Abstract
The transition metal cadmium is an environmental teratogen. In addition, cadmium and retinoic acid can act synergistically to induce forelimb malformations. The molecular mechanism underlying the teratogenicity of cadmium and the synergistic effect with retinoic acid has not been addressed. An evolutionarily conserved gene, β,β-carotene 15,15′-monooxygenase (BCMO), which is involved in retinoic acid biosynthesis, was studied in both Caenorhabditis elegans and murine Hepa 1–6 cells. In C. elegans, bcmo-1 was expressed in the intestine and was cadmium inducible. Similarly, in Hepa 1–6 cells, Bcmo1 was induced by cadmium. Retinoic acid-mediated signaling increased after 24-h exposures to 5 and 10 μm cadmium in Hepa 1–6 cells. Examination of gene expression demonstrated that the induction of retinoic acid signaling by cadmium may be mediated by overexpression of Bcmo1. Furthermore, cadmium inhibited the expression of Cyp26a1 and Cyp26b1, which are involved in retinoic acid degradation. These results indicate that cadmium-induced teratogenicity may be due to the ability of the metal to increase the levels of retinoic acid by disrupting the expression of retinoic acid-metabolizing genes.
The transition metal cadmium is a persistent toxicant that exists ubiquitously in the environment. It is ranked number 7 on the 2007 CERCLA Priority List of Hazardous Substances (1). Cadmium is introduced into the environment mainly through anthropogenic activities, such as copper and zinc mining, fossil fuel combustion, and the manufacturing of cadmium-containing products. For the general population, the primary routes of cadmium exposure are through ingestion of contaminated food and water, and cigarette smoking, which doubles the daily intake of cadmium, compared with the non-smoking population (2). Occupational exposure to cadmium is mainly through inhalation of cadmium fumes generated during heating or welding of cadmium-containing materials or inhalation of cadmium-containing dust (3).
Due to its stability in the environment and long retention time in the human body (half-life ∼15–20 years) (4), cadmium can accumulate and cause a variety of adverse effects and diseases. Cadmium has been classified as a category 1 human carcinogen (3). It is also a potent animal teratogen. Teratogenic effects of cadmium have been reported in a variety of laboratory species, including chicken, frog, rat, and mouse (5–8). Maternal exposure to cadmium in rodents causes a spectrum of birth defects, including fetal limb malformations, hydrocephalus, cleft palate, and neural tube defects, depending on the dose and the embryonic stage of development at the time of exposure (7, 9–11). Among these defects, limb malformations caused by administration of cadmium during early or middle gestation have been well documented (10, 12–14). When C57BL/6J mice were given a single injection of 4 mg/kg cadmium chloride on gestational day 9, limb defects, predominated by forelimb reduction deformities, were observed in ∼80% of the surviving fetuses (10, 15). In humans, maternal exposure to cadmium has been associated with preterm deliveries and low birth weight (16–18).
Although toxicological studies have established a relationship between cadmium exposure and birth defects, the molecular mechanisms of cadmium teratogenesis remain largely unknown. Following administration, cadmium is mainly distributed in maternal organs, such as liver, kidney, and placenta, while less reaches the fetus. This suggests that cadmium may exert teratogenic effects through an indirect mechanism (8, 19–21). Recent studies, however, have shown that cadmium exposure can alter signaling pathways that determine limb patterning (22–24).
Retinoic acid (RA)2 is an essential hormone-like molecule that regulates cell differentiation and proliferation during embryogenesis of vertebrates, through binding to RA receptors that control the transcription of a battery of genes (25). Embryonic exposure to excessive RA or retinoids can cause a spectrum of malformations, including those observed following cadmium exposure such as cleft lip, cleft palate, brachygnathia, and limb malformation (26). Cadmium has been shown to act synergistically with RA in the induction of forelimb ectrodactyly in mice (27). When C57BL/6 mice were co-administered sub-threshold doses of cadmium (0.5 mg/kg) and RA (1 mg/kg), the combined treatment resulted in forelimb ectrodactyly in 19% of the offspring. Moreover, co-administration of 1 mg/kg cadmium and 5 mg/kg RA showed a synergistic effect: 92% of the fetuses were found with the forelimb defect, as opposed to 10% if the response was additive (27). Although it has been suggested that cadmium and RA may share a common teratogenic mechanism, the molecular mechanism for the interactive effects has not been fully addressed (23, 27).
In our previous study of cadmium-regulated transcription in the nematode Caenorhabditis elegans, an evolutionarily conserved gene, Y46G5A.24, which encodes the nematode homolog of mammalian β,β-carotene 15,15′-monooxygenase (BCMO), was found to be highly cadmium-responsive (28). The predicted C. elegans protein encoded by Y46G5A.24, now designated bcmo-1, shares >95% sequence identity with the human (BCMO1) and mouse (Bcmo1) homologs. BCMO is a key enzyme in the transformation of β-carotene to retinal. In the present study, we show that bcmo-1 and the murine homolog are cadmium-responsive. In addition, cadmium affects other components in RA metabolism in mouse cells. Based on these results a mechanism for cadmium teratogenicity is proposed in which cadmium exposure causes an increase in the levels of RA by increasing RA synthesis and decreasing RA degradation, which subsequently leads to developmental abnormalities.
EXPERIMENTAL PROCEDURES
Preparation of bcmo-1::GFP Transgenic C. elegans
A reporter transgene was prepared using the PCR-fusion based approach (29). Briefly, ∼1.8 kb that is immediately upstream of the start codon of bcmo-1 was prepared from C. elegans genomic DNA by PCR, using the 5′ primer (primer A) and 3′ primer (Primer B) (Table 1). A second fragment of ∼1.6 kb that contains the coding region of a green fluorescent protein (GFP) was prepared from the plasmid pPD 95.67 (Addgene, Cambridge, MA), using Primers C and D (Table 1). Then, the two amplified fragments were combined and used as templates to obtain the fusion PCR product, bcmo-1::GFP, using Primer A′ (a primer nested to Primer A) and Primer D′ (a primer nested to Primer D) (Table 1).
TABLE 1.
Primer sequences used for C. elegansbcmo-1 ::GFP reporter gene construction
| Primer | Sequence (5′ to 3′) |
|---|---|
| Primer A | CAAAAGTAAATGCTTCATTTTTCTGC |
| Primer B | GGACAACTCCAGTGAAAAGTTCTTCTCCTTTACTCATT TCAGCTTCCATTTTTTAGAACA |
| Primer A′ | TTTTTCTGCAAAATATCGAGCG |
| Primer C | ATGAGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCC |
| Primer D | CCACTGAGCCTCAAACCCAAACCTTCTTCCG |
| Primer D′ | ATCTTTCTTGCATCGTGCTCATC |
Wild-type C. elegans (N2 Bristol) were transformed by microinjecting the mixture of the reporter transgene, bcmo-1::GFP (100 ng/μl), and a plasmid containing the selectable marker gene rol-6(su1006) (100 ng/μl) into the gonads of young adults. Injected C. elegans were rescued by slow recovery and then transferred to individual K-agar plates with food (30, 31). Transgenic nematodes were picked and maintained by selecting nematodes that expressed the roller phenotype.
Expression Analysis of bcmo-1::GFP
GFP expression was monitored in nematodes at different developmental stages. Age-synchronized nematodes were generated by collecting L1 transgenic C. elegans in K-medium and then allowing them to develop on K-agar plates (30). Synchronized transgenic nematodes were then transferred into S-medium (32) containing 100 μm cadmium (CdCl2), or onto K-agar plates containing 100 μm cadmium. Escherichia coli OP50 was supplied as food during both exposures. After a 5-h exposure, treated and control, non-exposed, transgenic nematodes were examined under a fluorescence microscope (Zeiss LSM 510 confocal microscope, Carl Zeiss MicroImaging Inc., Thornwood, NY).
For image acquisition, adult nematodes were anesthetized with 170 mm sodium azide and mounted on 2% agarose pads. Larvae (L1–L4) were placed in a drop of sodium azide solution, which was placed directly in the center of a printed microscope slide, and then covered with a glass coverslip.
Growth and Collection of Synchronized C. elegans
Wild-type (N2 Bristol) C. elegans were synchronized as previously described (33). L1 larvae were collected, transferred to K-agar plates with food, and allowed to grow at 20 °C. At the beginning of each larval stage (3 h after collection from buffer for L1s; 16 h for L2s; 25 h for L3s; 35 h for L4s; and 45 h for young adults) nematodes were transferred to S-medium containing food and 100 μm cadmium and incubated for 5 h at 20 °C. After exposure, nematodes were collected, washed twice with 0.1 m NaCl, rapidly frozen as pellets in liquid nitrogen, and stored at −80 °C. To obtain cadmium-treated embryos, gravid adults (56 h after transfer to K-agar plates) were cultured in S-medium with 100 μm cadmium for 12 h, and subsequently collected. Embryos were then prepared as previously described (33).
Isolation of RNA and qRT-PCR
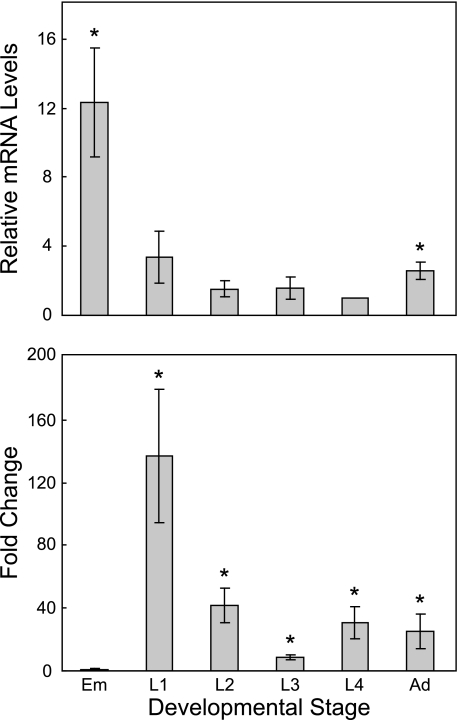
To examine the expression of bcmo-1, total RNA was prepared from age-synchronized nematodes using RNase-Free DNase Set and RNeasy Midi Kit (Qiagen) as previously described (28). qRT-PCR was performed using bcmo-1-specific oligonucleotide primers (forward primer, 5′-AGGGCATCGAGGAGGATGAT-3′; reverse primer, 5′-AGCATGAAATCCAAGTGGAA-3′) with a QuantiTect SYBR Green RT-PCR Kit (Qiagen) following the manufacture's instructions in an ABI Prism 7900H (Applied Biosystems, Foster City, CA). To compare bcmo-1 expression during different developmental stages, measurements were compared with that of L4 larvae, because L4 has the lowest absolute bcmo-1 mRNA level (Fig. 3A). To show the induction of bcmo-1 by cadmium in different developmental stages, measurements were compared with that of untreated nematodes at the same developmental stage (Fig. 3B). Final results were presented as mean fold change ± standard error (n = 3). A t-test was performed for significance of bcmo-1 induction following cadmium treatment.
FIGURE 3.
Developmental and cadmium responsive bcmo-1 transcription in C. elegans. Total RNA was extracted from cadmium-treated or control C. elegans at the indicated developmental stages, and the mRNA levels of bcmo-1 were measured. (upper panel) bcmo-1 transcription at different developmental stages in C. elegans not treated with cadmium. Levels of bcmo-1 mRNA were normalized to the bcmo-1 mRNA level in L4 larvae. (lower panel) effects of 5-h exposure to 100 μm cadmium on bcmo-1 mRNA levels at different developmental stages. mRNA levels were normalized to those observed in non-exposed, control nematodes at identical developmental stages and are presented as the fold change in the levels of expression. The results are mean fold change ± standard error (n = 3); *, indicates significantly different (p < 0.05) from L4, upper panel or from untreated nematodes, lower panel. Em, embryos; Ad, adults.
Mammalian Cell Culture
Mouse Hepa 1–6 cells, a hepatoma cell line derived from a C57L mouse liver tumor, were maintained in Dulbecco's modified Eagle's medium, supplemented with 4 mm l-glutamine, 1.5 g/liter sodium bicarbonate, 4.5 g/liter glucose, and 10% fetal bovine serum (GIBCO® Invitrogen, Carlsbad, CA) at 37 °C in a 5% CO2 atmosphere. For RA treatments, cells were maintained in charcoal-stripped fetal bovine serum. Cells were sub-cultured every 2–3 days, and passages 6–18 were used for experiments.
Transient Transfection and Dual Luciferase Reporter Assay
To measure the effect of cadmium on RA signaling, Hepa 1–6 cells were transfected with the reporter plasmid RARE-TK-Luc (34), which controls the expression of firefly luciferase, and the transfection control plasmid, phRL-TK, which controls the expression of Renilla luciferase (Promega, Madison, WI). RARE-TK-Luc contains three repeats of the RA response element (RARE), which is the binding site of the RA receptor (RAR) (a generous gift from Dr. Michael J. Spinella, Dartmouth University). Transfection was carried out using Lipofectamine Plus Reagent (Invitrogen) following manufacturer's instructions. Briefly, on day 0, cells were seeded in Dulbecco's modified Eagle's medium containing charcoal-stripped serum in 24-well culture plates at a density of 5 × 104 cells per well. On day 1, cells were transfected with 200 ng of reporter plasmid and 2 ng of control plasmid. On day 2, 24 h after transfection, cells were washed with phosphate-buffered saline and then treated with all-trans-RA and/or cadmium. All-trans-RA (Sigma-Aldrich) was dissolved in DMSO at a concentration of 0.1 m and stored at −20 °C for less than 1 month. Retinoic acid solutions were light-protected, and a final concentration of 0.1% of DMSO was used as a vehicle control.
Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's instructions. Briefly, after the exposure, cells were washed with phosphate-buffered saline, then lysed with passive lysis buffer, and incubated for 15 min at room temperature. Cell lysate was then transferred into an opaque 96-well plate and then LAR II and Stop & Glo Reagents were sequentially added by auto injection. Firefly luciferase activity was measured in a FLUOstar OPTIMA plate reader (BMG LABTECH, Durham, NC), normalized to Renilla luciferase activity, and then compared with that measured for untreated controls. A two-way ANOVA was used for significance tests of cadmium effects. Multiple comparisons (least significant difference procedure) were used to compare normalized luciferase activity between the control and individual cadmium treatments at the same RA level (35).
Isolation of RNA from Hepa 1–6 Cells and qRT-PCR
To measure the effects of cadmium on the expression of mouse genes, Bcmo1, Cyp26a1, and Cyp26b1, total RNA was collected from cadmium-treated and untreated Hepa 1–6 cells, and qRT-PCR was performed. Cells were plated in complete Dulbecco's modified Eagle's medium in triplicate at a density of 5 × 104 cells per well on a 24-well tissue culture plate and incubated for 48 h. Cells were then washed with phosphate-buffered saline, and fresh, complete Dulbecco's modified Eagle's medium containing 1, 5, or 10 μm cadmium was added. Following the indicated exposure times, cells of the same treatment were pooled together, and total RNA was extracted using a Qiagen RNeasy Mini Kit according to the manufacturer's instructions. Experiments were conducted in triplicate, and three RNA samples were isolated for each treatment.
Two-step qRT-PCR was performed employing SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) and TaqMan Gene Expression Assays (Applied Biosystems), according to the manufacturer's instructions. Briefly, cDNA was prepared from 0.5 μg of total RNA. PCR was then carried out using TaqMan Universal PCR Master Mix and TaqMan Gene Expression Assays (Applied Biosystems) in an ABI 7900H. TaqMan primers were Bcmo1, Mm00502437_m1; Cyp26a1, Mm00514486_m1; Cyp26b1, Mm00558507_m1; and glyceraldehyde-3-phosphate dehydrogenase, 4352932E. Each sample was measured in triplicate, and all measurements were normalized to glyceraldehyde-3-phosphate dehydrogenase. The fold change of each gene following cadmium exposure was compared with that observed in the untreated samples. Final results were presented in mean fold induction ± standard error. A one-way ANOVA was used for significance tests of cadmium effects. Multiple comparisons (least significant difference procedure) were used to compare expression levels after cadmium treatments with that of the untreated for each gene (35).
RESULTS
Expression of bcmo-1 in Transgenic C. elegans
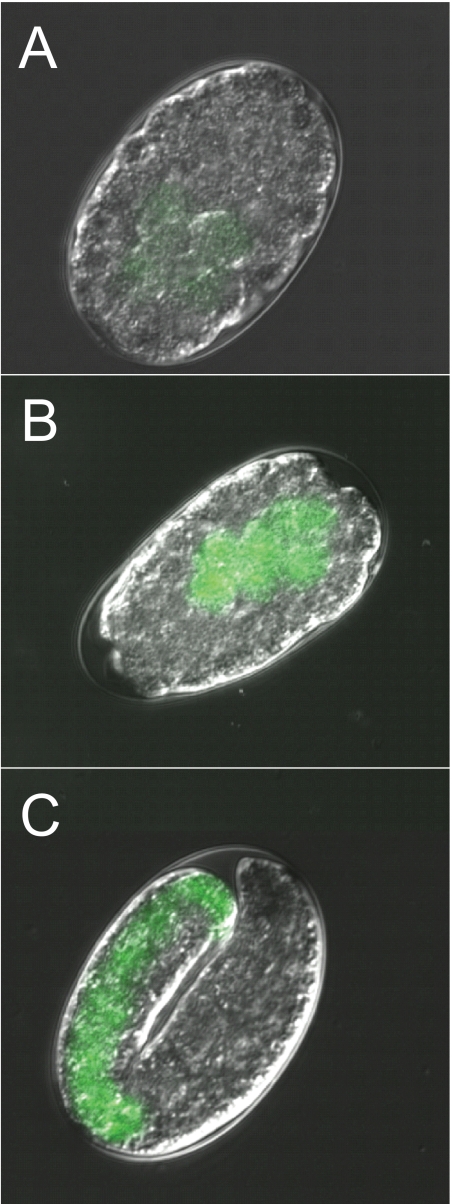
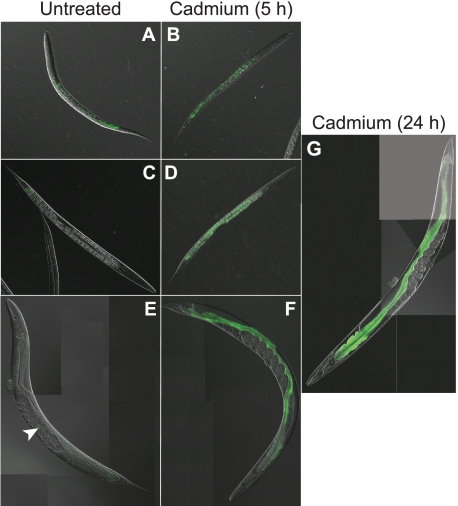
To study the expression pattern of bcmo-1 in C. elegans, a transgenic strain was generated by fusing the GFP coding sequence to the bcmo-1 promoter. GFP expression was greatest in embryos, and it decreased throughout the larval stages into adults, suggesting that the transcription of this gene was developmentally regulated (Figs. 1 and 2). The earliest detectable GFP signal was observed in the intestinal cells of embryos, after the development of the intestinal cell lineage (E cells) (36) (Fig. 1A). Constitutive bcmo-1 transcription was observed in all embryonic E cells (Fig. 1, B and C). The constitutive GFP expression decreased in post-embryonic stages of development. It was greatest in the intestines of L1 larvae (Fig. 2A), and the lowest level of GFP expression was observed in the L4 and adult nematodes (Fig. 2, C and E, respectively).
FIGURE 1.
bcmo-1::GFP expression in untreated C. elegans embryos. A, ∼125 min post-fertilization; B, ∼200 min post-fertilization; C, ∼700 min post-fertilization. Embryos were obtained from nematodes not exposed to cadmium (magnification, 1200×).
FIGURE 2.
bcmo-1::GFP expression in post-embryonic C. elegans. Images of untreated C. elegans (A, C, and E), and nematodes exposed to 100 μm cadmium for 5 h (B, D, and F) and 24 h (G). L1 (A and B) and L4 (C and D) larvae are at 400× and 200× magnification, respectively. Adult C. elegans (E, F, and G) are composites of images acquired at 200× magnification. Arrowhead, indicates the location of a bcmo-1::GFP expressing embryo in a gravid adult nematode.
After a 5-h exposure to 100 μm cadmium, bcmo-1 intestinal transcription increased in all larval stages and adults (Fig. 2, B, D, and F). The highest level of induction of bcmo-1 transcription was observed in animals that were exposed to cadmium for 24 h (Fig. 2G), which was consistent with previously reported microarray results (28). No obvious changes in the embryonic bcmo-1 expression were observed following cadmium exposure. This may be due to lower cadmium exposures in embryos, which are protected by the egg shell.
The response of bcmo-1 expression to other metals was also examined by exposing adult bcmo-1::GFP transgenic nematodes to copper (CuSO4, 100 μm), nickel (NiSO4, 100 μm), zinc (ZnSO4, 100 μm), and silver (AgNO3, 100 μm) for 5 or 24 h (results not shown). Among all the metals tested, only zinc caused increased bcmo-1 expression compared with the untreated nematodes. The bcmo-1 expression following zinc exposure occurred exclusively in intestinal cells, but the levels of GFP expression were less than that caused by cadmium. In addition, bcmo-1 transcription was not observed in any non-intestinal cells following cadmium or other metal treatments. These results suggest that the response of bcmo-1 is not cadmium-specific.
Expression of bcmo-1 in Wild-type C. elegans and in Response to Cadmium
The level of bcmo-1 mRNA at different developmental stages and the level of response to cadmium exposure were also examined in wild-type nematodes by qRT-PCR. Without cadmium, the highest bcmo-1 mRNA level was observed in embryos and the level decreased during development, which is consistent with the GFP expression in bcmo-1::GFP transgenic nematodes (Fig. 3). After a 5-h exposure to cadmium, bcmo-1 mRNA levels significantly increased in all postembryonic developmental stages (Fig. 3).
The results using qRT-PCR agreed with those from the bcmo-1::GFP transgenic C. elegans. In the transgenic strain, however, L4 and adult nematodes had similar, low levels of constitutive GFP expression (Fig. 2, C and E), whereas the qRT-PCR results indicated a relatively higher mRNA level in adults compared with that in L4s (2.6-fold, Fig. 3). This was likely due to the presence of developing embryos in the adult gonad (Fig. 2E, arrow). This could also contribute to the observation that the highest induction of GFP expression by cadmium in transgenic nematodes occurs in adults (Fig. 2, F and G), whereas highest -fold change in bcmo-1 mRNA in wild-type nematodes was in L1s, according to qRT-PCR results (Fig. 3). The high constitutive levels of bcmo-1 mRNA in developing embryos reduces the magnitude of the fold change observed in adult nematodes, although the absolute level of bcmo-1 mRNA was greatest in cadmium-treated adults.
Cadmium Induces RA Signaling in Hepa 1–6 Cells
Because the evolutionarily conserved RA biosynthesis gene, BCMO, was induced by cadmium in C. elegans, the effect of cadmium on RA signaling was examined by measuring the RARE-mediated transcription following cadmium exposure in Hepa 1–6 cells. Exposures between 10 and 1000 nm of all-trans-RA resulted in a dose-dependent increase in the RARE-TK-Luc expression following 6- and 24-h exposures. A 6-h exposure to 100 or 1000 nm RA produced a 2.4-fold increase in RARE reporter gene activity (results not shown).
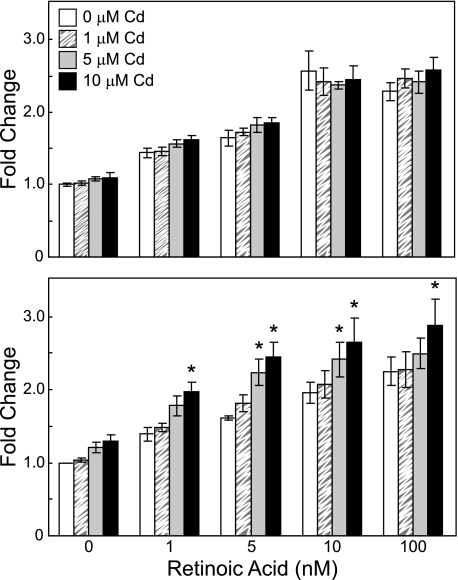
Co-exposure of cells to 1–10 μm cadmium for 6 h did not further increase RARE activity in RA-exposed cells (two-way ANOVA, p > 0.05, Fig. 4). Co-exposure to 5 or 10 μm cadmium for 24 h, however, resulted in a significant increase in RARE-mediated transcription, compared with that of RA exposure alone (two-way ANOVA, p < 10−7, Fig. 4). The increased RARE-mediated transcription caused by cadmium exposure was time- and dose-dependent, regardless of RA levels. This suggests that cadmium can induce the transcription of genes regulated via RA signaling.
FIGURE 4.
Effects of cadmium and all-trans-RA on RARE-TK-Luc reporter activity in Hepa 1–6 cells. Cells were co-transfected with RARE-TK-Luc and phRL-TK for 24 h and then exposed to 1 μm (striped box), 5 μm (gray box), or 10 μm (black box) cadmium and RA at the indicated concentrations for 6 h (upper panel) or 24 h (lower panel). Firefly luciferase activity was normalized to Renilla luciferase activity and then compared with the control (white box) (no cadmium and no RA treatments). Results are mean fold induction ± S.E., relative to control cells (n = 3). (*, significant difference from the non-cadmium-treated samples at the same RA level, p < 0.05.)
Cadmium Induces the Expression of Bcmo1 in Hepa 1–6 Cells
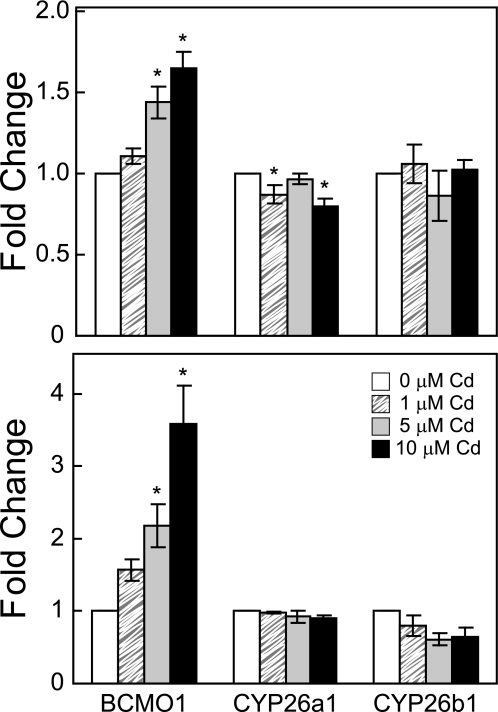
The effects of cadmium exposure on the steady-state mRNA levels of Bcmo1, which is involved in the synthesis of RA, and two genes that are involved in RA degradation, Cyp26a1 and Cyp26b1, were measured by qRT-PCR. The levels of Bcmo1 mRNA significantly increased following 6- and 24-h cadmium exposures (one-way ANOVA, p < 0.001 for 6 h, and p < 0.01 for 24-h exposure, Fig. 5). Bcmo1 levels increased by 1.4- and 1.7-fold following a 6-h exposure to 5 and 10 μm cadmium, respectively. A higher level of induction was observed following a 24-h exposure: 2.2-fold by 5 μm and 3.6-fold by 10 μm cadmium (Fig. 5), indicating the effect of cadmium on Bcmo1 transcription is both time- and dose-dependent.
FIGURE 5.
Effect of cadmium on Bcmo1, Cyp26a1, and Cyp26b1 mRNA expression. Cells were exposed to 1 μm (striped box), 5 μm (gray box), or 10 μm (black box) cadmium at the indicated concentrations for 6 h (upper panel) or 24 h (lower panel). Total RNA was extracted, and mRNA levels measured from untreated (white box) or cadmium-treated Hepa 1–6 cells. All measurements were normalized to the mRNA levels of glyceraldehyde-3-phosphate dehydrogenase before being compared with untreated cells. Results are displayed in mean -fold induction ± S.E., relative to untreated cells (n = 3). (*, significant difference from the control, p < 0.05.)
The expression of Cyp26a1 decreased 1.3-fold following a 6-h exposure to 10 μm cadmium (one-way ANOVA, p < 0.05, Fig. 5), and the expression of Cyp26b1 decreased 1.6-fold following 24-h exposures to 5 and 10 μm cadmium (one-way ANOVA, p = 0.11, Fig. 5). The simultaneous induction of Bcmo1 and inhibition of Cyp26a1 and Cyp26b1 by cadmium may cause increased RA synthesis and decreased RA degradation, which together could result in increased RA levels.
DISCUSSION
β,β-Carotene 15,15′-Monooxygenase Is an Evolutionarily Conserved Cadmium-responsive Gene
Through a comparative genomics approach, a conserved response to cadmium exposure in C. elegans and mouse cells that is mediated by BCMO was discovered. BCMO catalyzes the transformation of β-carotene to retinal, which can then be transformed into RA (37). BCMO is evolutionarily conserved throughout the animal kingdom (38). In mammals, BCMO is predominantly expressed in the intestine and liver, although enzymatic activity of BCMO has also been reported in lung, kidney, testis, and brain (39, 40). The expression pattern of bcmo-1::GFP in transgenic C. elegans indicates that bcmo-1 is also expressed in the intestine. In addition, its expression decreases as C. elegans develops and is minimal once the nematode reaches adulthood. RNA interference of bcmo-1 is associated with several developmental phenotypes, including slow growth, problems with postembryonic development, embryonic lethality, and abnormal morphology (41–43).
It has been reported that intestine-specific transcription in C. elegans is controlled by GATA-like motifs (TGATAA) located in the promoter regions (44). Sequence analysis of the bcmo-1 promoter identified a conserved GATA motif located −219 bp to −214 bp upstream of the predicted start codon. Following cadmium exposure, bcmo-1 expression remained intestine-specific, however, the expression levels significantly increased in all postembryonic and adult stages.
BCMO has been studied in many animal models because of its importance in the biosynthesis of RA (45–51). It has been reported that BCMO is essential for pattern formation and differentiation during zebrafish embryogenesis (52). However, little was known about the response of this gene to environmental stressors. In addition, there is a paucity of information on the relationship between BCMO expression and cadmium exposure. Microarray analysis of cadmium-exposed C. elegans indicated that BCMO is an early cadmium-responsive gene and is highly cadmium inducible (28). Bcmo1 was also significantly induced by cadmium following 6- and 24-h exposure in Hepa 1–6 cells. Because BCMO is highly conserved across many species, the transcriptional response to cadmium may also be conserved.
Effects of Cadmium on RA Signaling and Bcmo1 Transcription in Hepa 1–6 Cells
The effects of cadmium on RA signaling were examined at several RA concentrations using a RARE-driven reporter system. In most cases, the increase in normalized luciferase activity caused by cadmium was slight compared with RA exposure alone. The largest effect was observed when cells were treated with 5 μm cadmium and 5 nm RA for 24 h (Fig. 4). Cadmium caused a 1.5-fold further increase in the normalized luciferase activity (total fold induction of 2.4 in co-exposure versus 1.6-fold induction by exposure to RA alone). The overall effects of cadmium and RA on the normalized luciferase activity were additive rather than synergistic. However, RA-mediated transcriptional activity is not synonymous to the teratogenic effect, which may be caused by the abnormal activity of RA signaling. It is possible that small increases in the overall activity of RA signaling may cause larger downstream teratogenic effects during embryogenesis.
The transcriptional activity of RA signaling is mediated by ligand binding of cognate nuclear receptors, including RA receptors RAR and retinoid X receptor. It has been reported that cadmium does not change the binding property of RA for RAR or the RARE element (53). However, microarray data of metal-treated mouse liver indicates that cadmium could affect the expression of many RA-related genes, including Aldh1 and RAR, depending on the mouse strain and cadmium dose.3 Cadmium has also been shown to induce retinoid X receptor-γ in HeLa cells (54). Further examination of expressional change on these and other RA-related genes will help better understand the effect of cadmium on RA-mediated signal transduction pathways.
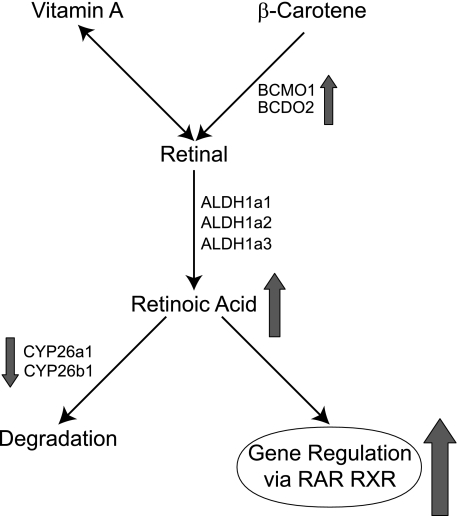
Lee et al. (27) reported that cadmium could act synergistically with RA in the formation of limb-bud malformations in mice. They hypothesized that cadmium and RA, as well as other teratogens that can cause forelimb ectrodactyly, may interfere with a final common pathway in digital development. Liao et al., further proposed that cadmium and RA may target different areas of the developing limb bud and a late convergence of the two chemical signaling pathways could result in a synergistic effect (23). Our results indicate that cadmium can induce the transcriptional activity mediated via RA signaling suggesting an early convergence of the two pathways. Moreover, qRT-PCR measurements of several RA metabolic genes in Hepa 1–6 cells indicate that RA synthesis may be induced while RA degradation may be inhibited by cadmium exposure. Similar gene expression changes were observed in the livers of cadmium-treated mice.4 These results suggest that cadmium may modify the steady-state levels of RA by regulating the expression of genes involved in RA metabolism. Based on these results, a molecular mechanism for cadmium-induced teratogenesis has been developed and is presented in Fig. 6. Specifically, RA levels are regulated by synthesis enzymes (Bcmo1, Bcdo2, Aldh1a1, Aldh1a2, and Aldh1a3) and catabolic enzymes (Cyp26a1 and Cyp26b1). Under normal situations, excessive RA will be degraded by catabolic enzymes, Cyp26a1 and Cyp26b1, which is controlled by a feedback mechanism, so that RA will be maintained at a steady-state level (55). Upon cadmium exposure, Bcmo1 expression is up-regulated while Cyp26a1 and Cyp26b1 are down-regulated, which could result in an elevation in RA levels. This subsequently leads to an increase in RA-signaling activity and changes in transcriptional activation, ultimately leading to abnormal development. Similar effects were observed in murine liver from cadmium-treated mice. Transcriptome analysis of liver mRNA from C57BL/6 mice exposed to cadmium reveals significant suppression of the steady-state mRNA levels for Cyp26a1 and Cyp26b1. In addition, there was a significant increase in the levels of Aldh1a1, RARγ, and Aldh1a2 mRNAs.4
FIGURE 6.
Mechanism for cadmium-induced teratogenicity. Retinoic acid is converted from β-carotene and vitamin A by catabolic enzymes (Bcmo1, Bcdo2, Aldh1a1, Aldh1a2, and Aldh1a3). Excessive retinoic acid can be degraded by P450s (Cyp26a1 and Cyp26b1). Upon cadmium exposure, Bcmo1 levels increase, while Cyp26a1 and Cyp26b1 levels decrease, which causes an increase in the level of RA (arrows). This leads to an increase in retinoid X receptor (RXR) and RAR-mediated transcription.
The mechanism by which cadmium alters the levels of expression of the genes involved in RA metabolism is unknown. It has been reported that UV irradiation, which induces oxidative stress, will increase BCMO activity (56). Cadmium also induces intracellular oxidative stress (57).
Cyp26a1 is an important enzyme that is responsible for RA degradation. In addition, the RA-inducible expression of Cyp26a1 is a feedback mechanism that controls RA levels (58). Cyp26a1 was inhibited by 1 and 10 μm cadmium after 6-h exposure (Fig. 5). This suggests a conflict between our model for the cadmium-mediated increase in RA levels (Fig. 6) and the observation that RA can induce Cyp26a1 expression. RA-independent expression of Cyp26a1 has been reported in mouse and zebrafish embryos, suggesting this gene is regulated by additional mechanisms (59, 60). In zebrafish hindbrain development, spatial expression of Cyp26a1 is activated by Fgf signaling and collaboration between RA and Fgf signaling in the anterior-posterior axis of zebrafish embryos is the key to forming the RA gradient system, which appears to be a common motif in pattern formation during vertebrate embryogenesis (61, 62). Although it is not known how cadmium inhibits Cyp26a1 transcription, it has been reported that cadmium can inhibit Fgf8 and Fgf4 expression in C57BL/6N mice during early limb development (23). A comparative study between C57BL/6N mouse strain and SWV strain also demonstrated that the differential strain response to cadmium-induced forelimb digital loss may be due to a polymorphic interference with Fgf signaling and others (22). These data suggest that Cyp26a1 transcription is regulated by a complex mechanism, which includes at least RA and Fgf signaling. It is possible that cadmium or a byproduct of cadmium exposure (e.g. reactive oxygen species, activation of other signal transduction pathways), interferes with multiple Cyp26a1 regulatory processes to ultimately inhibit transcription in the presence of RA.
The finding that cadmium interacts with RA signaling provides new insights in understanding the teratogenicity of cadmium. Although cadmium causes birth defects in a variety of animal models and has been associated with low birth weight in humans, the underlying molecular mechanisms remain largely unknown. The study in the mouse cell line, Hepa 1–6, suggests that cadmium may exert teratogenicity through regulating RA signaling. Because RA signaling is an essential regulator in embryonic development, cell differentiation, and apoptosis in the maintenance of adult organs, the proposed mechanism for cadmium teratogenesis may also provide insights into the understanding of other cadmium-induced diseases as well as cadmium-regulated gene expression.
Acknowledgments
We thank Dr. Terrance Kavanagh and his laboratory members at the University of Washington, Seattle, for the mouse liver transcriptome data.
This work was supported, in whole or in part, by National Institutes of Health Grant U19ES011375 and R01ES009949 (to J. H. F.) and by the Intramural Research Program of NIH and NIEHS (Grant Z01ES102045).
Y. Cui and J. H. Freedman, unpublished observation.
T. J. Kavanagh and J. H. Freedman, unpublished observation.
- RA
- retinoic acid
- RAR
- RA receptor
- RARE
- RA response element
- BCMO
- β,β-carotene 15,15′-monooxygenase
- GFP
- green fluorescent protein
- qRT
- quantitative reverse transcription
- ANOVA
- analysis of variance
- FgF
- fibroblast growth factor.
REFERENCES
- 1.Agency for Toxic Substances and Disease Registry ( 2007) CERCLA Priority List of Hazardous Substances, Atlanta, GA [Google Scholar]
- 2.Waalkes M. P., Coogan T. P., Barter R. A. (1992) Crit. Rev. Toxicol. 22, 175–201 [DOI] [PubMed] [Google Scholar]
- 3.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans ( 1993) Beryllium, Cadmium, Mercury, and Exposures in the Glass Manufacturing Industry, International Agency for Research on Cancer (IARC); distributed for the IARC by the Secretariat of the World Health Organization, Lyon, France [Google Scholar]
- 4.Jin T., Lu J., Nordberg M. (1998) Neurotoxicology 19, 529–535 [PubMed] [Google Scholar]
- 5.Thompson J., Bannigan J. (2001) Teratology 64, 87–97 [DOI] [PubMed] [Google Scholar]
- 6.Sunderman F. W., Jr., Plowman M. C., Hopfer S. M. (1992) IARC Sci. Publ. 249–256 [PubMed] [Google Scholar]
- 7.Holt D., Webb M. (1987) Arch. Toxicol. 59, 443–447 [DOI] [PubMed] [Google Scholar]
- 8.De S. K., Dey S. K., Andrews G. K. (1990) Toxicology 64, 89–104 [DOI] [PubMed] [Google Scholar]
- 9.Soukupová D., Dostál M. (1991) Funct. Dev. Morphol. 1, 3–9 [PubMed] [Google Scholar]
- 10.Messerle K., Webster W. S. (1982) Teratology 25, 61–70 [DOI] [PubMed] [Google Scholar]
- 11.Hovland D. N., Jr., Machado A. F., Scott W. J., Jr., Collins M. D. (1999) Teratology 60, 13–21 [DOI] [PubMed] [Google Scholar]
- 12.Padmanabhan R., Hameed M. S. (1990) Reprod. Toxicol. 4, 291–304 [DOI] [PubMed] [Google Scholar]
- 13.Milaire J. (1985) Teratology 32, 433–451 [DOI] [PubMed] [Google Scholar]
- 14.Layton W. M., Jr., Layton M. W. (1979) Teratology 19, 229–235 [DOI] [PubMed] [Google Scholar]
- 15.Hovland D. N., Jr., Cantor R. M., Lee G. S., Machado A. F., Collins M. D. (2000) Genomics 63, 193–201 [DOI] [PubMed] [Google Scholar]
- 16.Piasek M., Blanusa M., Kostial K., Laskey J. W. (2001) Reprod. Toxicol. 15, 673–681 [DOI] [PubMed] [Google Scholar]
- 17.Nishijo M., Nakagawa H., Honda R., Tanebe K., Saito S., Teranishi H., Tawara K. (2002) Occup. Environ. Med. 59, 394–396; discussion, 397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronco A. M., Arguello G., Muñoz L., Gras N., Llanos M. (2005) Biometals 18, 233–241 [DOI] [PubMed] [Google Scholar]
- 19.Mahalik M. P., Hitner H. W., Prozialeck W. C. (1995) Toxicol. Lett. 76, 195–202 [DOI] [PubMed] [Google Scholar]
- 20.Goyer R. A. (1991) Fundam. Appl. Toxicol. 16, 22–23 [DOI] [PubMed] [Google Scholar]
- 21.Levin A. A., Kilpper R. W., Miller R. K. (1987) Teratology 36, 163–170 [DOI] [PubMed] [Google Scholar]
- 22.Elsaid A. F., Délot E. C., Collins M. D. (2007) Mol. Genet. Metab. 92, 258–270 [DOI] [PubMed] [Google Scholar]
- 23.Liao X., Lee G. S., Shimizu H., Collins M. D. (2007) Toxicol. Appl. Pharmacol. 225, 47–60 [DOI] [PubMed] [Google Scholar]
- 24.Scott W. J., Jr., Schreiner C. M., Goetz J. A., Robbins D., Bell S. M. (2005) Reprod. Toxicol. 19, 479–485 [DOI] [PubMed] [Google Scholar]
- 25.Blomhoff R., Blomhoff H. K. (2006) J. Neurobiol. 66, 606–630 [DOI] [PubMed] [Google Scholar]
- 26.Ross S. A., McCaffery P. J., Drager U. C., De Luca L. M. (2000) Physiol. Rev. 80, 1021–1054 [DOI] [PubMed] [Google Scholar]
- 27.Lee G. S., Liao X., Cantor R. M., Collins M. D. (2006) Birth Defects Res. A Clin. Mol. Teratol. 76, 19–28 [DOI] [PubMed] [Google Scholar]
- 28.Cui Y., McBride S. J., Boyd W. A., Alper S., Freedman J. H. (2007) Genome Biol. 8, R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobert O. (2002) BioTechniques 32, 728–730 [DOI] [PubMed] [Google Scholar]
- 30.Anderson G. L., Boyd W. A., Williams P. L. (2001) Environ. Toxicol. Chem. 20, 833–838 [PubMed] [Google Scholar]
- 31.Evans T. C. ed. Transformation and microinjection ( April6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.108.1, http://www.wormbook.org [Google Scholar]
- 32.Sulston J., Hodgkin J. (1988) in The Nematode Caenorhabditis elegans ( Wood W. B. ed) pp. 587–606, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33.Stiernagle T.Maintenance of C. elegans ( February11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.101.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umesono K., Murakami K. K., Thompson C. C., Evans R. M. (1991) Cell 65, 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsey F. L., Schafer D. W. (2002) The Statistical Sleuth: A Course in Methods of Data Analysis, 2nd Ed., pp. 162–163, Duxbury/Thomson Learning, Pacific Grove, CA [Google Scholar]
- 36.Sulston J. (1988) in The Nematode Caenorhabditis elegans ( Wood W. B. ed.) pp. 123–155, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37.Kiefer C., Hessel S., Lampert J. M., Vogt K., Lederer M. O., Breithaupt D. E., von Lintig J. (2001) J. Biol. Chem. 276, 14110–14116 [DOI] [PubMed] [Google Scholar]
- 38.Wyss A. (2004) J. Nutr. 134, 246S–250S [DOI] [PubMed] [Google Scholar]
- 39.Franssen-van Hal N. L., Bunschoten J. E., Venema D. P., Hollman P. C., Riss G., Keijer J. (2005) Arch. Biochem. Biophys. 439, 32–41 [DOI] [PubMed] [Google Scholar]
- 40.During A., Nagao A., Hoshino C., Terao J. (1996) Anal. Biochem. 241, 199–205 [DOI] [PubMed] [Google Scholar]
- 41.Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Welchman D. P., Zipperlen P., Ahringer J. (2003) Nature 421, 231–237 [DOI] [PubMed] [Google Scholar]
- 42.Sönnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A. M., Artelt J., Bettencourt P., Cassin E., Hewitson M., Holz C., Khan M., Lazik S., Martin C., Nitzsche B., Ruer M., Stamford J., Winzi M., Heinkel R., Röoder M., Finell J., Häntsch H., Jones S. J., Jones M., Piano F., Gunsalus K. C., Oegema K., Gönczy P., Coulson A., Hyman A. A., Echeverri C. J. (2005) Nature 434, 462–469 [DOI] [PubMed] [Google Scholar]
- 43.Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S., Hirozane-Kishikawa T., Vandenhaute J., Orkin S. H., Hill D. E., van den Heuvel S., Vidal M. (2004) Genome Res 14, 2162–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGhee J. D., Sleumer M. C., Bilenky M., Wong K., McKay S. J., Goszczynski B., Tian H., Krich N. D., Khattra J., Holt R. A., Baillie D. L., Kohara Y., Marra M. A., Jones S. J., Moerman D. G., Robertson A. G. (2007) Dev. Biol. 302, 627–645 [DOI] [PubMed] [Google Scholar]
- 45.Takitani K., Zhu C. L., Inoue A., Tamai H. (2006) Eur. J. Nutr. 45, 320–326 [DOI] [PubMed] [Google Scholar]
- 46.Poliakov E., Gentleman S., Cunningham F. X., Jr., Miller-Ihli N. J., Redmond T. M. (2005) J. Biol. Chem. 280, 29217–29223 [DOI] [PubMed] [Google Scholar]
- 47.Paik J., During A., Harrison E. H., Mendelsohn C. L., Lai K., Blaner W. S. (2001) J. Biol. Chem. 276, 32160–32168 [DOI] [PubMed] [Google Scholar]
- 48.Gong X., Tsai S. W., Yan B., Rubin L. P. (2006) BMC Mol. Biol. 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.During A., Fields M., Lewis C. G., Smith J. C. (2000) Br. J. Nutr. 84, 117–124 [PubMed] [Google Scholar]
- 50.During A., Albaugh G., Smith J. C., Jr. (1998) Biochem. Biophys. Res. Commun. 249, 467–474 [DOI] [PubMed] [Google Scholar]
- 51.Boulanger A., McLemore P., Copeland N. G., Gilbert D. J., Jenkins N. A., Yu S. S., Gentleman S., Redmond T. M. (2003) FASEB J. 17, 1304–1306 [DOI] [PubMed] [Google Scholar]
- 52.Lampert J. M., Holzschuh J., Hessel S., Driever W., Vogt K., von Lintig J. (2003) Development 130, 2173–2186 [DOI] [PubMed] [Google Scholar]
- 53.Mostböck S., Fickova M., Macejová D., Baranová M., Kotyzová D., Micková V., Eybl V., Thalhamer J., Brtko J. (2002) Gen. Physiol. Biophys. 21, 443–456 [PubMed] [Google Scholar]
- 54.Koizumi S., Yamada H. (2003) J. Occup. Health 45, 331–334 [DOI] [PubMed] [Google Scholar]
- 55.Perlmann T. (2002) Nat. Genet. 31, 7–8 [DOI] [PubMed] [Google Scholar]
- 56.Takeda A., Morinobu T., Takitani K., Kimura M., Tamai H. (2003) J. Nutr. Sci. Vitaminol. 49, 69–72 [DOI] [PubMed] [Google Scholar]
- 57.Ercal N., Gurer-Orhan H., Aykin-Burns N. (2001) Curr. Top. Med. Chem. 1, 529–539 [DOI] [PubMed] [Google Scholar]
- 58.Reijntjes S., Blentic A., Gale E., Maden M. (2005) Dev. Biol. 285, 224–237 [DOI] [PubMed] [Google Scholar]
- 59.Dobbs-McAuliffe B., Zhao Q., Linney E. (2004) Mech. Dev. 121, 339–350 [DOI] [PubMed] [Google Scholar]
- 60.Sirbu I. O., Gresh L., Barra J., Duester G. (2005) Development 132, 2611–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White R. J., Schilling T. F. (2008) Dev. Dyn. 237, 2775–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White R. J., Nie Q., Lander A. D., Schilling T. F. (2007) PLoS Biol 5, e304. [DOI] [PMC free article] [PubMed] [Google Scholar]