Abstract
Elevated sphingolipids have been associated with increased cardiovascular disease. Conversely, atherosclerosis is reduced in mice by blocking de novo synthesis of sphingolipids catalyzed by serine palmitoyltransferase (SPT). The SPT enzyme is composed of the SPTLC1 and -2 subunits, and here we describe a novel protein-protein interaction between SPTLC1 and the PDZ protein Par3 (partitioning defective protein 3). Mammalian SPTLC1 orthologs have a highly conserved C terminus that conforms to a type II PDZ protein interaction motif, and by screening PDZ domain protein arrays with an SPTLC1 C-terminal peptide, we found it bound the third PDZ domain of Par3. Overlay and immunoprecipitation assays confirmed this interaction and indicate Par3 is able to associate with the SPTLC1/2 holoenzyme by binding the C-terminal SPTLC1 PDZ motif. The physiologic existence of the SPTLC1/2-Par3 complex was detected in mouse liver and macrophages, and short interfering RNA inhibition of Par3 in human THP-1 monocytes significantly reduced SPT activity and de novo ceramide synthesis by nearly 40%. Given monocyte recruitment into inflamed vessels is thought to promote atherosclerosis, and because Par3 and sphingolipids have been associated with polarized cell migration, we tested whether the ability of THP-1 monocytes to migrate toward MCP-1 (monocyte chemoattractant protein 1) depended upon Par3 and SPTLC1 expression. Knockdown of Par3 significantly reduced MCP1-induced chemotaxis of THP-1 monocytes, as did knockdown of SPTLC1, and this Par3 effect depended upon SPT activity and was blunted by ceramide treatment. In conclusion, protein arrays were used to identify a novel SPTLC1-Par3 interaction that associates with increased monocyte serine palmitoyltransferase activity and chemotaxis toward inflammatory signals.
Sphingolipids are a structurally diverse class of lipids that play correspondingly diverse roles in membrane structure, cell proliferation, immune function, and skin physiology (1–4). De novo sphingolipid synthesis is initiated by serine palmitoyltransferase (SPT),2 an enzyme that condenses serine and palmitoyl-CoA forming the biosynthetic intermediate 3-ketodihydrosphingosine that is subsequently converted to ceramide, sphingomyelin, and other sphingolipids (5). SPT is a heterodimer composed of the SPTLC1 and -2 subunits (6, 7), which may form higher order multimeric structures that can include a third subunit, SPTLC3 (8, 9). Both the SPTLC2 and -3 subunits are catalytically active and contain conserved lysines that act as Schiff bases during the condensation reaction (5, 8). In contrast, SPTLC1 does not contain the conserved catalytically active lysine, but is important for stabilizing the SPTLC2 subunit and anchoring the SPT holoenyzme on the cytosolic face of the endoplasmic reticulum (10, 11).
Expression and regulation of the SPTLC1/2 holoenyzme are of interest because its activity controls de novo synthesis of sphingomyelin, and increased plasma levels of this sphingolipid have been correlated with an increased incidence of cardiovascular disease in humans (12, 13). Conversely, inhibition of SPT activity with myriocin, a fungal metabolite, strongly inhibits atherosclerotic development in ApoE−/− mice (14–18). Moreover, the increased atherosclerosis seen in ApoE−/− mice has been associated with a post-translational increase in liver SPT activity (19). How SPT activity and sphingolipids may act to promote the progression of atherosclerosis is unclear, but the data do suggest analysis of factors that regulate SPT activity should provide mechanistic insight into the link between de novo sphingolipid synthesis and atherosclerosis. In this regard we have found that SPTLC1 can interact with the ABCA1 transporter and inhibit its ability to transfer cholesterol to apoA-I, a mechanism that would be expected to promote atherosclerosis (20). Thus, along with playing a direct role in the synthesis of sphingolipids, SPTLC1 may also have evolved as an SPT subunit whose function is to regulate SPT activity in response to the cellular demand for sphingolipids and other membrane constituents such as cholesterol. To play such a role, SPTLC1 may engage additional protein-protein interactions that integrate input from signaling pathways and allow SPTLC1 to modulate SPT activity in response to altered demand for sphingolipids.
Here we have explored this hypothesis by first conducting a protein array screen for SPTLC1 interacting factors. Consistent with the potential to engage cellular factors in protein-protein interactions, sequence alignment of the SPTLC1 C terminus indicates it has been strongly conserved in mammals as a type II PDZ domain binding motif. Moreover, because topology studies indicate the SPTLC1 C terminus resides in the cytoplasm where it could be bound by PDZ proteins, we used protein arrays spotted with 123 PDZ domains from 73 different proteins to screen for interactions with the SPTLC1 C terminus. This screen indicated the SPTLC1 C terminus directly interacts with the third PDZ domain of PARD3 (partitioning defective protein 3). PARD3, also known as Par3, is a scaffolding factor that recruits signaling molecules, including atypical protein kinase C and Cdc42 into multiprotein complexes that regulate formation of membrane microdomains required for apical/basal polarity and for directed cell migration (21–25). Mutation analysis confirmed the SPTLC1-Par3 interaction depended upon the SPTLC1 C-terminal PDZ motif, and immunoprecipitation assays indicate Par3 is able to associate with the SPTLC1/2 holoenyzme by binding the SPTLC1 C-terminal PDZ motif. The Par-3 interaction with the SPTLC1/2 holoenyzme was detected in the liver, a major site of SPT activity and sphingolipid synthesis. Given SPT activity is proatheroslerotic, and because we have also detected SPTLC1 expression in macrophages, a cell type that plays a central role in the progression of atherosclerosis, we tested and found that Par3 was expressed and interacted with the SPT holoenyzme in primary mouse macrophages. Significantly, loss of Par3 expression in human THP-1 monocyte macrophages reduced SPT activity and inhibited their ability to migrate toward MCP-1 (monocyte chemotactic protein-1). Likewise, shRNA suppression of SPTLC1 reduced monocyte migration toward MCP-1, as did myriocin inhibition of SPT activity, an effect that was blunted by loss of Par3 expression. In aggregate, our work has identified a novel protein-protein interaction between SPTLC1 and Par3 that is associated with an increase in SPT activity and the promotion of polarized cell migration in response to an inflammatory signal.
EXPERIMENTAL PROCEDURES
Reagents, Cells, and DNA Constructs
The following reagents were purchased from the indicated suppliers: human poly(A)+ mRNA tissue blot (OriGene); myriocin and C16-ceramide (Biomol); anti-Par3 antibody (Upstate); anti-HA and Myc tag antibodies (Covance). The SPTLC1, SPTLC2, and SPTLC3 antibodies have been described previously (8, 9). The LY-B cell line was obtained from Dr. Kentaro Hanada via the RIKEN BRC cell bank. The human THP-1 monocyte and 293HEK cell lines were obtained from the American Type Culture Collection. Primary bone marrow-derived macrophages were obtained from C57/BL6 mice and cultured as described previously (20). Plasmids containing the cDNAs for SPTLC1, SPTLC2, and the 150-kDa isoform of Par-3 were obtained from Open BioSystems and Dr. Ramnik Xavier of the Center for Computational and Integrative Biology, respectively. Integrity of the cDNA open reading frames was confirmed by sequencing, and as needed the cDNAs were modified to generate N-terminal FLAG, HA, and Myc-tagged versions of the encoded proteins using standard cloning techniques.
SPTLC1, -2, and -3 Ortholog Alignments
The Ensembl genome data base was used to align the orthologs of SPTLC1, SPTLC2, and SPTLC3 (26). The SPTLC1 C-terminal residues in all 17 sequenced placental mammalian genomes were found to be conserved as a type II PDZ motif characterized by hydrophobic residues at positions 0 and −2 (XΦ−2X−1Φ0-COOH). In SPTLC1 orthologs from other phyla these sequences were more variable and did not conform to a known PDZ-binding motif with the exception of SPTLC1 from Gasterosteus aculeatus and Drosophila melanogaster. Alignments of the SPTLC2 and SPTLC3 orthologs showed these subunits have been conserved but that their C termini do not conform to any known PDZ-binding motifs.
PDZ Array Screen and Overlay Assays
To screen for C-terminal SPTLC1 protein-protein interactions, we synthesized an N-terminally biotinylated SPTLC1 peptide (biotin-EEELQRAASTIREAAQAVLL-COOH). The peptide was purified to >95% homogeneity using high pressure liquid chromatography as determined by mass spectrometry. The peptide was incubated with protein arrays spotted in duplicate with 123 PDZ domains from 73 PDZ domain-containing proteins (150 nm in blocking buffer, Panomics). After washing, peptide binding was detected using a horseradish peroxidase-avidin conjugate and enhanced chemiluminescence. Arrays were imaged and quantified using an Alpha Innotech FluorChem 8800 instrument as described previously (27). The binding of full-length Par3 protein (150-kDa isoform) to the SPTLC1 C terminus was determined using overlay assays as described previously (28). In brief, sequences representing the C-terminal 75 amino acids of SPTLC1 were expressed and purified from bacteria as His-tagged polypeptides with or without an intact PDZ motif (wild type, Δ3, VLL-AAA). The polypeptides were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with a lysate from 293-EBNA-T cells expressing full-length HA-tagged Par3. After washing the blot (1× phosphate-buffered saline, 0.1% Tween 20), the binding of Par3 was detected using the anti-HA antibody. The Coomassie staining of gels run in parallel was used to demonstrate the equal loading of His-tagged SPTLC1 polypeptides.
Immunological, RNA, and shRNA Methods
To assess the tissue distribution of the Par3 protein, lysates (1% Triton X-100, 10% glycerol, 140 mm NaCl, 3 mm MgCl2, 50 mm HEPES, pH 7.4, protease inhibitor mixture, Sigma) were generated from the indicated mouse C57/BL6 tissues, and the samples (20 μg of total protein) were separated by SDS-PAGE. After transfer to nitrocellulose membranes, Par3 expression in the samples was detected by immunoblotting with the anti-Par3 antibody, and to test for SPTLC1 expression the blot was stripped and re-probed with the anti-SPTLC1 antibody. To assess the tissue distribution of Par3 mRNA versus that of FRMPD4 mRNA, a human mRNA tissue blot (OriGene) was sequentially probed with radiolabeled DNA derived from the cDNAs for Par3, FRMPD4, and β-actin. Images of the resulting Northern blots were captured and quantified using a Typhoon PhosphorImager (GE Healthcare). To analyze the role that the SPTLC1 PDZ motif plays in allowing Par3 to associate with the SPTLC1/2 holoenzyme, FLAG-SPTLC1 and the indicated FLAG-SPTLC1 C-terminal PDZ motif mutants were expressed in 293-EBNA-T cells in the presence of HA-Par3 and Myc-SPTLC2 using transient transfection methods (Lipofectamine 2000, Invitrogen). 36 h post-transfection Triton X-100 lysates were prepared, and 0.5 mg of the lysates was precipitated with 35 μl of anti-FLAG antibody beads (Sigma), anti-HA-antibody, or anti-Myc antibody as indicated in the specific experiments. After washing and elution of the precipitate proteins, the amount of Par3, SPTLC1, and SPTLC2 was determined by immunoblotting using the appropriate primary antibodies and a secondary TrueBlot antibody (eBiosciences). Immunoprecipitation assay using mouse liver and bone marrow-derived macrophages lysate were carried out as above using anti-SPTLC1 and SPTLC2 antibodies and TrueBlot anti-rabbit IgG beads (eBiosciences). For shRNA knockdown of Par3, primary screening of OpenBiosystems lentiviral constructs targeting human Par3 showed clones NM_019619.2-2017s1c1 and NM_019619.2-2782s1c1 (denoted F12 and G1 in the figures) had the greatest efficacy to reduce expression of the Par3 protein as assessed by transient transfection of the human Par3 cDNA with these clones. Particles generated by packaging these lentiviral constructs were used to infect THP-1 monocytes, and stable clones expressing the shRNAs targeting Par3 were obtained by selection with puromycin (4 μg/ml, 48 h post-infection for 72 h). Likewise, THP-1 cells expressing the shRNA targeting SPTLC1 siRNA or a control shRNA targeting green fluorescent protein (GFP) were also generated by puromycin selection as described previously (20).
Cell Migration Assay
To measure the importance of Par3 and SPTLC1 for THP-1 monocyte migration cells were suspended in RPMI 1640 medium (0.1% bovine serum albumin) at 2 × 106 cells/ml. 50 μl of the cell suspensions were transferred to the upper wells of a ChemoTx 96-well assay plate with an 8-μm pore filter (Neuroprobe). The lower chamber contained 30 μl of media with or without 10 nm MCP-1, Fractalkine, or fMLP (R & D Systems). The plates were incubated at 37 °C for 2 h, and after washing the filter with phosphate-buffered saline, the plates were spun at 400 × g for 5 min. Cells that had migrated into the lower camber were subsequently quantitated using the Cell Titer-Glo assay (Promega). Additional assays for viability were conducted using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay as described previously (20).
Serine Palmitoyltransferase Activity
THP-1 cells were harvested by centrifugation (5 min, 900 × g) and lysed in a hypotonic buffer (250 mm sucrose, 10 mm HEPES, pH 7.5, protease inhibitor mixture), cell debris was removed by centrifugation (10 min, 900 × g), and a microsome membrane fraction was obtained by centrifugation (1 h, 100,000 × g). The resulting membrane pellet was resuspended in reaction buffer (50 mm HEPES buffer, pH 8.0, 0.5 mm l-serine, 50 μm palmitoyl-CoA, 20 μm pyridoxal 5′-phosphate) with 1 μCi of l-[14C]serine at 0.2 mg of microsomal protein, 0.2 ml of reaction buffer. After incubation for 20 min at 37 °C, the reaction was stopped by adding 0.5 ml of 100 mm KOH in methanol/chloroform (4:1), followed by addition of 0.5 ml of chloroform. Then 0.5 ml of 2 mm ammonium and 0.1 ml of 2 n NH4OH were added, and the reaction was vortexed and then centrifuged. The resulting upper aqueous layer was aspirated, and the lower organic phase was washed two times with 0.9 ml of 0.002 n ammonium hydroxide. 400 μl of the resulting organic phase was mixed with scintillation fluid, and the amount of l-[14C]serine-containing lipids was quantified using a scintillation counter (Brinkmann Instruments).
Metabolic Labeling of Cellular Sphingolipids
To measure de novo sphingolipid synthesis, THP-1 cells (5 × 106 cells) were suspended in RPMI 1640 medium (10% fetal bovine serum). The cells were either pretreated with vehicle or 10 μm myriocin (Biomol) for 2 h and then were labeled with 5 μCi/ml of [14C]serine for 24 h at 37 °C. Cells were harvested by centrifugation, washed with 1× phosphate-buffered saline, and lipids extracted 1× with 0.75 ml of chloroform/methanol (1:2 v/v) and 1× with 0.25 ml of chloroform, 0.2 ml of distilled H2O. After centrifugation the combined organic layer was washed and dried under a stream of nitrogen gas. The samples were resuspended in chloroform/methanol (1:2 v/v) and applied to 10 × 16-cm thin layer silica gel 60F plates (Merck). The plates were developed sequentially with chloroform/methanol/water (14:6:1,v/v) and chloroform/methanol/acetic acid (190:9:1, v/v) as described previously (29). Radioactivity was quantified using a Typhoon PhosphorImager (GE Healthcare).
Statistical Analysis
Data sets were found to have equal variance and were further compared by a two-tailed Student's t test using the SigmaStat software package. Statistical significance was defined by a p value <0.05.
RESULTS
SPTLC1 Is Bound by Par3 through a Type II PDZ Protein Interaction
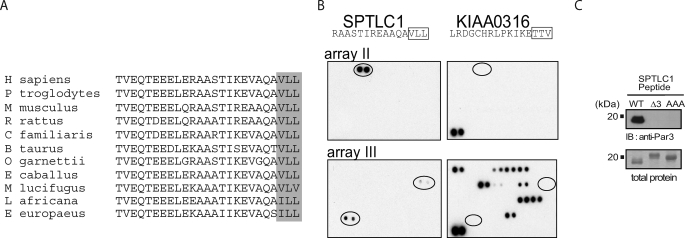
SPT activity in mammalian cells localizes to the cytosolic face of the endoplasmic reticulum, and topology studies have shown that mammalian SPTLC1 contains a single transmembrane domain and a cytoplasmic C terminus (10, 11, 30). Given the orientation of the C terminus, this domain may allow SPTLC1 to interact with cytoplasmic proteins. PDZ proteins are cytoplasmic factors that contain one or more 90-amino acid modules that bind the C termini of other proteins, and this capacity along with additional protein-binding domains allows PDZ proteins to assemble signaling and membrane microdomain complexes. Ortholog alignment of the C-terminal residues of a protein can predict its potential to interact with PDZ domains (31, 32), thus using the Ensembl data base we aligned the C termini of SPTLC1 orthologs from a variety of species (Fig. 1A). This showed the SPTLC1 C terminus to have been strongly conserved in the mammalian lineage as a type II PDZ-binding motif, characterized by hydrophobic residues at positions −2 and 0 (XΦ−2X−1Φ0-COOH). The hydrophobic tail of SPTLC1 is unique compared with the C termini of SPTLC2 and -3, which did not conform to any known PDZ-binding motifs (data not shown). Next, the capacity of SPTLC1 to bind PDZ domains was assessed by probing a series of protein arrays spotted in duplicate with 123 PDZ domains from 73 proteins with a biotinylated 20-mer SPTLC1 C-terminal peptide. Shown are the two arrays containing PDZ domains that bound the SPTLC1 peptide (Fig. 1B). That the binding of the SPTLC1 peptide to these domains was driven by a type II interaction was indicated by probing additional arrays with a peptide (KIAA0316) containing a type I motif, characterized by a serine or threonine at the −2 position (X(S/T)−2X−1Φ0-COOH), which bound a distinct set of PDZ domains compared with the SPTLC1 peptide (Fig. 1B). Of the domains that the SPTLC1 peptide bound, the most intense signal was generated by the single PDZ domain from a poorly characterized protein (FRMPD4), followed by the signal from the third PDZ domain of Par3 (PARD3), a protein that functions to establish polarized membrane domains and to control directional cell migration (21–25). The third domain of PTPN13, a protein-tyrosine phosphatase that regulates cell growth, differentiation, and oncogenic transformation, was also found to weakly interact with the SPTLC1 peptide (33, 34).
FIGURE 1.
SPTLC1 contains a conserved C-terminal PDZ motif that interacts with the third PDZ domain of Par3. A, alignment of the C-terminal 25 amino acids of SPTLC1 from the indicated species is shown, and the final three conserved hydrophobic residues that conform to a class II PDZ motif (XΦ−2X−1Φ0-COOH) are boxed. B, SPTLC1 C terminus is able to bind to the third PDZ domain of Par3 as identified by the binding of a biotinylated 20-mer C-terminal SPTLC1 peptide to protein arrays that are spotted in duplicate with 123 PDZ domains from 73 proteins. Shown in the left panels are the two arrays of four containing PDZ domains that bound the SPTLC1 peptide (signal intensity FRMPD4 > Par3 > PTPN13). That binding of the SPTLC1 peptide to these PDZ domains was driven by a type II interaction was indicated by the binding pattern of a control biotinylated peptide (KIAA0316), which conforms to a class I PDZ motif (X(S/T)−2X−1-Φ0-COOH) and interacts with a distinct set of PDZ domains (right panels). C, binding of full-length Par3 (150-kDa isoform) to the SPTLC1 C terminus requires the final three residues that comprise the SPTLC1 type II PDZ motif as determined by overlay assays. Purified bacterial polypeptides encoding the wild type SPTLC1 C terminus or the C terminus with the PDZ motif deleted (Δ3) or substituted with alanines (AAA) were separated by SDS-PAGE and transferred to nitrocellulose. Incubation of the membrane with a lysate containing HA-tagged Par3 expressed in 293-EBNA-T cells only bound WT SPTLC1 as determined by immunoblotting with an anti-HA antibody. Gels run in parallel and Coomassie-stained for total SPTLC1 protein indicate the failure of Par3 to bind the mutant SPTLC1 polypeptides was not due to unequal loading in these assays.
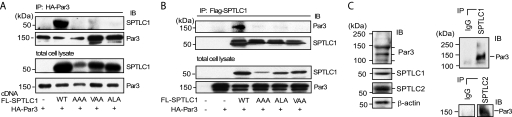
SPTLC1 mRNA has a reported wide tissue distribution, and the Par3 mRNA distribution was also found to be more widely distributed compared with that of FRMPD4, which was principally expressed in the brain and the heart (supplemental Fig. 1A). At the protein level, expression of Par3 and SPTLC1 was also found to be widespread and largely coincident (supplemental Fig. 1B). Given the more ubiquitous expression of Par3 and the involvement of Par3 in establishing polarized cell behavior, which also has been shown to require sphingolipids and SPT activity (35, 36), we further investigated the interaction of Par3 and SPTLC1. Significance of the interaction of the SPTLC1 peptide with FRMPD4 and PTPN13 PDZ domains detected in our protein array screen remains to be investigated and will be reported elsewhere. For Par3, we used overlay assays to test whether the full-length Par3 could interact with SPTLC1, and if so whether binding required the SPTLC1 C-terminal PDZ motif. Purified bacterial polypeptides encoding the wild type SPTLC1 C terminus or mutant SPTLC1 polypeptides with a disrupted PDZ motif (Δ3, VLL-AAA) were subjected to SDS-PAGE and transferred to nitrocellulose, and a 293-EBNA-T cell lysate containing HA-tagged Par3 (150-kDa isoform) was incubated with the membranes. We found Par3 strongly bound the wild type SPTLC1 C terminus and that this binding required the type II PDZ motif (Fig. 1C). Coomassie-stained gels run in parallel showed similar amounts of the SPTLC1 proteins were used in these assays, which demonstrated that the lack of Par3 binding to the mutant peptides was not explained by loading artifacts. However, because the mutant SPTLC1 peptides (particularly the Δ3 construct) ran anomalously despite being reduced and denatured, we conducted further assays where both full-length SPTLC1 and Par3 were expressed in 293HEK cells, and their interaction was assessed by immunoprecipitation. To conduct these experiments, we generated a series of SPTLC1 constructs containing either an N-terminal FLAG or HA tag, a position that has been previously found to preserve SPTLC1 protein interactions and SPT activity (7, 11). We confirmed these results by showing our FLAG- and HA-SPTLC1 constructs, like WT SPTLC1, bind SPTLC2 stabilizing its expression and stimulating SPT activity in LY-B cells, which lack endogenous SPTLC1 expression (supplemental Fig. 2, A and B). Having established the utility of our constructs, we generated a series of mutants that again disrupted the C-terminal PDZ motif. Interestingly, either deletion or substitution to alanines of the three residues that make up the PDZ motif produced mutants that expressed substantially less SPTLC1 protein but not mRNA suggesting that these sequences are important for maintaining SPTLC1 protein stability (Fig. 2A and supplemental Fig. 3A). But the lower expression of these mutants made comparison of their ability to bind Par3 difficult. Thus, additional mutants were constructed that selectively replaced the −2 and 0 positions with alanines (SPTLC1-ALA, -VLA, and -ALL), substitutions that produced mutants that were expressed at levels more comparable with WT SPTLC1 (supplemental Fig. 3A). Using the AAA and ALA mutants, as well as a third mutant (VAA), we tested by immunoprecipitation assays whether their interaction with Par3 was disrupted. Whereas Par3 co-immunoprecipitated wild type SPTLC1, little, if any, mutant SPTLC1 protein associated with Par3 (Fig. 2A, IP, top panel). Failure of the mutants to interact was not explained by the amount of Par3 precipitated in these assays because as much or more Par3 was present in the precipitates derived from the lysates expressing mutant SPTLC1 (Fig. 2A, IP, bottom panel). Furthermore, low expression of the ALA and VAA mutants could not explain their failure to interact with Par3 because total expression of these mutants was near that of WT SPTLC1 in these assays (Fig. 2A, total lysate, top panel). These results were confirmed in converse immunoprecipitations where the FLAG-SPTLC1 proteins were captured with limiting amounts of anti-FLAG antibody beads and eluted with a FLAG peptide. Again, mutation of the SPTLC1 C terminus disrupted the ability of SPTLC1 to co-precipitate Par3, despite similar levels of the SPTLC1 mutants being captured by the anti-FLAG beads and similar levels of Par3 being co-expressed with the SPTLC1 mutants (Fig. 2B). These results indicate that Par3 interacts with SPTLC1 in a cellular context and that their binding requires the SPTLC1 C-terminal PDZ motif.
FIGURE 2.
Par3 binding of SPTLC1 requires the SPTLC1 type II PDZ motif and occurs at physiologic expression levels in the mouse liver. A, immunoprecipitation (IP) of HA-Par3 out of 293-EBNA-T cells expressing FLAG (FL)-tagged SPTLC1 or the indicated FLAG-SPTLC1 mutants showing the SPTLC1 C-terminal PDZ motif is required for the binding of SPTLC1 and Par3 in a cellular context. The top two panels show immunoblots (IB) of the precipitated SPTLC1 and Par3, and the lower two panels show the amount of total SPTLC1 and Par3 expressed in the transiently transfected 293HEK cells. B, converse immunoprecipitations of the FLAG-SPTLC1 proteins confirm the Par3 interaction with SPTLC1 requires the SPTLC1 C-terminal PDZ motif. C, Par3 150-kDa isoform is expressed in the mouse liver along with the SPTLC1 and SPTLC2 subunits as determined by immunoblots (left panels) and interacts with SPTLC1 and SPTLC2 as determined by immunoprecipitation of the liver lysates with normal IgG or antibodies against SPTLC1 (top right panel) or SPTLC2 (lower right panel).
Having demonstrated the ability of SPTLC1 and Par3 to interact in a cellular setting, we tested whether the complex could be detected at endogenous expression levels in a physiologic context. We investigated the expression of Par3 in the liver, a tissue where SPTLC1 and SPT activity are important for the incorporation of sphingolipids into lipoproteins and where SPT activity is post-translationally regulated by ApoE expression (19). Immunoblots of mouse liver detected Par3 as a series of isoforms ranging from ∼100 to 150 kDa as has been described for Par3 expression in other tissues, and as expected the liver expressed significant amounts of SPTLC1 and SPTLC2 (Fig. 2C, left panels). Initially, immunoprecipitations were attempted with the commercially available Par3 antibody and the liver lysates. However, a high level of binding of the secondary antibody to the 50-kDa heavy chain of the precipitating antibody obscured detection of SPTLC1, which is also 50 kDa in size. To circumvent this problem additional assays were conducted using an SPTLC1 antibody to precipitate the liver lysates, and it was found that the 150-kDa Par3 isoform prominently co-precipitated with SPTLC1 (Fig. 2C, right, top panel). This result indicates Par3 and SPTLC1 can interact at endogenous expression levels in the physiologic setting of the liver. Because the SPT holoenzyme is formed by a complex between SPTLC1 and SPTLC2, we also sought to test whether Par3 would also be co-precipitated using an SPTLC2 antibody, which was found to be the case (Fig. 2C, right, bottom panel). This result suggested Par3 could either directly interact with SPTLC2 or that it was associating with the SPT holoenzyme by binding the SPTLC1 C-terminal PDZ motif.
Par3 Can Bind the SPTLC1 C Terminus When SPTLC1 Is Associated with SPTLC2, but the SPTLC1 PDZ Motif Is Not Required for Holoenzyme Formation
Our data indicated that in the liver Par3 was able to interact with both SPTLC1 and SPTLC2, and thus we sought to distinguish if this was due to an ability to bind the SPTLC1 C-terminal PDZ motif in the context of the SPT holoenzyme or whether Par3 could also directly interact with SPTLC2. To address this question we conducted additional immunoprecipitation assays using an N-terminal Myc-tagged SPTLC2 construct that we verified could interact with SPTLC1 in a normal manner and catalyze SPT activity, as has been reported previously (supplemental Fig. 2B). Myc-SPTLC2 was expressed in 293-EBNA-T cells alone, in the presence of Par3, or in the presence of Par3 and either SPTLC1 or the SPTLC1-ALA mutant. Myc-SPTLC2 was precipitated out of lysates from the transfected cells, and probing these precipitates detected the presence of Par3 in the lysates of cells that expressed WT SPTLC1 but not in the lysates of cells expressing the SPTLC1-ALA mutant or in lysates of cells expressing only SPTLC2 (Fig. 3, top left panel). Probing the blot for SPTLC2 showed similar amounts Myc-SPTLC2 were precipitated out of each of the lysates from the Myc-SPTLC2-transfected cells (Fig. 3, bottom left panel). Thus, the lack of Par3 in the precipitates from the cells expressing the ALA mutant was not explained by a failure to precipitate myc-SPTLC2. Likewise, probing for the amount of SPTLC1 and SPTLC1-ALA that co-precipitated with SPTLC2 indicated SPTLC2 still captured a significant amount of the SPTLC1-ALA mutant (Fig. 3, middle left panel). Finally, probing for the total amount of Par3 in the cell lysates used to perform these immunoprecipitations showed it was expressed at similar levels when expressed in the presence of SPTLC2, or in the presence of SPTLC2 and SPTLC1, or the ALA mutant (Fig. 3, top right panel). These results indicate SPTLC2 alone is not able interact with Par3 but that in the presence of SPTLC1 carrying an intact C-terminal PDZ motif SPTLC2 can co-precipitate Par3.
FIGURE 3.
Par3 interacts with the SPTLC1/2 holoenyzme through an interaction with the SPTLC1 PDZ motif. Myc-tagged SPTLC2 was co-transfected into 293-EBNA-T cells alone or in the presence of HA-Par3 and the indicated FLAG-SPTLC1 constructs. SPTLC2 was immunoprecipitated (IP) with an anti-Myc antibody, and the amount of Par3, SPTLC1, and SPTLC2 that precipitated was determined by immunoblotting (IB) (left panels), and immunoblotting the input protein lysates shows the total amount of Par3, SPTLC1, and SPTLC2 that was expressed in the cells (right panels).
However, because Myc-SPTLC2 was still able to co-precipitate SPTLC1-ALA, this indicates the association of SPTLC1 and SPTLC2 does not have a critical dependence upon the SPTLC1 PDZ motif. To further verify that the SPTLC1 PDZ motif mutants could bind SPTLC2, we conducted additional co-expression studies and quantitated the ability of SPTLC1 and the SPTLC1-AAA and ALA mutants to stabilize SPTLC2 protein expression. Again, when the amount of SPTLC2 protein expression was normalized to the amount of expressed WT or mutant SPTLC1 protein, the mutants showed little or no deficit in their ability to stabilize SPTLC2 protein expression confirming that they are expressed as normally folded proteins capable of binding and stabilizing SPTLC2 (supplemental Fig. 3C). In aggregate, these results indicate Par3 can interact with the SPTLC1/2 holoenzyme at physiologic expression levels in the liver and that the association of Par3 with the holoenzyme is mediated by its ability to bind the SPTLC1 PDZ motif.
Par3 Modulates THP-1 Monocyte SPT Activity and Ceramide Levels
Given that Par3 interacted with the SPTLC1/2 holoenzyme, we sought to test whether Par3 could influence SPT activity. As in the liver, SPTLC1 is also expressed in cells of the monocyte-macrophage lineage, a cell type whose migration to the subendothelial space of inflamed coronary arteries plays a key role in the development of atherosclerosis (37). Moreover, because both Par3 and sphingolipids have been implicated in polarized cell behavior, we tested whether Par3 was expressed in the monocyte-macrophage cell lineage and, if so, whether Par3 influenced SPTLC1 expression and function. To address this possibility we probed for Par3 expression in human THP-1 monocyte-macrophages and found these cells do express Par3 protein as undifferentiated monocytes, as well as when differentiated to the adherent macrophage phenotype induced by the phorbol ester phorbol 12-myristate 13-acetate (Fig. 4A and supplemental Fig. 4A). Likewise, primary mouse bone marrow macrophages also were found to express Par3 and immunoprecipitation of the SPT holoenzyme using either SPTLC1 or SPTLC2 antibodies co-precipitated by Par3 (supplemental Fig. 4B). These results indicate Par3 is expressed in cells of the monocyte-macrophage lineage and can associate with the SPT holoenyzme.
FIGURE 4.
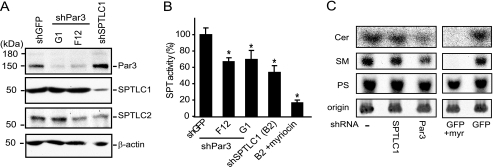
shRNA suppression of Par3 in human THP-1 monocytes inhibits serine palmitoyltransferase activity. A, THP-1 monocytes express Par3, SPTLC1, and SPTLC2 as determined by immunoblotting, and lentivirus-mediated expression of shRNAs targeting Par3 (G1 and F12) or SPTLC1 specifically repress Par3 and SPTLC1 protein expression, respectively. B, lentivirus delivered shRNAs targeting Par3, and SPTLC1 inhibits SPT activity by ∼40% in THP-1 monocytes as determined by measuring the condensation of [14C]serine and palmitoyl-CoA catalyzed by microsomal cell membrane lysates. Pretreatment of the SPTLC1-targeted lysate with the SPT inhibitor myriocin further inhibited activity to 90% (B2 + myriocin, n = 3, ±S.D., *, p < 0.05). C, de novo synthesis of ceramide (Cer) and sphingomyelin (SM) is reduced in cells expressing shRNAs targeting Par3 or SPTLC1 expression as compared with uninfected control cells, or cells expressing a lentivirus-delivered shRNA targeting GFP. As with the Par3- and SPTLC1-targeted cells, myriocin treatment of the control GFP-targeted cells reduced incorporation of [14C]serine into ceramide (Cer) and sphingomyelin (SM) but not phosphatidylserine (PS). Cells were incubated with [14C]serine (5 μCi) for 24 h at 37 °C, and cellular lipids were isolated by chloroform/methanol extraction and separated on TLC plates along with purified standards for ceramide, sphingomyelin, and phosphatidylserine. Labeled lipids remaining at the plate origin was similar demonstrating equal loading of the samples (results representative of two or more independent experiments, and quantification of labeled ceramide and sphingomyelin is shown in supplemental Fig. 5B).
To test whether Par3 influences SPTLC1 function, lentiviral vectors were used to introduce shRNAs into THP-1 cells that suppress Par3 mRNA expression. Compared with THP-1 monocytes infected with a control lentivirus expressing an shRNA targeting GFP, cells expressing shRNAs targeting Par3 mRNA showed a pronounced decrease in Par3 protein expression (Fig. 4A, top panel). Similarly, in cells expressing an shRNA targeting SPTLC1, protein levels of SPTLC1 were markedly depleted (Fig. 4A, 2nd panel). Probing this blot for SPTLC2 showed lower SPTLC2 protein levels in the lysate from cells expressing the SPTLC1 shRNA, as well as in the lysates from cells expressing the F12 Par3 shRNA (Fig. 4A, 3rd panel). However, after normalization to β-actin in a series of blots, it was found that loss of Par3 did not have a significant impact on expression levels of either SPTLC1 or SPTLC2 (supplemental Fig. 4C). Likewise, knockdown of SPTLC1 did not significantly alter Par3 protein levels, but it did reduce levels of SPTLC2 as has been previously reported (supplemental Fig. 4C). These results indicate Par3 expression is not critical for maintaining SPTLC1 protein stability and suggest the poor expression observed for the SPTLC1-Δ3 and AAA mutants is likely not accounted for by an inability to bind Par3 in 293HEK cells.
Having established THP-1 cells that stably suppress Par3 or SPTLC1 protein expression, we next quantitated levels of SPT activity in lysates from these cells. Compared with lysates from the cells expressing the GFP shRNA, lysates from cells expressing shRNAs against Par3 or SPTLC1 had significant reductions in SPT activity of ∼40 and 50%, respectively (Fig. 4B). This was an intermediate level of inhibition compared with that induced by treating the cells with the SPT inhibitor myriocin (10 μm), which inhibited activity by ∼90%. To rule out that the loss of SPT activity in the Par3 or SPTLC1 lysates was due to a general inhibitory effect on cell viability or metabolism, we measured the ATP levels in the cell lines and their ability to metabolize tetrazolium salts as a measure of mitochondrial function, and we found no significant differences in these parameters (supplemental Fig. 4D). Additionally, because our results suggest Par3 binding to SPTLC1 may act to recruit the SPT holoenzyme to Par3-associated microdomains, we further tested whether such an effect may alter the amount of SPTLC1 that partitions into the microsomal membranes derived from Par3-suppressed THP-1 cells and thus could explain the loss of SPT activity seen in these lysates, but this was not found to be the case because SPT activity when normalized to SPTLC1 contained in the microsomal fraction was still found to be significantly reduced (supplemental Fig. 5A).
Having confirmed all the cell lines were equally viable, we next used [14C]serine to assess whether Par3 modulated de novo synthesis of sphingolipids that are generated by SPTLC1 and SPT activity. Here THP-1 cells expressing the Par3 or SPTLC1 shRNAs were compared with either uninfected THP-1 cells or THP-1 cells expressing the shRNA targeting GFP. Equal numbers of cells (5 × 106) were incubated in media containing 5 μCi of [14C]serine, and to control for lipids whose synthesis depended upon SPT activity additional cells expressing the GFP shRNA were preincubated for 2 h with 10 μm myriocin to block SPT activity. Then after 24 h of additional incubation at 37 °C, [14C]serine-labeled lipids were extracted from the cells with chloroform/methanol and separated on TLC plates along with purified standards for ceramide, sphingomyelin, and phosphatidylserine. Phosphorimages of labeled ceramide, sphingomyelin, and phosphatidylserine are shown in Fig. 4C. As was found in the cell-free assays, knockdown of either SPTLC1 or Par3 reduced the de novo synthesis of ceramide and sphingomyelin but had little effect on the level of incorporation of [14C]serine into phosphatidylserine. Like the effect of the SPTLC1 and Par3 siRNAs, myriocin treatment of the control cells expressing the GFP siRNA strongly reduced incorporation of [14C]serine into ceramide and sphingomyelin but not phosphatidylserine (Fig. 4C, right panels). Quantification of the Par3 siRNA effect showed a significant 30 and 40% decrease in the de novo synthesis of sphingomyelin and ceramide, respectively (supplemental Fig. 5B). Thus, as analyzed by both cell-free assays for SPT activity and by [14C]serine labeling of de novo synthesized sphingolipids, loss of Par3 expression in THP-1 cells inhibited the activity of the SPTLC1/2 holoenzyme.
Par3 and SPTLC1 Are Required for MCP-1-induced Chemotaxis of THP-1 Monocytes
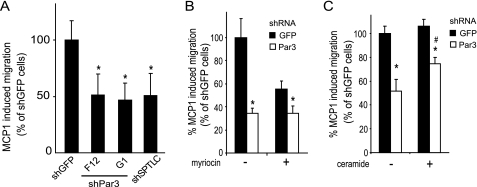
Given that Par3 modulated THP-1 monocyte SPT activity and sphingolipid synthesis, we further tested if Par3 and SPTLC1 may influence the migratory behavior of these cells. Because Par3 has recently been shown to be required for polarized migration of keratinocytes and because inhibition of SPT activity has also been shown to inhibit neuronal cell migration (25, 36), we speculated that Par3 and SPTLC1 could play a role in the ability of monocytes to migrate toward an inflammatory signal. To explore this possibility we tested whether loss of either Par3 or SPTLC1 expression in THP-1 monocytes inhibited the ability of these cells to migrate toward a gradient of MCP-1 using the Boyden chamber assay. Compared with control cells expressing the shRNA directed against GFP, cells expressing either shRNAs targeting Par3 or SPTLC1 had a significant 50% reduction in their ability to migrate in response to the MCP-1 signal (Fig. 5A). As indicated above this inhibition of migration was not accounted for by a difference in viability as determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays and by measurement of ATP production in these cells (supplemental Fig. 4D). Furthermore, the defect imparted by loss of Par3 or SPTLC1 was specific for the polarized response to MCP1 because the number of cells that randomly migrated into the lower chamber in the absence of the chemokine was similar for all the cell lines (supplemental Fig. 5C). To test whether the defect in MCP-1-induced migration seen in the cells with suppressed Par3 expression was acting through a mechanism that involved the ability of Par3 to modulate SPT activity, control and Par3 knockdown cells were first pretreated for 24 h with vehicle or myriocin to inhibit SPT. Then the ability of the cells to migrate in response to MCP-1 was again tested. As before, the MCP-1-induced migration of the cells expressing the Par3 shRNA was reduced by over 60% compared with the control cells expressing the GFP shRNA (Fig. 5B). Additionally, pretreatment of the control GFP shRNA-expressing cells with myriocin inhibited MCP-1-induced migration by ∼40%, but myriocin pretreatment did not further affect the residual MCP-1-induced migration of cells with suppressed Par3 expression (Fig. 5B). This result suggested the ability of myriocin to modulate migration in response to MCP-1 was acting through a mechanism that depends upon the expression of Par3. Because the cells with suppressed Par3 expression were found to have a defect in the de novo synthesis of ceramide, and because ceramide has previously been reported to be important in the induction of neuronal cell migration (36), we tested whether supplying exogenous ceramide to cells with suppressed Par3 expression improved their response to MCP-1. This was found to be the case in that ceramide treatment of Par3-suppressed cells significantly improved their migratory response to MCP-1 (Fig. 5C). In composite, these data indicate Par3 modulated THP-1 migration through a mechanism that depended upon SPT activity and likely the de novo synthesis of ceramide. However, because ceramide treatment did not completely restore the migration defect of the Par3-suppressed cells, it is possible that the synthesis of sphingomyelin or other bioactive lipids may also be involved.
FIGURE 5.
shRNA suppression of Par3 and SPTLC1 inhibits polarized migration of THP-1 cells in response to monocyte chemoattractant protein 1. A, THP-1 monocytes expressing shRNAs against Par3 or SPTLC1 have an ∼50% reduction in their ability to migrate toward a gradient of MCP-1 (10 nm) compared with control cells expressing an shRNA targeting GFP as determined by Boyden chamber assays. B, myriocin pretreatment of control cells expressing the GFP shRNA reduces their MCP-1-induced migration by 44% but does affect the residual MCP-1-induced migration of THP-1 monocytes with suppressed Par3 expression. C, ceramide pretreatment of Par3-suppressed THP-1 monocytes significantly improves their MCP-1-induced migration defect (n = 3, ± S.D., *, p < 0.05 versus shGFP untreated; #, p < 0.05 versus shPar3 untreated; results representative of two or more independent experiments).
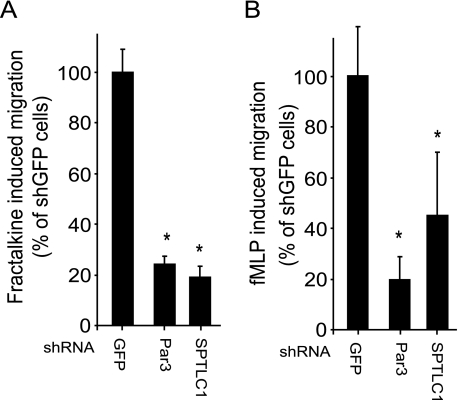
To explore whether THP-1 monocytes more broadly require Par3 and SPTLC1 to respond to inflammatory stimuli, we measured the ability of the Par3- and SPTLC1 shRNA-suppressed cells to respond to Fractalkine and fMLP. Fractalkine is a CX3C chemokine, a class distinct from MCP-1, but like MCP-1 Fractalkine is also important for recruiting monocytes to sites of vascular inflammation and has also been found to be pro-atherosclerotic (38, 39). fMLP is a formylated tripeptide released by bacteria at sites of infection that, like MCP-1 and Fractalkine, induces chemotaxis by signaling through G-protein-coupled receptors (40). As with MCP-1, the response of the cells with suppressed Par3 or SPTLC1 expression showed significant deficits in their ability to migrate toward a gradient of Fractalkine or fMLP-1, although the response of SPTLC1-targeted cells toward fMLP-1 was found to be more variable (Fig. 6, A and B). In composite, our results indicate Par3 and SPTLC1 are broadly required for the ability of THP-1 monocytes to respond to a number of inflammatory signals.
FIGURE 6.
Loss of Par3 or SPTLC1 expression inhibits THP-1 chemotaxis induced by structurally diverse chemokines including Fractalkine and the fMLP bacterially formylated tripeptide. A, Fractalkine (10 nm)-induced migration of THP-1 cells expressing shRNAs against Par3 or SPTLC1 is reduced by ∼80% compared with control cells expressing an shRNA targeting GFP. B, fMLP (10 nm)-induced migration of THP-1 cells expressing shRNAs against SPTLC1 or Par3 is reduced ∼60 and 80%, respectively, as compared with the control GFP shRNA cells (n = 3, ±S.D., *, p < 0.05; results are representative of two or more independent experiments).
DISCUSSION
Here we have identified a protein-protein interaction between SPTLC1 and Par3, a PDZ protein. In mammals the SPTLC1 C terminus has been conserved as a type II PDZ-binding motif, and our array screen showed a C-terminal SPTLC1 peptide bound the third PDZ domain of Par3. Overlay assays confirmed the 150-kDa isoform of Par3 interacts with the SPTLC1 C terminus and that this binding required the SPTLC1 PDZ motif. Immunoprecipitation assays show Par3 is able to form a complex with the SPTLC1/2 holoenzyme by binding the SPTLC1 PDZ motif, and we found Par3 interacted with the holoenzyme at endogenous expression levels in the liver and in primary macrophages. Par3 is expressed in both human THP-1 monocytes and macrophages, and shRNA suppression of Par3 in THP-1 monocytes was found to inhibit holoenzyme serine palmitoyltransferase activity and de novo sphingolipid synthesis. Par3 or SPTLC1 shRNA suppression also significantly impaired polarized THP-1 monocyte migration in response to gradients of inflammatory chemokines but did not alter cell viability or random migration. Likewise, pharmacologic inhibition of the SPT holoenzyme modulated MCP-1-induced migration through a mechanism that depended upon Par3 expression, and ceramide treatment significantly improved the Par3 defect in migration. These results have uncovered a novel physical and functional link between SPTLC1 and Par3 that is associated with increased SPT activity and polarized migration toward an inflammatory signal.
Our work provides new insight into the role that Par3 protein-protein interactions play in controlling polarized migration toward a chemotactic signal. Recently, the migration of keratinocytes induced by epidermal growth factor has also been shown to depend upon Par3 (25). As for the more established role of Par3 in mediating tight junction formation that delineates apical/basolateral polarity of epidermal tissues, this regulation of keratinocyte migration is characterized by the Par3 recruitment of atypical PKC to membrane microdomains. Atypical PKC recruitment is of interest because migration of neuronal precursor cells and establishment of polarity in the primitive ectoderm have also been found to depend on de novo synthesis of ceramide and the recruitment of atypical PKC to ceramide-rich membrane microdomains (35, 36). Our work extends these observations by showing Par3 and SPTLC1 modulate the migratory response of cells of the monocyte lineage to gradients of inflammatory chemokines. In addition to MCP-1, we found the ability of THP-1 monocytes to respond to other chemokines, including Fractalkine and fMLP, also had a dependence upon Par3 and SPTLC1. Because the Par3 polarity complex has recently been shown to be required for T-cell polarization and migration toward stromal cell-derived factor-1α (21), another chemokine that triggers G-protein-coupled signaling, these and our observations indicate Par3 plays a broad role in mediating chemokine-induced polarized migration of immune cells. Assuming SPTLC1 also plays a similarly broad role in mediating the migratory responses to inflammatory signals, our results may provide mechanistic explanation for studies that have associated the ability of the SPT inhibitor myriocin to reduce atherosclerosis in ApoE−/− mice with an ability to reduce the macrophage content of their atherosclerotic lesions (41).
What is the relationship between the binding of Par3 and SPTLC1 and the ability of Par3 to stimulate SPT activity and monocyte migration? Although deleting or mutating the SPTLC1 C-terminal PDZ motif to alanines was found to destabilize SPTLC1 protein expression, more subtle substitutions of this motif that disrupted the ability of Par3 to bind SPTLC1 did not have a strong effect on the stability of SPTLC1. This suggests the binding of SPTLC1 by Par3 does not play a critical role in stabilizing SPTLC1 protein expression, a result further confirmed by SPTLC1 protein levels not being significantly altered in THP-1 cells expressing Par3 targeted shRNAs. SPTLC2 expression in the Par3-suppressed cells was also similar to that of control cells, and it was found that the mutation of the SPTLC1 PDZ motif did not prevent the SPTLC1 binding and stabilization of SPTLC2. Because a third SPT subunit has been identified, it is possible that Par3 could modulate the interaction of SPTLC1 with this subunit (8). However, this seems unlikely to explain the ability of Par3 to modulate SPT activity in THP-1 monocytes because we were unable to detect SPTLC3 expression in these cells.3 In aggregate, our results indicate the SPTLC1 PDZ motif and Par3 binding of this motif do not play a critical role in the ability of SPTLC1 to bind and stabilize SPTLC2.
Because Par3 modulated SPT activity but did not alter the protein levels of SPTLC1 and -2, and because Par3 is known to recruit and activate atypical PKC, it is possible that Par3 may be modulating the phosphorylation status of the SPT holoenzyme. This possibility is intriguing because a number a reports have suggested SPT activity is regulated at the post-translational level (19, 42, 43), and analysis of SPTLC1 and -2 orthologs indicate both have well conserved potential phosphorylation sites. Additionally, the migration of SPTLC2 on two-dimensional gels is altered by phosphatase treatment suggesting it is indeed a phosphoprotein.4 In such a model the Par 3 binding of the SPTLC1 PDZ motif may be a mechanism to alter the phosphorylation status of the SPT holoenzyme as a means to modulate the de novo synthesis of sphingolipids, possibly in response to an inflammatory stimulus. Whether this could occur in polarized microdomains organized by Par3 remains to be clarified, and our co-expression studies suggest this may be the case when, expressed in the presence of Par3, SPTLC1 was found to co-localize with Par3 in the cell periphery near sites of cell-cell contact and in cellular protrusions.4 These results suggest Par3 may be able to dynamically alter the distribution of SPTLC1 and give insight into a recent report by Wei et al. (44) that suggests SPTLC1 can associate with focal adhesion complexes in the cell periphery. In summary, we have identified a protein interaction between Par3 and SPTLC1 that is associated with increased serine palmitoyltransferase activity and migration of THP-1 monocytes toward a number of inflammatory chemokines. Although the mechanism of this association needs to be clarified, the results lend additional support for a model in which SPTLC1 acts as a regulator of SPT activity by interacting with Par3 and other proteins such as ABCA1 to couple signals that integrate cellular demand for the de novo synthesis of sphingolipids with the biogenesis of membrane lipid domains (20). Further investigation of this hypothesis is warranted, particularly because additional small subunits of the SPT holoenzyme have been recently identified by Han et al. (45) that alter acyl-CoA substrate specificity and appear to be required for maximal enzyme activity.
Supplementary Material
Acknowledgments
We acknowledge the technical advice provided by the laboratory of Dr. Kathryn J. Moore for the cell migration assays and the helpful comments provided by Dr. Mason W. Freeman.
This work was supported, in part, by National Institutes of Health Grant HL074136 from NHLBI (to M. L. F.). This work was also supported by an American Heart Association Grant-in-Aid 09GRNT2260352 (to M. L. F.) and by fellowship training grants from the Uehara Memorial Foundation, the Nagai Foundation of Tokyo, and the Japan Heart Foundation (to N. T.), the American Heart Association (to Z. M.), and the Massachusetts Biomedical Research Corporation (to S. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
N. Tamehiro and M. L. Fitzgerald, unpublished observations.
T. Hornemann unpublished observations.
- SPT
- serine palmitoyltransferase
- Par3
- partitioning defective protein 3
- HA
- hemagglutinin
- shRNA
- short hairpin RNA
- GFP
- green fluorescent protein
- fMLP
- formylmethionylleucylphenylalanine
- siRNA
- short interfering RNA
- WT
- wild type
- PKC
- protein kinase C.
REFERENCES
- 1.Rivera J., Proia R. L., Olivera A. (2008) Nat. Rev. Immunol. 8, 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Meer G., Voelker D. R., Feigenson G. W. (2008) Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wymann M. P., Schneiter R. (2008) Nat. Rev. Mol. Cell Biol. 9, 162–176 [DOI] [PubMed] [Google Scholar]
- 4.Zuo Y., Zhuang D. Z., Han R., Isaac G., Tobin J. J., McKee M., Welti R., Brissette J. L., Fitzgerald M. L., Freeman M. W. (2008) J. Biol. Chem. 283, 36624–36635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanada K. (2003) Biochim. Biophys. Acta 1632, 16–30 [DOI] [PubMed] [Google Scholar]
- 6.Hanada K., Hara T., Fukasawa M., Yamaji A., Umeda M., Nishijima M. (1998) J. Biol. Chem. 273, 33787–33794 [DOI] [PubMed] [Google Scholar]
- 7.Hanada K., Hara T., Nishijima M. (2000) J. Biol. Chem. 275, 8409–8415 [DOI] [PubMed] [Google Scholar]
- 8.Hornemann T., Richard S., Rütti M. F., Wei Y., von Eckardstein A. (2006) J. Biol. Chem. 281, 37275–37281 [DOI] [PubMed] [Google Scholar]
- 9.Hornemann T., Wei Y., von Eckardstein A. (2007) Biochem. J. 405, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandon E. C., Ehses I., Rother J., van Echten G., Sandhoff K. (1992) J. Biol. Chem. 267, 11144–11148 [PubMed] [Google Scholar]
- 11.Yasuda S., Nishijima M., Hanada K. (2003) J. Biol. Chem. 278, 4176–4183 [DOI] [PubMed] [Google Scholar]
- 12.Jiang X. C., Paultre F., Pearson T. A., Reed R. G., Francis C. K., Lin M., Berglund L., Tall A. R. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 2614–2618 [DOI] [PubMed] [Google Scholar]
- 13.Nelson J. C., Jiang X. C., Tabas I., Tall A., Shea S. (2006) Am. J. Epidemiol. 163, 903–912 [DOI] [PubMed] [Google Scholar]
- 14.Glaros E. N., Kim W. S., Rye K. A., Shaymon J. A., Garner B. (2008) J. Lipid Res. 49, 1677–1681 [DOI] [PubMed] [Google Scholar]
- 15.Glaros E. N., Kim W. S., Wu B. J., Suarna C., Quinn C. M., Rye K. A., Stocker R., Jessup W., Garner B. (2007) Biochem. Pharmacol. 73, 1340–1346 [DOI] [PubMed] [Google Scholar]
- 16.Hojjati M. R., Li Z., Zhou H., Tang S., Huan C., Ooi E., Lu S., Jiang X. C. (2005) J. Biol. Chem. 280, 10284–10289 [DOI] [PubMed] [Google Scholar]
- 17.Park T. S., Panek R. L., Mueller S. B., Hanselman J. C., Rosebury W. S., Robertson A. W., Kindt E. K., Homan R., Karathanasis S. K., Rekhter M. D. (2004) Circulation 110, 3465–3471 [DOI] [PubMed] [Google Scholar]
- 18.Park T. S., Panek R. L., Rekhter M. D., Mueller S. B., Rosebury W. S., Robertson A., Hanselman J. C., Kindt E., Homan R., Karathanasis S. K. (2006) Atherosclerosis 189, 264–272 [DOI] [PubMed] [Google Scholar]
- 19.Jeong Ts., Schissel S. L., Tabas I., Pownall H. J., Tall A. R., Jiang X. (1998) J. Clin. Invest. 101, 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamehiro N., Zhou S., Okuhira K., Benita Y., Brown C. E., Zhuang D. Z., Latz E., Hornemann T., von Eckardstein A., Xavier R. J., Freeman M. W., Fitzgerald M. L. (2008) Biochemistry 47, 6138–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gérard A., Mertens A. E., van der Kammen R. A., Collard J. G. (2007) J. Cell Biol. 176, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joberty G., Petersen C., Gao L., Macara I. G. (2000) Nat. Cell Biol. 2, 531–539 [DOI] [PubMed] [Google Scholar]
- 23.Lin D., Edwards A. S., Fawcett J. P., Mbamalu G., Scott J. D., Pawson T. (2000) Nat. Cell Biol. 2, 540–547 [DOI] [PubMed] [Google Scholar]
- 24.Nishimura T., Kaibuchi K. (2007) Dev. Cell 13, 15–28 [DOI] [PubMed] [Google Scholar]
- 25.Pegtel D. M., Ellenbroek S. I., Mertens A. E., van der Kammen R. A., de Rooij J., Collard J. G. (2007) Curr. Biol. 17, 1623–1634 [DOI] [PubMed] [Google Scholar]
- 26.Hubbard T. J., Aken B. L., Ayling S., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Clarke L., Coates G., Fairley S., Fitzgerald S., Fernandez-Banet J., Gordon L., Graf S., Haider S., Hammond M., Holland R., Howe K., Jenkinson A., Johnson N., Kahari A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Rios D., Schuster M., Slater G., Smedley D., Spooner W., Spudich G., Trevanion S., Vilella A., Vogel J., White S., Wilder S., Zadissa A., Birney E., Cunningham F., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Kasprzyk A., Proctor G., Smith J., Searle S., Flicek P. (2009) Nucleic Acids Res. 37, D690–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuhira K., Fitzgerald M. L., Sarracino D. A., Manning J. J., Bell S. A., Goss J. L., Freeman M. W. (2005) J. Biol. Chem. 280, 39653–39664 [DOI] [PubMed] [Google Scholar]
- 28.Fitzgerald M. L., Okuhira K., Short G. F., 3rd, Manning J. J., Bell S. A., Freeman M. W. (2004) J. Biol. Chem. 279, 48477–48485 [DOI] [PubMed] [Google Scholar]
- 29.Doering T., Holleran W. M., Potratz A., Vielhaber G., Elias P. M., Suzuki K., Sandhoff K. (1999) J. Biol. Chem. 274, 11038–11045 [DOI] [PubMed] [Google Scholar]
- 30.Han G., Gable K., Yan L., Natarajan M., Krishnamurthy J., Gupta S. D., Borovitskaya A., Harmon J. M., Dunn T. M. (2004) J. Biol. Chem. 279, 53707–53716 [DOI] [PubMed] [Google Scholar]
- 31.Chen J. R., Chang B. H., Allen J. E., Stiffler M. A., MacBeath G. (2008) Nat. Biotechnol. 26, 1041–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stiffler M. A., Chen J. R., Grantcharova V. P., Lei Y., Fuchs D., Allen J. E., Zaslavskaia L. A., MacBeath G. (2007) Science 317, 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T., Irie S., Kitada S., Reed J. C. (1995) Science 268, 411–415 [DOI] [PubMed] [Google Scholar]
- 34.Zhu J. H., Chen R., Yi W., Cantin G. T., Fearns C., Yang Y., Yates J. R., 3rd, Lee J. D. (2008) Oncogene 27, 2525–2531 [DOI] [PubMed] [Google Scholar]
- 35.Krishnamurthy K., Wang G., Silva J., Condie B. G., Bieberich E. (2007) J. Biol. Chem. 282, 3379–3390 [DOI] [PubMed] [Google Scholar]
- 36.Wang G., Krishnamurthy K., Chiang Y. W., Dasgupta S., Bieberich E. (2008) J. Neurochem. 106, 718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park Y. M., Febbraio M., Silverstein R. L. (2009) J. Clin. Invest. 119, 136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boring L., Gosling J., Cleary M., Charo I. F. (1998) Nature 394, 894–897 [DOI] [PubMed] [Google Scholar]
- 39.Lesnik P., Haskell C. A., Charo I. F. (2003) J. Clin. Invest. 111, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevenson N. J., Haan S., McClurg A. E., McGrattan M. J., Armstrong M. A., Heinrich P. C., Johnston J. A. (2004) J. Immunol. 173, 3243–3249 [DOI] [PubMed] [Google Scholar]
- 41.Park T. S., Rosebury W., Kindt E. K., Kowala M. C., Panek R. L. (2008) Pharmacol. Res. 58, 45–51 [DOI] [PubMed] [Google Scholar]
- 42.Herget T., Esdar C., Oehrlein S. A., Heinrich M., Schütze S., Maelicke A., van Echten-Deckert G. (2000) J. Biol. Chem. 275, 30344–30354 [DOI] [PubMed] [Google Scholar]
- 43.Perry D. K., Carton J., Shah A. K., Meredith F., Uhlinger D. J., Hannun Y. A. (2000) J. Biol. Chem. 275, 9078–9084 [DOI] [PubMed] [Google Scholar]
- 44.Wei J., Yerokun T., Leipelt M., Haynes C. A., Radhakrishna H., Momin A., Kelly S., Park H., Wang E., Carton J. M., Uhlinger D. J., Merrill A. H., Jr. (2009) Biochim. Biophys. Acta 1791, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han G., Gupta S. D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R. H., Jr., Harmon J. M., Dunn T. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8186–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.