were added and incubated for an additional hour. The immunoprecipitates were washed with Cdk kinase buffer (50 mm Hepes, pH 7.4, 10 mm MgCl2, 10 mm MnCl2, and 1 mm dithiothreitol) and mixed in a 50-μl reaction containing 50 mm Hepes, pH 7.4, 40 mm MgCl2, 25 mm ATP, 2.5 μCi of [γ-32P]ATP (3000 Ci/mmol), and 1 μg of histone H1 at 37 °C for 30 min. Reactions were terminated by the addition of SDS sample buffer, and the samples were subjected to 10% SDS-PAGE. The gels were dried, and phosphorylation of histone H1 was detected with autoradiography.
Preparation of RNA and Northern Hybridization Analysis
The preparation of total RNA by the guanidine isothiocyanate-cesium chloride method and northern hybridization analyses were carried out as described previously (24). The cDNA fragments for human CcnA2 and Cdk2 were prepared by reverse transcriptase-PCR using a set of primers corresponding to sequences for human CcnA2 (GenBankTM accession number NM001237) and Cdk2 (GenBankTM accession number NM001798). The PCR products were subcloned into pBluescript (Stratagene, La Jolla, CA), and the nucleotide sequences were confirmed. Some northern blots were quantified using a Molecular Imager (Bio-Rad).
Real-time Quantitative RT-PCR (qRT-PCR)
qRT-PCR was performed on the LightCycler 480 real-time PCR system (Roche Diagnostics) using the TaqMan® probe (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Briefly, total RNA (500 ng) was reverse-transcribed with random hexamers using the High Capacity cDNA reverse transcription kit (Applied Biosystems) in a 20-μl reaction volume. After the reverse transcriptase reaction, qRT-PCR was performed using the TaqMan® Gene Expression Master Mix reagents, the TaqMan® gene expression assay (c-myc, assay ID Hs00153408_m1), and the endogenous control (18 S rRNA) in a final volume of 20 μl. Reactions were performed and analyzed using the LightCycler 480 system (Roche Diagnostics). Each mRNA level was normalized to that of the 18 S rRNA, and the relative expression level was determined using the standard curve method for multiplex PCR. The transcript abundance was calculated as the percentages of the control or experimental sample values normalized to the 18 S rRNA.
RESULTS
IFNγ Differentially Inhibits the Growth of Human Oral Squamous Cell Carcinomas
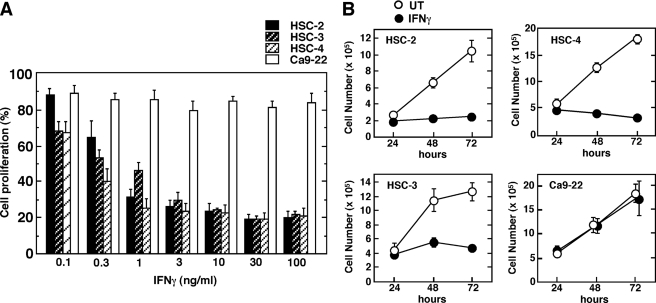
IFNγ has been shown to inhibit the growth of a wide variety of tumor cells. We initially examined the effects of various doses of IFNγ on the growth of four asynchronously proliferating human OSCC lines (Fig. 1A). Treatment of HSC-2, HSC-3, and HSC-4 cells with IFNγ inhibited proliferation in a dose-dependent fashion. However, IFNγ had only a marginal effect on the proliferation of Ca9–22 cells, even at high concentrations (100 ng/ml). In separate experiments, a higher concentration of IFNγ (300 ng/ml) similarly showed no significant inhibitory effect on the proliferation of Ca9–22 cells (data not shown). The differential sensitivity of cell proliferation to IFNγ was also determined by direct cell counting at different times during culture (Fig. 1B). IFNγ inhibited the growth of HSC-2, HSC-3, and HSC-4 cells, with a significant inhibition observed after 48 h of IFNγ treatment. By contrast, IFNγ failed to inhibit the growth of Ca9–22 cells at any of the time points examined.
FIGURE 1.
Effect of IFNγ on the growth of human oral squamous cell carcinoma lines. A, OSCC cell lines were seeded in 96-well plates and incubated for 20 h followed by treatment with or without various concentrations of IFNγ for 96 h. Cell proliferation was determined using a cell counting kit, as described under “Experimental Procedures.” Each column and bar represents the mean ± S.E. of four independent experiments. B, cells were seeded in a 6-cm dish and incubated for 20 h before stimulation with IFNγ (10 ng/ml). After cultivation with or without IFNγ (untreated (UT)) for the indicated time periods, cell numbers were determined using a hemocytometer. Data correspond to the mean ± S.E. of four independent experiments.
Different Sensitivities of OSCC Cells to IFNγ Are Independent of STAT1 Activation
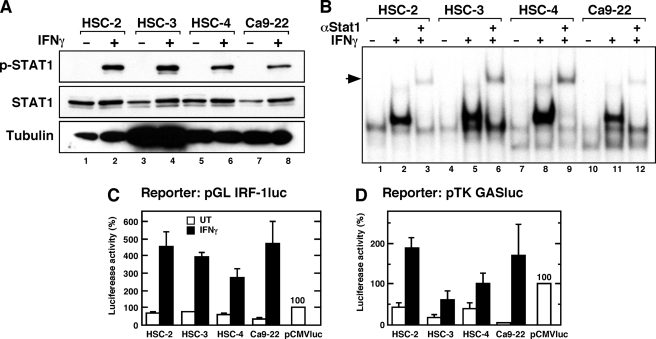
IFNγ-mediated biological activities are mediated by the STAT1-dependent signaling pathway (32, 33). To determine whether the differential antiproliferative effect of IFNγ is because of a defect in STAT1-dependent function, we analyzed the functional integrity of IFNγ-induced STAT1 activation in these oral carcinoma cells. Initially, we assessed tyrosine phosphorylation of STAT1 (Try-701) in the OSCC cells by Western blotting (Fig. 2A). IFNγ caused phosphorylation of STAT1 on Tyr-701, and all of the OSCC cells showed similar levels of phosphorylated STAT1. We next analyzed the DNA binding activity of STAT1 in nuclear extracts from cells treated with IFNγ (Fig. 2B). Gel-shift assays using nuclear extracts from OSCC cells showed that in all cell lines IFNγ induced the formation of a prominent DNA binding complex on the probe for the IRF-1 GAS, which contains a high affinity STAT1 binding motif (26, 30, 34). The presence of STAT1 in the DNA binding complexes was demonstrated by a supershift assay with anti-STAT1 antibody.
FIGURE 2.
IFNγ-induced STAT1-dependent transcriptional activity is intact in the OSCC lines. A, levels of STAT1 phosphorylation on tyrosine 701 in IFNγ-stimulated OSCC cells. Cells were stimulated with IFNγ (10 ng/ml) for 1 h before the preparation of total cell lysates. The lysates were subjected to electrophoresis (20 μg per lane) followed by Western blotting with the indicated antibodies. B, DNA binding activity of STAT1 in nuclear extracts from IFNγ-stimulated cells. The cells were treated with IFNγ (10 ng/ml) for 1 h before the preparation of nuclear extracts. Samples (10 μg) of each nuclear extract were analyzed for GAS binding activity by electrophoretic mobility shift assay. Super-shifted complexes containing the anti-STAT1 antibody are shown. Similar results were obtained in three separate experiments. C and D, STAT1-dependent transcriptional activities in IFNγ-stimulated OSCC lines. The cells were transiently transfected with the indicated luciferase reporter construct. Twenty-four hour after transfection, the cells were either left untreated (UT) or were treated with IFNγ (10 ng/ml) for 8 h, after which luciferase activity was measured. The relative luciferase activity is shown as a percentage of the activity from pCMVluc-transfected cells. Each column and bar represents the mean ± S.E. of three independent experiments.
We further analyzed IFNγ-induced STAT1-dependent transcriptional activity using luciferase reporter constructs containing 1.3 kb of the 5′-flanking sequence from the IRF-1 gene (Fig. 2C), which is regulated by STAT1 (26, 30), and a heterologous promoter construct containing four copies of the GAS motif from the IRF-1 gene placed upstream of the herpes simplex virus-TK promoter (31) (Fig. 2D). IFNγ markedly induced the luciferase activity driven by the IRF-1 promoter and the TK heterologous promoter in all OSCC cells. These results indicate that IFNγ-induced STAT1-dependent transcriptional activity is intact in these OSCC cells and that activation of STAT1 by itself may be insufficient to mediate the antiproliferative effect of IFNγ.
Recent studies have shown that IFNγ-activated STAT3 functions as a component of the STAT1-independent signaling pathway (35). To determine whether the differential antiproliferative effect of IFNγ is because of IFNγ-induced STAT3, we analyzed the levels of tyrosine-phosphorylated STAT3 (Tyr-705) in the OSCC cells by Western blotting (supplemental Fig. 1). Although constitutively active tyrosine-phosphorylated STAT3 was observed in all OSCC cells, IFNγ failed to further enhance the phosphorylated Tyr-705 STAT3. These results indicate that STAT3 is not involved in the differences observed in the antiproliferative effect of IFNγ on the different OSCC cells.
IFNγ has been shown to activate the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway, which mediates some biological activities of IFNγ (36). To examine whether IFNγ differentially affects the PI3K/Akt signaling pathway in HSC-2 and Ca9–22 cells, we measured the levels of the phosphorylated forms of Akt over time using Western blotting techniques (supplemental Fig. 2). Both HSC-2 and Ca9–22 cells constitutively expressed phosphorylated forms of Akt on Thr-308 and Ser-473. However, IFNγ failed to up-regulate these phosphorylated forms of Akt. Consistent with these results, phosphorylation of PDK1 (3-phosphoinositide-dependent protein kinase-1) (37, 38), an upstream kinase responsible for the phosphorylation of Akt Thr-308, was also unchanged in the cells treated with IFNγ. These results do not implicate the PI3K/Akt pathway in the observed differential sensitivity to IFNγ.
IFNγ Induces Cell Cycle Arrest in S-phase in OSCC Cells
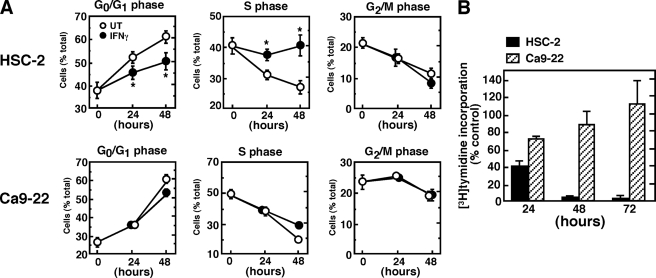
To understand the underlying mechanism of the IFNγ-induced differential growth inhibition of OSCC cells, we next analyzed the cell-cycle distribution in response to IFNγ treatment using flow cytometry (Fig. 3). Quantitative analysis of the distribution of cells in the cell cycle indicates that treatment of HSC-2 cells with IFNγ blocks S-phase progression concomitant with a decrease in the relative proportion of cells in the G0/G1 phase (Fig. 3A). By contrast, Ca9–22 cells showed only a slight change in cell cycle distribution after IFNγ treatment. To confirm that the S-phase cell cycle arrest in HSC-2 cells correlated with the antiproliferative effect of IFNγ, we assessed the rate of DNA synthesis by measuring [3H]thymidine incorporation into DNA (Fig. 3B). A marked inhibition of [3H]thymidine incorporation was observed in HSC-2 cells after 48 h of IFNγ treatment.
FIGURE 3.
Effect of IFNγ on the proportions of HSC-2 and Ca9–22 cells at different stages of the cell cycle. A, asynchronously growing HSC-2 or Ca9–22 cells were treated with or without IFNγ (10 ng/ml) for the indicated time periods. The cells were then harvested, and flow cytometric analysis for DNA content was performed, as described under “Experimental Procedures.” Flow cytometric data were analyzed using WinCycle software to determine the proportion of cells in each stage of the cell cycle. Data shown are the means ± S.E. of 8–11 independent experiments. Asterisks denote a statistically significant difference from untreated (UT) cultures (*, p < 0.05, Student's t test). B, inhibition of DNA synthesis in IFNγ-treated HSC-2 cells. The cells were cultured in the presence or absence of IFNγ (10 ng/ml) for the indicated time periods, and the rate of DNA synthesis was determined by pulse labeling with [3H]thymidine for the last hour of incubation. Each column and bar represents the mean ± S.E. of three independent experiments.
IFNγ Inhibits CcnA2-associated Kinase Activity and Down-regulates CcnA2 and Cdk2 Expression
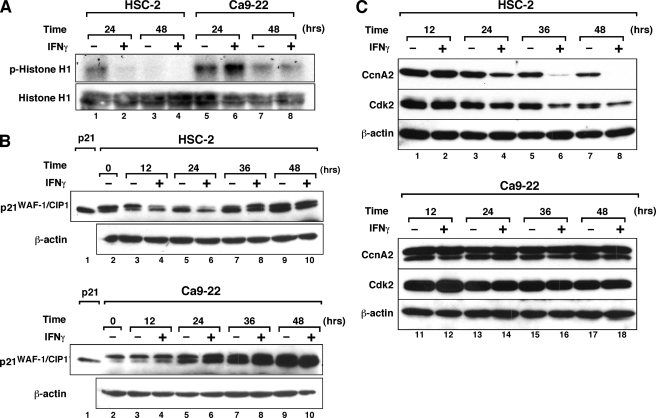
Because DNA replication and S-phase progression are regulated by CcnA2-associated kinase (39), we analyzed the activity of this kinase using histone H1 as a substrate (Fig. 4A). Although CcnA2-associated kinase activity decreased in a time-dependent fashion in untreated cultures (lanes 1 and 3), treatment of HSC-2 cells with IFNγ markedly inhibited the kinase activity (lanes 2 and 4). By contrast, no inhibitory effect of IFNγ on the kinase activity was observed in Ca9–22 cells. These results indicate that IFNγ inhibits CcnA2-associated kinase activity in HSC-2 cells but not in Ca9–22 cells.
FIGURE 4.
IFNγ inhibits CcnA2-associated Cdk activity and down-regulates CcnA2 and Cdk2 protein expression in HSC-2 cells. A, CcnA2-associated Cdk activity in HSC-2 and Ca9–22 cells. Cells were treated with or without IFNγ (10 ng/ml) for the indicated time periods, after which total cell lysates were prepared. CcnA2 was immunoprecipitated from the lysate with anti-CcnA2 antibody, and CcnA2-associated Cdk activity was assayed by the in vitro kinase assay using histone H1 as substrate. Input histone H1 was assessed by Western blotting. B, effect of IFNγ on p21WAF-1/CIP1 protein expression in HSC-2 and Ca9–22 cells. Cells were either left untreated or were treated with IFNγ (10 ng/ml) for the indicated time periods before total cell extracts were prepared. Equal amounts of cellular protein (40 μg/lane) were loaded on the gel and analyzed by Western blotting using the indicated antibodies. Lysate from HEK293 cells transfected with the expression plasmid encoding human p21 cDNA (a kind gift from Dr. Bert Vogelstein, The Johns Hopkins Oncology Center) was used as a positive control for p21 protein (lane 1). C, Western blotting analysis of CcnA2 and Cdk2 expression levels in IFNγ-treated OSCC lines. Cells were either left untreated or treated with IFNγ for the indicated time periods, as described above. Equal amounts of cellular protein (40 μg/lane) were loaded in the gel and analyzed by Western blotting using the indicated antibodies. Similar results were obtained in three separate experiments.
CcnA2-associated kinase activity is regulated by Cdk inhibitors such as p21WAF1/Cip1 (8, 9, 40), and IFNγ is known to up-regulate p21WAF1/Cip1 expression in a wide variety of cell types (3–7). However, using Western blotting, we were unable to detect any appreciable induction of p21WAF1/Cip1 protein levels in response to IFNγ in HSC-2 cells (Fig. 4B), although constitutive expression of p21 was observed in both cell types. We did not observe any increase in p27Kip1 expression in HSC-2 and Ca9–22 cells after 24 h of IFNγ treatment (data not shown).
We next assessed the levels of CcnA2 and Cdk2 proteins in HSC-2 and Ca9–22 cells. As shown in Fig. 4C, IFNγ treatment significantly decreased the levels of CcnA2 protein in HSC-2 cells after 24 h of IFNγ treatment, and the expression levels of CcnA2 became essentially undetectable after 48 h in culture. IFNγ also affected the Cdk2 expression levels in HSC-2 cells. By contrast, these protein levels were not affected by IFNγ treatment of Ca9–22 cells.
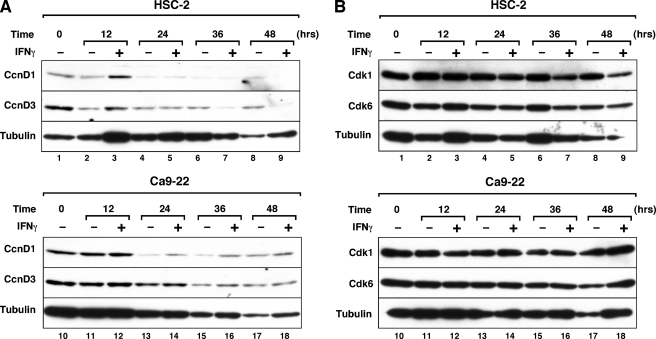
To determine the specificity of IFNγ-mediated down-regulation of CcnA2 and Cdk2 expression, we examined the expression levels of other cyclins and Cdk proteins in HSC-2 and Ca9–22 cells. As shown in Fig. 5, the expression of cyclin D3 protein in HSC-2 cells was inhibited by IFNγ after 36 h of treatment, whereas IFNγ had no inhibitory effect on the expression of Cdk6 protein (Fig. 5B). Although the levels of cyclin D1 and Cdk1 protein were inhibited after 48 h of IFNγ treatment, these proteins were refractory to IFNγ treatment until 36 h. Because the onset of cell growth inhibition was observed after 24 h of IFNγ treatment (Fig. 3), which correlates with the onset of inhibition of CcnA2 protein expression (Fig. 4C), the down-regulation of these proteins at later time periods of IFNγ treatment may not be directly implicated in the growth inhibition. The expression of cyclin D2, cyclin E, and Cdk4 proteins was undetectable in HSC-2 and Ca9–22 cells by Western blotting (data not shown). These results indicate that IFNγ down-regulates a subset of cell cycle regulatory proteins in HSC-2 cells.
FIGURE 5.
Effect of IFNγ on cyclin D and Cdk protein expression levels in HSC-2 and Ca9–22 cells. Cells were either left untreated or were treated with IFNγ (10 ng/ml) for the indicated time periods, as described in the legend for Fig. 4. Equal amounts of cellular protein (40 μg/lane) were loaded in the gel and analyzed by Western blotting using the indicated antibodies. Similar results were obtained in three separate experiments.
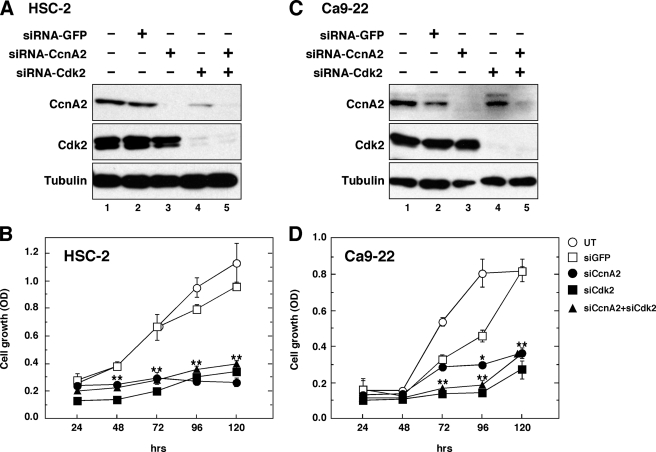
CcnA2 or Cdk2 Is Required for Cell Growth Progression in OSCC Cells
To determine whether CcnA2 and/or Cdk2 are necessary for the growth of OSCC cells, we individually knocked down CcnA2 and Cdk2 with siRNA and examined the effects on cell proliferation (Fig. 6). The effectiveness of siRNA to reduce CcnA2 and Cdk2 expression was examined by Western blotting (Fig. 6, A and C), and siRNA-green fluorescent protein was used as a control. The cell proliferation assay demonstrated that down-regulation of either CcnA2 or Cdk2 significantly inhibited the growth of HSC-2 cells (Fig. 6B). The introduction of Cdk2 siRNA into Ca9–22 cells significantly impaired cell proliferation at 72 h, and CcnA2 siRNA showed a similarly significant inhibitory effect on cell proliferation in the exponential growth phase (96–120 h). These results indicate that CcnA2 and Cdk2 are required for the proliferation of these OSCC cells and that down-regulation of CcnA2 and Cdk2 by IFNγ correlates with the inhibition of cell growth.
FIGURE 6.
CcnA2 and Cdk2 are required for the growth of HSC-2 and Ca9–22 cells. Cells were transfected with siRNA as described under “Experimental Procedures.” siRNA-green fluorescent protein (GFP) was used as a negative control. After 48 h of transfection, total cellular lysates were prepared and subjected to electrophoresis followed by Western blotting with the indicated antibodies (A and C). The cells transfected with siRNA were re-seeded at a density of 2 × 103 cells in 96-well plates and cultured for the indicated times (B and D). Cell proliferation was determined using a cell counting kit, as described under “Experimental Procedures.” The data shown are the means ± S.E. of quadruplicate determinations from a representative experiment that was repeated three times with similar results. Asterisks denote a statistically significant difference compared with cultures with siRNA-green fluorescent protein (*, p < 0.05; **, p < 0.01, Student's t test). UT, untreated.
IFNγ Inhibits CcnA2 and Cdk2 mRNA Expression via Down-regulation of Transcriptional Activities and the Destabilization of Their mRNAs
To determine the molecular mechanism involved in the IFNγ-mediated down-regulation of CcnA2 and Cdk2 expression, we assessed possible regulatory pathways for the expression of CcnA2 and Cdk2. Because cyclins and Cdks have been shown to be regulated by ubiquitin-proteasome pathways and because IFNγ stimulates proteasome-dependent protein degradation (41), the reduction in CcnA2 and Cdk2 protein expression may be because of proteasome-dependent degradation. To test this possibility, we examined the effect of the proteasome inhibitor MG132 on the levels of CcnA2 and Cdk2 protein in the presence or absence of IFN γ (supplemental Fig. 3). Although MG132 partially restored the IFNγ-mediated reduction of CcnA2 protein levels in HSC-2 cells (lane 4), the inhibitor also enhanced the levels of CcnA2 protein in untreated HSC-2 cells as well as in Ca9–22 cells (lanes 2, 6, and 8). Because the increase in the levels of CcnA2 protein observed in MG132-treated control cultures is comparable with that observed in cultures treated with IFNγ, a marked decrease in CcnA2 protein levels in IFNγ-treated HSC-2 cells does not account for the increase in the rate of proteasome-dependent protein degradation. Thus, these results suggest that IFNγ directly inhibits the expression of the CcnA2 and Cdk2 genes.
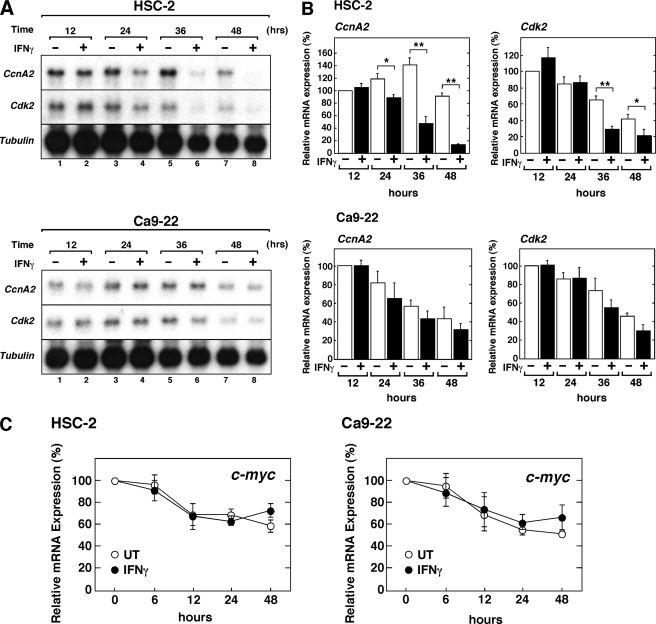
To examine the effect of IFNγ on CcnA2 and Cdk2 gene expression, we assessed the steady-state level of these mRNAs by northern hybridization (Fig. 7A). IFNγ markedly decreased the levels of CcnA2 and Cdk2 mRNA in HSC-2 cells. Quantitative analysis showed that significant inhibition of mRNA expression was observed after 24 h of IFNγ treatment for CcnA2 and after 36 h for Cdk2 (Fig. 7B). By contrast, IFNγ only marginally affected the expression of CcnA2 and Cdk2 mRNA in Ca9–22 cells.
FIGURE 7.
IFNγ down-regulates CcnA2 and Cdk2 mRNA in HSC-2 cells. A, HSC-2 or Ca9–22 cells were either left untreated or were treated with IFNγ (10 ng/ml) for the indicated time periods before preparation of total RNA and analysis of specific mRNA levels by northern hybridization. A sample (10 μg) of each total RNA was analyzed in each lane. B, Northern blots were quantified by phosphorimaging analysis, and relative mRNA levels for CcnA2 or Cdk2 are presented as a percentage of the expression in untreated cells cultured for 12 h. The data shown represent the means ± S.E. of three independent experiments. Asterisks denote a statistically significant difference compared with the untreated cultures (*, p < 0.05; **, p < 0.01, Student's t test). C, qRT-PCR analysis of c-myc mRNA expression in HSC-2 and Ca9–22 cells treated with IFNγ. Total RNA was prepared as described above and was used for qRT-PCR analysis of c-myc mRNA. The levels of c-myc mRNA are expressed as the percentage of the levels obtained at day 0 and were normalized to those of the 18 S rRNA used as an internal control. The data shown represent means ± S.E. of three independent experiments. UT, untreated.
c-Myc has been implicated in cell growth regulation (42), and IFNγ has been shown to regulate the expression of the c-myc gene via STAT1-independent mechanisms (43, 44). To determine the specificity of the IFNγ-mediated down-regulation of CcnA2 and Cdk2 expression levels, we quantitatively measured the steady state levels of c-myc mRNA using real-time qRT-PCR. As shown in Fig. 7C, although the expression of c-myc mRNA decreased in a time-dependent manner, IFNγ had no suppressive effect on the steady state level of c-myc mRNA expression in both cell types. These results indicate that IFNγ selectively inhibits the expression of a subset of cell cycle regulator genes but does not have a global inhibitory effect.
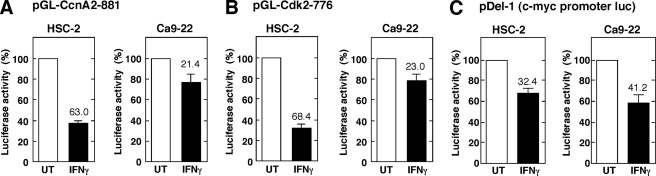
To explore the mechanism involved in the IFNγ-mediated down-regulation of CcnA2 and Cdk2 mRNA expression, we first examined the effect of IFNγ on the promoter activity of the CcnA2 and Cdk2 genes using a luciferase reporter assay (Fig. 8). Cells were transfected with the luciferase reporter construct pGL-CcnA2–881, which contains the 0.8-kb 5′-flanking sequence of the human CcnA2 gene, and the cells were then treated with IFNγ for 24 h. The results demonstrate a marked decrease in the CcnA2 promoter activity in response to IFNγ in HSC-2 cells (Fig. 8A, 63.0 ± 3.0% inhibition). However, IFNγ had only a marginal effect on the promoter activity in Ca9–22 cells (21.4 ± 6.1%). A similar inhibitory effect of IFNγ on the activity of the Cdk2 promoter was observed in HSC-2 cells (Fig. 8B, 68.4 ± 4.1%) but not in Ca9–22 cells (23.0 ± 8.0%). Previous studies demonstrated that IFNγ represses the transcriptional activity of the c-myc gene via a STAT1-dependent mechanism (43, 44). To determine whether IFNγ represses the transcriptional activity of the c-myc gene in these OSCC cells, HSC-2 and Ca9–22 cells were transfected with the luciferase reporter construct pDel-1 (29), which contains the 2.5-kb 5′-flanking sequence of the human c-myc gene, and the cells were then treated with IFNγ. As shown in Fig. 8C, treatment with IFNγ modestly inhibited the c-myc promoter activity in both cell types. These results indicate that IFNγ differentially affects the promoter activity of the CcnA2 and Cdk2 genes and the c-myc gene.
FIGURE 8.
IFNγ down-regulates CcnA2 and Cdk2 promoter activities in HSC-2 cells. HSC-2 or Ca9–22 cells were transiently transfected with the indicated CcnA2 (A), Cdk2 (B), or c-myc (C) luciferase reporter constructs. At 24 h after transfection the cells were either left untreated (UT) or were treated with IFNγ (10 ng/ml) for 24 h before measuring luciferase activity. The relative luciferase activity is shown as a percentage of the activity in untreated cells, and the percentage of inhibition by IFNγ is indicated above the column. Each column and bar represents the mean ± S.E. of three independent experiments.
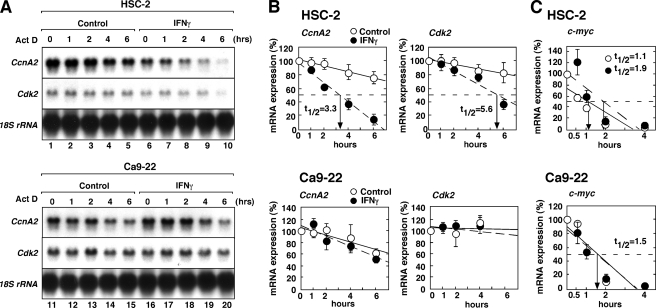
To further explore the mechanism underlying IFNγ-mediated down-regulation of CcnA2 and Cdk2 mRNA expression, we examined the effect of IFNγ on the stability of these mRNAs in HSC-2 and Ca9–22 cells (Fig. 9). For this experiment, the cells were cultured in the presence or absence of IFNγ for 24 h and then treated with actinomycin D to prevent further transcription. After additional incubation periods, total RNA was prepared and analyzed to assess the specific mRNA content by northern hybridization (Fig. 9A). In addition, the half-life (t½) of the mRNAs was quantified by phosphorimaging analysis (Fig. 9B). Although the CcnA2 and Cdk2 mRNAs were relatively stable in unstimulated cultures (t½ > 8 h), treatment with IFNγ markedly reduced the t½ of both CcnA2 mRNA (Fig. 9B, t½ = 3.3 h) and Cdk2 mRNA (t½ = 5.6 h). In contrast to HSC-2 cells, IFNγ had no effect on the decay of these mRNAs in Ca9–22 cells. As a control for specificity, we assessed the t½ of c-myc mRNA by real-time qRT-PCR (Fig. 9C). In contrast to CcnA2 and Ckd2 mRNAs, treatment with IFNγ did not stimulate the decay of c-myc mRNA in HSC-2 cells; rather, IFNγ somewhat prolonged the t½ of c-myc mRNA in HSC-2 cells. These results indicate that IFNγ selectively affects the mRNA stability of cell cycle regulator genes.
FIGURE 9.
IFNγ destabilizes CcnA2 and Cdk2 mRNA in HSC-2 cells. A, HSC-2 or Ca9–22 cells were either left untreated (control) or were treated with IFNγ for 24 h. Each culture was subsequently treated with actinomycin D (ActD, 5 mg/ml), and the incubation was continued for the indicated times. Untreated cultures were harvested before ActD treatment and are designated as time 0. Total RNA was prepared, and 10 μg of each sample was analyzed for specific mRNA levels by northern hybridization. B, Northern blots were quantified by phosphorimaging analysis, and the relative amounts of residual mRNA encoding CcnA2 or Cdk2 were calculated. Data shown represent the means ± S.E. of three independent experiments. C, qRT-PCR analysis of c-myc mRNA stability in HSC-2 and Ca9–22 cells treated with IFNγ. Total RNA was prepared as described above and was used for qRT-PCR analysis of c-myc mRNA. The levels of c-myc mRNA are expressed as the percentage of the levels at time 0 and were normalized to those of the 18 S rRNA used as an internal control. The data shown represent the means ± S.E. of three independent experiments. The half-lives of the mRNAs (t½) are shown.
Taken together, these results indicate that the down-regulation of CcnA2 and Cdk2 expression by IFNγ was mediated, at least partially, via the inhibition of the transcriptional activities of the cyclin A and Cdk2 genes as well as via the destabilization of their mRNAs. This suggests that the resistance of Ca9–22 cells to the antiproliferative effect of IFNγ results from a defect in the signaling component responsible for the IFNγ-induced down-regulation of CcnA2 and Cdk2 expression.
DISCUSSION
IFNγ has an antiproliferative effect on a variety of tumor cells. However, some types of tumor cells are resistant to IFNγ. The resistance to IFNγ has been attributed to deficiencies in components of the JAK-STAT signaling pathway (16, 17). It has also been suggested that STAT1-dependent signaling is insufficient for IFNγ-mediated growth inhibition (12, 19). In the present study we explored the mechanisms underlying the antiproliferative effect of IFNγ on human OSCC cells and their resistance to IFNγ-mediated growth inhibition. We found that the growth of Ca9–22 cells is not inhibited by IFNγ despite the presence of intact IFNγ-activated STAT1-dependent transcription. The defect in IFNγ-mediated growth inhibition results from a deficiency in the down-regulation of CcnA2-associated kinase activity that causes S-phase arrest in IFNγ-responsive HSC-2 cells. Furthermore, the IFNγ-induced S-phase arrest in HSC-2 cells occurs because of the down-regulation of CcnA2 and Cdk2 gene expression, whereas these genes are not inhibited by IFNγ in Ca9–22 cells. The essential roles of CcnA2 and Cdk2 for the growth of both cell types were demonstrated through siRNA knockdown of these proteins. These results indicate that the resistance of OSCC cells to the antiproliferative effect of IFNγ is not because of a deficiency in STAT1-dependent signaling but, rather, to a defective signaling component(s) that mediates the IFNγ-induced down-regulation of CcnA2 and Cdk2 gene expression. To the best of our knowledge, the present study is the first report to provide direct experimental evidence linking resistance to IFN with the expression of CcnA2 and Cdk2.
CcnA2-dependent kinase activity is required for the onset of DNA replication and subsequent progression through S-phase (39, 45, 46). The requisite role of CcnA2/Cdk2 in S-phase progression has been demonstrated in a number of previous studies. Phosphorylation of the E2F-1/DP-1 heterodimeric complex by CcnA2/Cdk2 leads to the inactivation of E2F-1/DP-1 DNA binding activity, which is required for orderly S-phase progression (46). Inhibition of CcnA2-dependent kinase activity has been shown to slow S-phase progression and to maintain E2F-1-dependent transcription, which ultimately leads to apoptosis via both p53-dependent and -independent pathways (47). In this way the IFNγ-mediated down-regulation of CcnA2-associated kinase leads to an arrest in S-phase and subsequent apoptosis. Indeed, treatment of HSC-2 cells with IFNγ increased the proportion of cells in S-phase after 24 h of treatment with IFNγ (Fig. 3A) and inhibited DNA synthesis (Fig. 3B). IFNγ has been shown to arrest the cell cycle at the G2-M phases in human mesothelioma (11). In human OSCC, IFNγ did not significantly change the proportion of cells in the G2/M phase, although a small but significant decrease in the G0/G1 phase population was observed (Fig. 3A). These results indicate that cells cultured in the absence of IFNγ progressed through the cell cycle and accumulated at G0/G1 phase, whereas cells treated with IFNγ arrested at S phase, stopped replicating their DNA, and subsequently underwent apoptosis.
The IFNγ-mediated inhibition of CcnA2-associated kinase activity in HSC-2 cells was because of the down-regulation of CcnA2 and Cdk2 expression. IFNγ directly inhibited CcnA2 and Cdk2 gene expression at both the transcriptional and the post-transcriptional levels (Figs. 8 and 9). In this regard, Sibinga et al. (48) previously reported that IFNγ inhibited the CcnA2 gene in vascular smooth muscle cells at the transcriptional level but not at the post-transcriptional level. In the present study, we extended the negative regulatory role of IFNγ to include Cdk promoter activity in human OSCC cells. We show that IFNγ also down-regulates the promoter activity of the Cdk2 gene in HSC-2 cells, although not in Ca9–22 cells. These results suggest that a common mechanism may be involved in down-regulating these promoter activities. In addition to the transcriptional regulation, we also demonstrated that IFNγ selectively destabilizes CcnA2 and Cdk2 mRNA but not c-myc mRNA in OSCC cells (Fig. 9). Multiple studies have demonstrated that adenylate/uridylate (AU)-rich elements (ARE) located in the 3′-untranslated region regulate mRNA stability, and several proteins have been implicated in mRNA stability by virtue of their sequence-specific binding to AREs (49). It is worth noting that the sequence motif in the 3′-untranslated regions of the CcnA2 and Cdk2 genes contains classical AU-rich motifs, such as the AUUUA pentanucleotide or AUUUUA, which is associated with the control of mRNA stability. In fact, CcnA2 is regulated by post-transcriptional mechanisms through the 3′-untranslated region ARE motif (50). Thus, it is conceivable that IFNγ induces the expression of ARE-binding proteins and destabilizes the mRNAs of CcnA2 and Cdk2 in HSC-2 cells.
Although IFNγ inhibits the expression of CcnA2 and Cdk2 in HSC-2 cells, it is possible that the ability of IFNγ to down-regulate CcnA2 and Cdk2 may be the effect of cell growth inhibition. To assess the specificity of IFNγ-mediated down-regulation of CcnA2 and Cdk2 expression, we examined the expression levels of other cyclins and Cdk proteins in these OSCC cells (Fig. 5). The results indicate that IFNγ down-regulates a subset of cell cycle regulatory proteins in HSC-2 cells. The expression of cyclin D3 protein was inhibited by IFNγ after 36 h of treatment, whereas IFNγ had no inhibitory effect on the expression of Cdk6 protein. Although the expression levels of cyclin D1 and Cdk1 protein were inhibited after 48 h of IFNγ treatment, the expression levels of these proteins were refractory to IFNγ treatment until 36 h. Because the onset of cell growth inhibition was observed after 24 h of IFNγ treatment (Fig. 3), which correlates with the onset of inhibition of CcnA2 protein expression (Fig. 4C, lane 4), it is less likely that the down-regulation of cyclin D1 and Cdk1 is directly implicated in the growth inhibition. To further assess the specificity of IFNγ, we also examined the effect of IFNγ on the transcription and mRNA stability of c-myc. c-Myc is involved in cell growth regulation (42), and IFNs have been shown to modulate the expression of the c-myc gene in a wide range of cell types (43, 44, 51–53). Previous studies have demonstrated that STAT1 functions as a negative regulator of c-myc expression in human fibrosarcoma cells and murine B cell lines (43, 44). In agreement with these previous findings, IFNγ modestly inhibited the promoter activity of the c-myc gene in both HSC-2 and Ca9–22 cells (Fig. 8C), suggesting that STAT1 is functional for inhibiting c-myc transcriptional activity. In addition to the promoter activity, we assessed the t½ of c-myc mRNA in OSCC cells treated with IFNγ (Fig. 9C). In contrast to CcnA2 and Cdk2 mRNA, IFNγ did not stimulate the decay of c-myc mRNA in HSC-2 cells. Rather, IFNγ prolonged the t½ of c-myc mRNA in HSC-2 cells. Although the mechanism involved in the IFNγ-mediated prolongation of c-myc mRNA is unknown, these results indicate that IFNγ differentially affects mRNA stability. Taken together, these results indicate that IFNγ selectively inhibits the expression of a subset of cell cycle regulatory genes, and the down-regulation of CcnA2 and Cdk2 is not because of cell growth inhibition.
Resistance to IFN treatment has been demonstrated in various types of tumors, and deficiencies in components of the JAK-STAT-signaling pathway have been identified in these tumor cells (13–17). It has also been reported that STAT1-independent signaling, such as the PI3K/Akt-signaling pathway, is implicated in the resistance to IFNs (20, 54). Previous studies demonstrated that sensitivity and resistance to IFNβ in colorectal cancer cells depends on the status of Akt activation after IFNβ treatment (20). In IFNβ-resistant colorectal cancer cells, treatment with IFNβ up-regulated the activation of Akt, which induced downstream anti-apoptotic pathways (20). In contrast to the positive regulatory role of the PI3K/Akt pathway, IFNγ-induced apoptosis and growth inhibition have been shown to be associated with a persistent suppression of the constitutive tyrosine-phosphorylated STAT3. Fully active mammalian target of rapamycin (mTOR), a serine/threonine kinase regulated by Akt (55), appears to be required for the suppression of STAT3 phosphorylation (54). In the present study, we analyzed the levels of phosphorylated Akt (Thr-308 and Ser-473) in OSCC cells. Although constitutive phosphorylation on these sites of Akt was observed in HSC-2 and Ca9–22 cells, IFNγ failed to further up-regulate the observed phosphorylation (supplemental Fig. 2). These results indicate that the PI3K/Akt pathway is not directly implicated in the differential sensitivity to IFNγ in OSCC cells. Although the precise molecular details involved in the differential sensitivity to IFNγ in these OSCC cells remain unknown, our data suggest the possibility that a gene product(s) or a signaling molecule(s) downstream of STAT1 or a STAT1-indepenent signaling pathway might be responsible for the observed effect.
In conclusion, our findings contribute to our understanding of the antiproliferative effects of IFNγ on tumor cells and elucidate the molecular mechanisms involved in the resistance of OSCC cells to IFNγ. Further analysis of the molecular mechanisms underlying the IFNγ-mediated down-regulation of CcnA2 and Cdk2 expression will shed new light on the additional signaling components that mediate the antiproliferative effect of IFNγ.
Supplementary Material
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan and from the Meikai University School of Dentistry.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- IFN
- interferon
- JAK-STAT
- Janus kinase-signal transducer activation of transcription
- GAS
- γ-IFN activation sequence
- IRF
- interferon regulatory factor
- OSCC
- oral squamous cell carcinoma
- CcnA2
- cyclin A2
- Cdk
- cyclin-dependent kinase
- TK
- thymidine kinase
- qRT-PCR
- real-time quantitative RT-PCR
- ARE
- adenylate/uridylate (AU)-rich elements
- siRNA
- small interfering RNA
- kb
- kilobase(s)
- PI3K
- phosphoinositide 3-kinase.
REFERENCES
- 1.Pestka S., Langer J. A., Zoon K. C., Samuel C. E. (1987) Annu. Rev. Biochem. 56, 727–777 [DOI] [PubMed] [Google Scholar]
- 2.Boehm U., Klamp T., Groot M., Howard J. C. (1997) Annu. Rev. Immunol. 15, 749–795 [DOI] [PubMed] [Google Scholar]
- 3.Chin Y. E., Kitagawa M., Su W. C., You Z. H., Iwamoto Y., Fu X. Y. (1996) Science 272, 719–722 [DOI] [PubMed] [Google Scholar]
- 4.Sangfelt O., Erickson S., Einhorn S., Grandér D. (1997) Oncogene 14, 415–423 [DOI] [PubMed] [Google Scholar]
- 5.Mandal M., Bandyopadhyay D., Goepfert T. M., Kumar R. (1998) Oncogene 16, 217–225 [DOI] [PubMed] [Google Scholar]
- 6.Kominsky S., Johnson H. M., Bryan G., Tanabe T., Hobeika A. C., Subramaniam P. S., Torres B. (1998) Oncogene 17, 2973–2979 [DOI] [PubMed] [Google Scholar]
- 7.Gooch J. L., Herrera R. E., Yee D. (2000) Cell Growth Differ. 11, 335–342 [PubMed] [Google Scholar]
- 8.El-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 9.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]
- 10.Harvat B. L., Jetten A. M. (1996) Cell Growth Differ. 7, 289–300 [PubMed] [Google Scholar]
- 11.Vivo C., Lévy F., Pilatte Y., Fleury-Feith J., Chrétien P., Monnet I., Kheuang L., Jaurand M. C. (2001) Oncogene 20, 1085–1093 [DOI] [PubMed] [Google Scholar]
- 12.Kortylewski M., Komyod W., Kauffmann M. E., Bosserhoff A., Heinrich P. C., Behrmann I. (2004) J. Invest. Dermatol. 122, 414–422 [DOI] [PubMed] [Google Scholar]
- 13.Xu B., Grandér D., Sangfelt O., Einhorn S. (1994) Blood 84, 1942–1949 [PubMed] [Google Scholar]
- 14.Wong L. H., Krauer K. G., Hatzinisiriou I., Estcourt M. J., Hersey P., Tam N. D., Edmondson S., Devenish R. J., Ralph S. J. (1997) J. Biol. Chem. 272, 28779–28785 [DOI] [PubMed] [Google Scholar]
- 15.Sun W. H., Pabon C., Alsayed Y., Huang P. P., Jandeska S., Uddin S., Platanias L. C., Rosen S. T. (1998) Blood 91, 570–576 [PubMed] [Google Scholar]
- 16.Kaplan D. H., Shankaran V., Dighe A. S., Stockert E., Aguet M., Old L. J., Schreiber R. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7556–7561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dovhey S. E., Ghosh N. S., Wright K. L. (2000) Cancer Res. 60, 5789–5796 [PubMed] [Google Scholar]
- 18.Clifford J. L., Walch E., Yang X., Xu X., Alberts D. S., Clayman G. L., El-Naggar A. K., Lotan R., Lippman S. M. (2002) Clin. Cancer Res. 8, 2067–2072 [PubMed] [Google Scholar]
- 19.Chawla-Sarkar M., Leaman D. W., Jacobs B. S., Tuthill R. J., Chatterjee-Kishore M., Stark G. R., Borden E. C. (2002) J. Interferon Cytokine Res. 22, 603–613 [DOI] [PubMed] [Google Scholar]
- 20.Lei H., Furlong P. J., Ra J. H., Mullins D., Cantor R., Fraker D. L., Spitz F. R. (2005) Cancer Biol. Ther. 4, 709–715 [DOI] [PubMed] [Google Scholar]
- 21.Kamata N., Chida K., Rikimaru K., Horikoshi M., Enomoto S., Kuroki T. (1986) Cancer Res. 46, 1648–1653 [PubMed] [Google Scholar]
- 22.Momose F., Araida T., Negishi A., Ichijo H., Shioda S., Sasaki S. (1989) J. Oral. Pathol. Med. 18, 391–395 [DOI] [PubMed] [Google Scholar]
- 23.Hiroi M., Ohmori Y. (2003) Biochem. J. 376, 393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohmori Y., Schreiber R. D., Hamilton T. A. (1997) J. Biol. Chem. 272, 14899–14907 [DOI] [PubMed] [Google Scholar]
- 25.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sims S. H., Cha Y., Romine M. F., Gao P. Q., Gottlieb K., Deisseroth A. B. (1993) Mol. Cell Biol. 13, 690–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henglein B., Chenivesse X., Wang J., Eick D., Bréchot C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5490–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiffman D., Brooks E. E., Brooks A. R., Chan C. S., Milner P. G. (1996) J. Biol. Chem. 271, 12199–12204 [DOI] [PubMed] [Google Scholar]
- 29.He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 30.Pine R., Canova A., Schindler C. (1994) EMBO J. 13, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohmori Y., Hamilton T. A. (1997) J. Immunol. 159, 5474–5482 [PubMed] [Google Scholar]
- 32.Meraz M. A., White J. M., Sheehan K. C., Bach E. A., Rodig S. J., Dighe A. S., Kaplan D. H., Riley J. K., Greenlund A. C., Campbell D., Carver-Moore K., DuBois R. N., Clark R., Aguet M., Schreiber R. D. (1996) Cell 84, 431–442 [DOI] [PubMed] [Google Scholar]
- 33.Durbin J. E., Hackenmiller R., Simon M. C., Levy D. E. (1996) Cell 84, 443–450 [DOI] [PubMed] [Google Scholar]
- 34.Decker T., Kovarik P., Meinke A. (1997) J. Interferon Cytokine Res. 17, 121–134 [DOI] [PubMed] [Google Scholar]
- 35.Qing Y., Stark G. R. (2004) J. Biol. Chem. 279, 41679–41685 [DOI] [PubMed] [Google Scholar]
- 36.Nguyen H., Ramana C. V., Bayes J., Stark G. R. (2001) J. Biol. Chem. 276, 33361–33368 [DOI] [PubMed] [Google Scholar]
- 37.Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 38.Stephens L., Anderson K., Stokoe D., Erdjument-Bromage H., Painter G. F., Holmes A. B., Gaffney P. R., Reese C. B., McCormick F., Tempst P., Coadwell J., Hawkins P. T. (1998) Science 279, 710–714 [DOI] [PubMed] [Google Scholar]
- 39.Girard F., Strausfeld U., Fernandez A., Lamb N. J. (1991) Cell 67, 1169–1179 [DOI] [PubMed] [Google Scholar]
- 40.Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. (1993) Nature 366, 701–704 [DOI] [PubMed] [Google Scholar]
- 41.Yang Y., Waters J. B., Früh K., Peterson P. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4928–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt E. V. (1999) Oncogene 18, 2988–2996 [DOI] [PubMed] [Google Scholar]
- 43.Asao H., Fu X. Y. (2000) J. Biol. Chem. 275, 867–874 [DOI] [PubMed] [Google Scholar]
- 44.Ramana C. V., Grammatikakis N., Chernov M., Nguyen H., Goh K. C., Williams B. R., Stark G. R. (2000) EMBO J. 19, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagano M., Pepperkok R., Verde F., Ansorge W., Draetta G. (1992) EMBO J. 11, 961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krek W., Xu G., Livingston D. M. (1995) Cell 83, 1149–1158 [DOI] [PubMed] [Google Scholar]
- 47.Furukawa Y., Nishimura N., Furukawa Y., Satoh M., Endo H., Iwase S., Yamada H., Matsuda M., Kano Y., Nakamura M. (2002) J. Biol. Chem. 277, 39760–39768 [DOI] [PubMed] [Google Scholar]
- 48.Sibinga N. E., Wang H., Perrella M. A., Endege W. O., Patterson C., Yoshizumi M., Haber E., Lee M. E. (1999) J. Biol. Chem. 274, 12139–12146 [DOI] [PubMed] [Google Scholar]
- 49.Ross J. (1995) Microbiol. Rev. 59, 423–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W., Caldwell M. C., Lin S., Furneaux H., Gorospe M. (2000) EMBO J. 19, 2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jonak G. J., Knight E., Jr. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 1747–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Einat M., Resnitzky D., Kimchi A. (1985) Nature 313, 597–600 [DOI] [PubMed] [Google Scholar]
- 53.Bennett M. R., Littlewood T. D., Hancock D. C., Evan G. I., Newby A. C. (1994) Biochem. J. 302, 701–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang P., Hwa V., Rosenfeld R. G. (2006) Exp. Cell Res. 312, 1229–1239 [DOI] [PubMed] [Google Scholar]
- 55.Navé B. T., Ouwens M., Withers D. J., Alessi D. R., Shepherd P. R. (1999) Biochem. J. 344, 427–431 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.