Abstract
STIM1 and ORAI1, the two limiting components in the Ca2+ release-activated Ca2+ (CRAC) signaling cascade, have been reported to interact upon store depletion, culminating in CRAC current activation. We have recently identified a modulatory domain between amino acids 474 and 485 in the cytosolic part of STIM1 that comprises 7 negatively charged residues. A STIM1 C-terminal fragment lacking this domain exhibits enhanced interaction with ORAI1 and 2–3-fold higher ORAI1/CRAC current densities. Here we focused on the role of this CRAC modulatory domain (CMD) in the fast inactivation of ORAI1/CRAC channels, utilizing the whole-cell patch clamp technique. STIM1 mutants either with C-terminal deletions including CMD or with 7 alanines replacing the negative amino acids within CMD gave rise to ORAI1 currents that displayed significantly reduced or even abolished inactivation when compared with STIM1 mutants with preserved CMD. Consistent results were obtained with cytosolic C-terminal fragments of STIM1, both in ORAI1-expressing HEK 293 cells and in RBL-2H3 mast cells containing endogenous CRAC channels. Inactivation of the latter, however, was much more pronounced than that of ORAI1. The extent of inactivation of ORAI3 channels, which is also considerably more prominent than that of ORAI1, was also substantially reduced by co-expression of STIM1 constructs missing CMD. Regarding the dependence of inactivation on Ca2+, a decrease in intracellular Ca2+ chelator concentrations promoted ORAI1 current fast inactivation, whereas Ba2+ substitution for extracellular Ca2+ completely abrogated it. In summary, CMD within the STIM1 cytosolic part provides a negative feedback signal to Ca2+ entry by triggering fast Ca2+-dependent inactivation of ORAI/CRAC channels.
The Ca2+ release-activated Ca2+ (CRAC)5 channel is one of the best characterized store-operated entry pathways (1–7). Substantial efforts have led to identification of two key components of the CRAC channel machinery: the stromal interaction molecule 1 (STIM1), which is located in the endoplasmic reticulum and acts as a Ca2+ sensor (8–10), and ORAI1/CRACM1, the pore-forming subunit of the CRAC channel (11–13). Besides ORAI1, two further homologues named ORAI2 and ORAI3 belong to the ORAI channel family (12, 14).
STIM1 senses endoplasmic reticulum store depletion primarily by its luminal EF-hand in its N terminus (8, 15), redistributes close to the plasma membrane, where it forms puncta-like structures, and co-clusters with ORAI1, leading to inward Ca2+ currents (12, 16–19). The STIM1 C terminus, located in the cytosol, contains two coiled-coil regions overlapping with an ezrin-radixin-moesin (ERM)-like domain followed by a serine/proline- and a lysine-rich region (2, 8, 20–22). Three recent studies have described the essential ORAI-activating region within the ERM domain, termed SOAR (Stim ORAI-activating region) (23), OASF (ORAI-activating small fragment) (24), and CAD (CRAC-activating domain) (25), including the second coiled coil domain and the following ∼55 amino acids. We and others have provided evidence that store depletion leads to a dynamic coupling of STIM1 to ORAI1 (26–28) that is mediated by a direct interaction of the STIM1 C terminus with ORAI1 C terminus probably involving the putative coiled-coil domain in the latter (27).
Furthermore, different groups have proven that the C terminus of STIM1 is sufficient to activate CRAC as well as ORAI1 channels independent of store depletion (22–25, 27, 29). We have identified that OASF-(233–474) or shorter fragments exhibit further enhanced coupling to ORAI1 resulting in 3-fold increased constitutive Ca2+ currents. A STIM1 fragment containing an additional cluster of anionic amino acids C-terminal to position 474 displays weaker interaction with ORAI1 as well as reduced Ca2+ current comparable with that mediated by wild-type STIM1 C terminus. Hence, we have suggested that these 11 amino acids (474–485) act in a modulatory manner onto ORAI1; however, their detailed mechanistic impact within the STIM1/ORAI1 signaling machinery has remained so far unclear.
In this study, we focused on the impact of this negative cluster on fast inactivation of STIM1-mediated ORAI Ca2+ currents. Lis et al. (30) have shown that all three ORAI homologues display distinct inactivation profiles, where ORAI2 and ORAI3 show a much more pronounced fast inactivation than ORAI1. Moreover, it has been reported (31) that different expression levels of STIM1 to ORAI1 affect the properties of CRAC current inactivation. Yamashita et al. (32) have demonstrated a linkage between the selectivity filter of ORAI1 and its Ca2+-dependent fast inactivation. Here we provide evidence that a cluster of acidic residues within the C terminus of STIM1 is involved in the fast inactivation of ORAI1 and further promotes that of ORAI3 and native CRAC currents.
MATERIALS AND METHODS
Molecular Cloning and Mutagenesis
Human ORAI1 (ORAI1; GenBankTM accession number NM_032790) was kindly provided by the A. Rao laboratory (Harvard Medical School, Cambridge, MA). Human ORAI3 (ORAI3; accession number NM_152288.1) were courtesy of the Lutz Birnbaumer laboratory (NIEHS, National Institutes of Health, Research Triangle Park, NC). N-terminally tagged ORAI1 constructs were cloned via SalI and SmaI restriction sites of pECFP-C1 and pEYFP-C1 expression vectors (Clontech). For N-terminally tagged ORAI3 constructs, the restriction sites BamHI and XbaI were used.
Human STIM1 (STIM1; accession number NM_003156) and hSTIM1 D76A N-terminally ECFP- and EYFP-tagged were kindly provided by the T. Meyer laboratory, Stanford University, Palo Alto, CA. pEYFP-STIM1 was used as template for the generation of pEYFP-STIM1 aa 1–450, aa 1–474, aa 1–485, and aa 1–535 by introducing a stop codon at amino acid positions 451, 475, 486, and 536, respectively, using the QuikChange XL site-directed mutagenesis kit (Stratagene). pEYFP-STIM1 7xA was generated via PCR by introducing codons for Ala (GCT or GCC) at amino acid positions 475, 476, 478, 479, 481, 482, and 483 consequently substituting the amino acids 475–483 from DDVDDMDEE to AAVAAMAAA.
The STIM1 C terminus (aa 233–685) was cloned into the T/A site of pcDNA3.1V5 His TOPO by PCR and subcloned into pECFP-C1 and pEYFP-C1 via their internal restriction sites KpnI and XbaI. pECFP-C1 and pEYFP-C1 STIM1 C terminus was used as template for the generation of the STIM1 fragments by introducing a stop codon at position 451 (aa 233–450), position 475 (aa 233–474), position 486 (aa 233–485), and position 536 (aa 233–535) using the QuikChange XL site-directed mutagenesis kit (Stratagene). The STIM1 C terminus fragments aa 333–474 and aa 344–449 were amplified by PCR and directly cloned into pECFP-C1 and pEYFP-C1 via KpnI and XbaI. pECFP-C1 and pEYFP-C1 STIM1-(233–685) and -(233–485) served as templates for the generation of pECFP-C1 and pEYFP-C1 STIM1-(233–685) 7xA and -(233–485) 7xA as described for hSTIM1 7xA. pECFP-C1 and pEYFP-C1 STIM1-(233–685) 3xA and STIM1-(233–685) 5xA were generated via PCR by introducing codons for Ala (GCT or GCC) at amino acid positions 481, 482, and 483 and positions 478, 479, 481, and 482, 483, respectively. The integrity of all resulting clones was confirmed by sequence analysis.
Electrophysiology
Transfection of HEK293 cells and electrophysiological recordings in the whole-cell configuration were carried out according to (33). In brief, after Orai/CRAC currents were fully activated, and rectangular voltage pulses from a holding potential of 0 mV to −90, −70, −50, and −30 mV of 2-s duration were applied. The internal pipette solution contained (in mm) 3.5 MgCl2, 145 cesium methane sulfonate, 8 NaCl, 10 HEPES, 20 EGTA (or 0.1 EGTA), pH 7.2. Extracellular solution consisted of (in mm) 145 NaCl, 5 CsCl, 1 MgCl2, 10 HEPES, 10 glucose, 10 CaCl2 (or 10 BaCl2), pH 7.4. Currents were leak-corrected. At the beginning of each rectangular pulse, 2.5 ms were blanked out so that residual capacitative artifacts could be eliminated according to Ref. 30. Overlay of average current traces was performed by their normalization to 1 at 2.6 ms. In some experiments where Orai1:STIM1 were varied at a plasmid ratio of 1:1 and 1:4, normalized CFP-Orai1:YFP-STIM1 (1–474) protein ratios were determined from fluorescence microscopy measurements on single cells as 3.00 ± 0.19 (n = 32) and 1.26 ± 0.15 (n = 33), respectively, consistent with Ref. 34.
Statistics
Mean ± S.E. values were shown throughout this report with significance determined by the two-tailed Mann-Whitney test.
RESULTS AND DISCUSSION
In our previous report (24), we have identified a modulatory domain for ORAI1 activation within the STIM1 C terminus. This domain comprises a stretch of 11 amino acids ranging from aa 474 to 485. Closer inspection of this region, to be termed CRAC modulatory domain (CMD), reveals a cluster of 7 negatively charged amino acids (2 glutamates, 5 aspartates), which are highly conserved among all STIM1 homologues of vertebrates; however, they are only partially retained in Drosophila STIM and missing in Caenorhabditis elegans STIM1 (supplemental Fig. S1).
In the following, we investigated whether this anionic cluster affects gating of ORAI1/CRAC channels, focusing on fast inactivation of ORAI1 channels, which represents one of their biophysical hallmarks (30, 32). Fast inactivation was examined during voltage steps from whole-cell recordings of ORAI1 Ca2+ currents activated by STIM1 or various STIM1 deletion mutants that were applied from a holding potential of 0 mV to −90, −70, −50, and −30 mV.
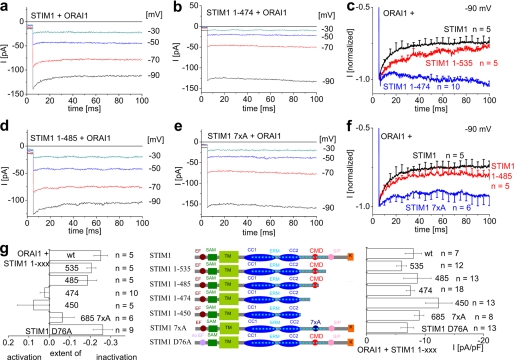
Fast Inactivation of ORAI1 Currents Is Preserved as Long as the Anionic Amino Acids Are Present within STIM1 (aa 475–483)
The STIM1-induced ORAI1 Ca2+ currents exhibited fast inactivation at voltage steps to −90 mV of about 25% within the first 100 ms (Fig. 1, a, c, and g), which was followed by a slight reactivation wave (data not shown) consistent with Lis et al. (30). Elucidation of the domain(s) mediating this inactivation was carried out by STIM1 mutants with increasing deletions from the C-terminal side, i.e. STIM1-(1–535), STIM1-(1–485), STIM1-(1–474), and STIM1-(1–450) (Fig. 1g, middle panel). All of these STIM1 deletion mutants induced comparable maximum ORAI1 Ca2+ current densities (Fig. 1g, right panel). STIM1-(1–535) (Fig. 1, c and g) as well as 1–485 (Fig. 1, d, f, and g) produced a similar extent of fast inactivation as full-length STIM1, whereas STIM1-(1–474) (Fig. 1, b, c, and g) and STIM1-(1–450) (Fig. 1g) displayed substantially reduced or even abolished inactivation. Recently, Scrimgeour et al. (31) have reported that a decrease of fast inactivation occurs when STIM1 is expressed at lower amounts than ORAI1. Nevertheless, increasing the plasmid ratio of STIM1-(1–474) to ORAI1 from the usual 1:1 to 4:1 ratio in our system did not increase the extent of inactivation (data not shown). Thus, the presence of CMD, i.e. the cluster of negative amino acids within region 474–485 of the STIM1 C terminus appears essential for fast inactivation of ORAI1 currents.
FIGURE 1.
Fast inactivation of ORAI1 currents is preserved as long as CMD is present in STIM1. a, b, d, and e, fast inactivation of STIM1- (a), STIM1-(1–474)- (b), STIM1-(1–485)- (d), and STIM1 7xA-mediated (e) ORAI1 Ca2+ currents upon voltage steps from a holding potential of 0 mV to −30, −50, −70, and −90 mV. c and f, mean fast inactivation of normalized current traces of STIM1, STIM1-(1–535), STIM1-(1–485), STIM1-(1–474), and STIM1 7xA upon voltage steps to −90 mV. g, scheme of various STIM1 deletion mutants (STIM1, STIM1-(1–535), STIM1-(1–485), STIM1-(1–474), STIM1-(1–450)) as well as STIM1 7xA and EF-hand mutant STIM1 D76A in comparison with the extent of inactivation of ORAI1 currents at t = 90 ms (p < 0.05 for STIM1 when compared with STIM1-(1–474), STIM1-(1–450), and STIM1 7xA) and maximum current densities. wt, wild type; SAM, sterile α motif; TM, transmembrane; pA, picoampere; pF, picofarad.
In a complementary approach, we mutated all 7 anionic residues within CMD to neutral alanines (STIM1 7xA) to support the concept of their requirement for transmitting fast inactivation onto ORAI1. STIM1 7xA-induced ORAI1 Ca2+ currents exhibited comparable maximum current densities as wild-type STIM1 (Fig. 1g). In contrast, fast inactivation was almost completely abolished in STIM1 7xA-induced Ca2+ currents (Fig. 1, e and g) similar to STIM1-(1–474) with CMD deleted (compare with Fig. 1, c and g). In addition, we investigated fast inactivation of STIM1 D76A EF-hand mutant. As reported previously (8, 9), this mutant induced constitutive ORAI1 currents (Fig. 1g, right panel) independent of store depletion, and these STIM1 D76A-induced constitutive ORAI1 Ca2+ currents inactivated similarly as those activated by wild-type STIM1 (Fig. 1g, left panel), suggesting that fast inactivation occurred independent of store depletion.
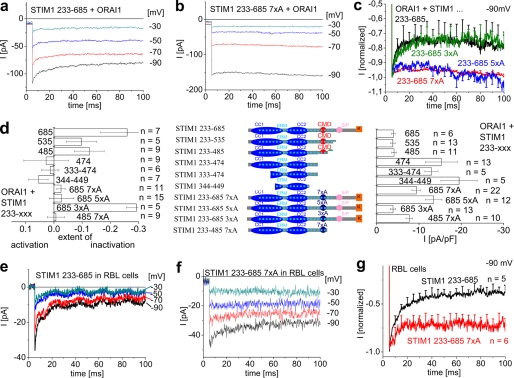
STIM1 Cytosolic Fragments Convey Fast Inactivation of Constitutive ORAI1 Currents as Long as the Anionic Amino Acids Are Present within CMD
In extension, we carried out comparable studies of fast inactivation with the STIM1 C terminus as well as its shorter cytosolic fragments (STIM1-(233–535), -(233–485), -(233–474), (333–474), (344–449); Fig. 2d, middle panel) that constitutively activated ORAI1 currents without store depletion. As we have recently reported (24), all fragments stimulated ORAI1 currents, with those including CMD or beyond displaying smaller currents, whereas shorter fragments without CMD (233–474 and all smaller ones) induced maximum ORAI1 Ca2+ currents (Fig. 2d, right panel). Fast inactivation of ORAI1 currents evoked by the STIM1 C terminus (233–685) or its shorter fragments was comparable with that of full-length STIM1 (Fig. 2, a, c, and d, left panel) as long as CMD was present (Fig. 2d, left panel). Deletion of CMD in fragments like OASF-(233–474) (24) or even shorter ones (333–474, 344–449) (23, 25) stimulated ORAI1 currents with abolished fast inactivation or even slight reactivation (Fig. 2d, left panel).
FIGURE 2.
Fast inactivation of ORAI1 currents is diminished after deletion of CMD in cytosolic STIM1 C-terminal fragments HEK 293 as well as RBL mast cells. a and b, fast inactivation of STIM1-(233–685)- (a) and STIM1-(233–685) 7xA-mediated (b) ORAI1 Ca2+ currents upon voltage steps from a holding potential of 0 mV to −30, −50, −70, and −90 mV. c, mean fast inactivation of normalized current traces of STIM1-(233–685), -(233–685) 7xA (D/E → A, aa 475, 476, 478, 479, 481–483), -(233–685) 5xA (D/E → A, aa 478, 479, 481–483), and -(233–685) 3xA (D/E → A, aa 481–483) upon voltage steps to −90 mV. d, scheme of various STIM1 C terminus deletion mutants (233–685, 233–535, 233–485, 233–474), STIM1-(233–685) 7xA, 5xA, 3xA, and STIM1-(233–485) 7xA in comparison with the extent of inactivation of ORAI1 currents t = 90 ms (p < 0.05 for STIM1-(233–685) when compared with STIM1-(233–474), STIM1-(233–450), STIM1-(233–685) 7xA, STIM1-(233–485) 7xA), and maximum current densities). ERM, ezrin-radixin-moesin. pA, picoampere; pF, picofarad. e and f, fast inactivation of STIM1- (e) and STIM1 7xA-mediated (f) RBL CRAC currents upon voltage steps from a holding potential of 0 mV to −30, −50, −70, or −90 mV. g, mean fast inactivation of normalized current traces of e and f upon voltage steps to −90 mV (t = 90 ms, p < 0.05).
In an analogous approach, as with full-length STIM1, the 7 negatively charged residues within CMD were mutated to alanines (7xA) in the STIM1 C terminus as well as STIM1-(233–485) fragments. Both 7xA STIM1 C-terminal fragments induced ORAI1 currents with an almost abolished inactivation similar to STIM1-(233–474) (Fig. 2, c and d, left panel) in contrast to their non-mutated isoforms. Accordingly, maximum ORAI1 activity evoked by these 7xA STIM1 C-terminal fragments reached 2-fold higher levels of current densities (Fig. 2d, right panel).
We additionally examined whether partial mutation of these negative residues would still lead to an inhibition of fast inactivation. Mutation of 5 residues (D478A,D479A,D481A,E482A, E483A) to alanines (5xA) was still sufficient to inhibit inactivation (Fig. 2, c and d), similar to the 7xA STIM1 C terminus, whereas mutation of only 3 aa (D481A,D482A,D483A; 3xA) led to an inactivation profile similar to wild type (Fig. 2, c and d). This suggested that the remaining 4 negative residues are sufficient to accomplish rapid inactivation. Hence, the cluster of negative residues within CMD essentially contributes to fast inactivation of ORAI1 Ca2+ currents either activated constitutively or activated by store depletion.
Fast Inactivation of Endogenous CRAC Currents Is Reduced by STIM1 Fragments Lacking CMD
The CMD region 474–485 within the STIM1 C terminus was further investigated for its impact onto fast inactivation of native CRAC currents of RBL-2H3 mast cells (3). We compared the extent of inactivation of Ca2+ currents from RBL cells expressing complete STIM1 C terminus, STIM1-(333–474), or the STIM1 C terminus 7xA. All three constructs constitutively activated endogenous CRAC currents in RBL cells, whereas those missing CMD stimulated Ca2+ currents to an about 2-fold higher level than the STIM1 C terminus (data not shown) as recently reported (24). STIM1 C terminus-induced CRAC currents displayed prominent fast inactivation by about 60% (Fig. 2, e and g), in line with several recent reports (3, 4, 35). This inactivation was substantially reduced when CRAC currents were stimulated by 7xA the STIM1 C terminus mutant (Fig. 2, f and g) or a shorter fragment (333–474) lacking CMD (data not shown). Hence, CMD indeed contributes to fast inactivation of both expressed ORAI1 as well as native CRAC channels. It is of note that inactivation of native CRAC is much more pronounced than that observed for ORAI1 currents in HEK 293 cells (compare Fig. 2, a and e). Hence, additional factors apparently contribute to inactivation of RBL CRAC currents. ORAI2 or ORAI3 proteins when expressed together with STIM1 in HEK293 cells indeed exhibit a stronger, fast inactivation than ORAI1 (30). Thus, heteromeric assemblies with ORAI1 possibly increasing the extent of inactivation are theoretically conceivable.
Considering this hypothesis, we investigated whether fast inactivation of ORAI3 channels would also be affected by STIM1 or its fragments when CMD is deleted. Although ORAI1 currents inactivated by about 25% within the first 100 ms of a voltage step to −90 mV, ORAI3 exhibited much stronger inactivation to 50% (supplemental Fig. S2, a and d) when activated via STIM1. Stimulation of ORAI3 with either STIM1-(1–474) (supplemental Fig. S2, b and d) or a shorter cytosolic fragment, both lacking CMD, similarly reduced inactivation of ORAI3 currents; however, reduction was not as complete as with ORAI1. Hence, ORAI3 channels may possess structures missing in ORAI1 that further promote fast inactivation in addition to CMD within STIM1.
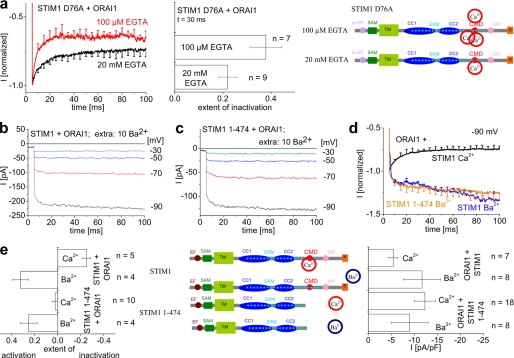
Fast Inactivation of ORAI1 Channels Occurs in a Ca2+-dependent Manner
In an attempt to establish the concept of ORAI current inactivation as a negative feedback mechanism to Ca2+ entry, we examined whether inactivation occurred in a Ca2+-dependent manner. We initially compared distinct intracellular capacities of Ca2+ buffers, either 20 mm EGTA or 100 μm EGTA. The latter buffer, mimicking physiological conditions (36, 37), led to a significant increase in the rate as well as extent of inactivation when compared with 20 mm EGTA (Fig. 3a). Moreover, we employed Ba2+ in the extracellular solution substituted for Ca2+ (together with 20 mm EGTA in the intracellular solution) to potentially eliminate Ca2+-dependent inactivation (32, 38) of STIM1-activated ORAI1 currents. The permeability of Ba2+ through ORAI1 channels is so far controversial as Yamashita et al. (32) as well as Scrimgeour et al. (34) have observed ORAI1 Ba2+ permeation in contrast to another recent study that has reported Ba2+ impermeability of ORAI1 (39). We were able to record Ba2+ currents through STIM1-evoked ORAI1 channels (Fig. 3, b, d, and e) that completely lacked fast inactivation in line with Ref. 31. Identical current traces were recorded from ORAI1 channels activated by the STIM1-(1–474) fragment lacking CMD (Fig. 3, c, d, and e). It is of note, however, that particularly the fast inactivation within the first ∼15 ms following repolarization was fully eliminated with Ba2+ but still slightly visible with Ca2+ and STIM1-(1–474) lacking CMD (Fig. 1c). This might suggest additional domains either in STIM1-(1–474) or in ORAI1 (32) contributing to that very fast inactivation.
FIGURE 3.
Fast inactivation of ORAI1 channels occurs in a Ca2+-dependent manner. a, fast inactivation profiles as well as block diagrams for t = 90 ms of STIM1 D76A-mediated constitutive ORAI1 currents in the presence of 20 mm EGTA in comparison with 100 μm EGTA intracellular solution (t = 30 ms, p < 0.05). The scheme on the right side represents STIM1 D76A in the presence of the two distinct Ca2+ buffering conditions. SAM, sterile α motif; TM, transmembrane. b and c, activation of STIM1- (b) and STIM1-(1–474)-mediated (c) ORAI1 Ba2+ currents upon voltage steps from a holding potential of 0 mV to −30, −50, −70, and −90 mV. d, mean activation of STIM1- and STIM1-(1–474)-mediated ORAI1 Ba2+ current in comparison with fast inactivation of STIM1- and STIM1-(1–474)-mediated ORAI1 Ca2+ currents. e, scheme of STIM1 deletion mutants in the presence of Ca2+ or Ba2+ in comparison with the extent of inactivation of ORAI1 currents t = 90 ms (p < 0.05 for STIM1 (Ca2+), when compared with STIM1-(1–474) (Ca2+), STIM1 (Ba2+), STIM1-(1–474) (Ba2+), and maximum current densities. pA, picoamperes; pF, picofarads.
Our results indicated that fast inactivation occurred in a Ca2+-dependent manner because it was enhanced/diminished upon weak/strong intracellular Ca2+ binding efficiency and eliminated in the presence of extracellular Ba2+. Thus, it is tempting to speculate that fast inactivation of ORAI channels involves electrostatic interactions of CMD that are specific for Ca2+ over Ba2+. Although the negative amino acids within CMD do not represent a canonical EF-hand motif (40), 7 acidic residues within a 9-amino-acid stretch may be capable of binding Ca2+. Thus, STIM1 that interacts with ORAI channels upon store depletion transmits fast inactivation onto ORAI channels via a Ca2+-induced conformational change involving CMD within the STIM1 C terminus. It remains to be determined whether this Ca2+-dependent process occurs via direct binding of Ca2+ to CMD or based on our finding that even 4 negative residues accomplish fast inactivation by the interplay with a third component, both requiring Ca2+ affinities in the μm range.
In conclusion, we identified the negatively charged amino acids within CMD of STIM1 as indispensable for fast inactivation of ORAI1 channels. ORAI3 and native CRAC of RBL mast cells, unlike ORAI1, still display inactivation when stimulated by STIM1 lacking CMD, which is further increased by the presence of CMD. Thus, CMD represents a common motif for all ORAI channels contributing to a general negative feedback mechanism controlling Ca2+ entry. The relevant structures that mediate inactivation of ORAI3 channels independent of CMD within STIM1 remain to be determined.
Acknowledgments
We thank S. Buchegger and B. Kenda for excellent technical assistance.
This work was supported by the Austrian Science Foundation (FWF) Project P19820 (to K. G.) and Project P21118 as well as Subproject 11 within W1201 (to C. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
This article was selected as a Paper of the Week.
- CRAC
- Ca2+ release-activated Ca2+
- CMD
- CRAC modulatory domain
- STIM
- stromal interaction molecule
- OASF
- ORAI-activating small fragment
- aa
- amino acids
- RBL
- rat basophilic leukemia.
REFERENCES
- 1.Berridge M. J., Lipp P., Bootman M. D. (2000) Nat. Rev. Mol. Cell Biol. 1, 11–21 [DOI] [PubMed] [Google Scholar]
- 2.Hoth M., Penner R. (1992) Nature 355, 353–356 [DOI] [PubMed] [Google Scholar]
- 3.Zweifach A., Lewis R. S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 6295–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parekh A. B., Putney J. W., Jr. (2005) Physiol. Rev. 85, 757–810 [DOI] [PubMed] [Google Scholar]
- 5.Lewis R. S. (2001) Annu. Rev. Immunol. 19, 497–521 [DOI] [PubMed] [Google Scholar]
- 6.Prakriya M., Lewis R. S. (2003) Cell Calcium 33, 311–321 [DOI] [PubMed] [Google Scholar]
- 7.Berridge M. J., Bootman M. D., Roderick H. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 517–529 [DOI] [PubMed] [Google Scholar]
- 8.Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spassova M. A., Soboloff J., He L. P., Xu W., Dziadek M. A., Gill D. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4040–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S. H., Tanasa B., Hogan P. G., Lewis R. S., Daly M., Rao A. (2006) Nature 441, 179–185 [DOI] [PubMed] [Google Scholar]
- 12.Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. (2006) Science 312, 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeromin A. V., Zhang S. L., Jiang W., Yu Y., Safrina O., Cahalan M. D. (2006) Nature 443, 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gwack Y., Srikanth S., Feske S., Cruz-Guilloty F., Oh-hora M., Neems D. S., Hogan P. G., Rao A. (2007) J. Biol. Chem. 282, 16232–16243 [DOI] [PubMed] [Google Scholar]
- 15.Zheng L., Stathopulos P. B., Li G. Y., Ikura M. (2008) Biochem. Biophys. Res. Commun. 369, 240–246 [DOI] [PubMed] [Google Scholar]
- 16.Luik R. M., Wu M. M., Buchanan J., Lewis R. S. (2006) J. Cell Biol. 174, 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu M. M., Buchanan J., Luik R. M., Lewis R. S. (2006) J. Cell Biol. 174, 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercer J. C., Dehaven W. I., Smyth J. T., Wedel B., Boyles R. R., Bird G. S., Putney J. W., Jr. (2006) J. Biol. Chem. 281, 24979–24990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frischauf I., Schindl R., Derler I., Bergsmann J., Fahrner M., Romanin C. (2008) Channels 2, 261–268 [DOI] [PubMed] [Google Scholar]
- 20.Baba Y., Hayashi K., Fujii Y., Mizushima A., Watarai H., Wakamori M., Numaga T., Mori Y., Iino M., Hikida M., Kurosaki T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16704–16709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smyth J. T., Dehaven W. I., Jones B. F., Mercer J. C., Trebak M., Vazquez G., Putney J. W., Jr. (2006) Biochim. Biophys. Acta 1763, 1147–1160 [DOI] [PubMed] [Google Scholar]
- 22.Huang G. N., Zeng W., Kim J. Y., Yuan J. P., Han L., Muallem S., Worley P. F. (2006) Nat. Cell Biol. 8, 1003–1010 [DOI] [PubMed] [Google Scholar]
- 23.Yuan J. P., Zeng W., Dorwart M. R., Choi Y. J., Worley P. F., Muallem S. (2009) Nat. Cell Biol. 11, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muik M., Fahrner M., Derler I., Schindl R., Bergsmann J., Frischauf I., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., Lewis R. S. (2009) Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barr V. A., Bernot K. M., Srikanth S., Gwack Y., Balagopalan L., Regan C. K., Helman D. J., Sommers C. L., Oh-Hora M., Rao A., Samelson L. E. (2008) Mol. Biol. Cell 19, 2802–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muik M., Frischauf I., Derler I., Fahrner M., Bergsmann J., Eder P., Schindl R., Hesch C., Polzinger B., Fritsch R., Kahr H., Madl J., Gruber H., Groschner K., Romanin C. (2008) J. Biol. Chem. 283, 8014–8022 [DOI] [PubMed] [Google Scholar]
- 28.Navarro-Borelly L., Somasundaram A., Yamashita M., Ren D., Miller R. J., Prakriya M. (2008) J. Physiol. 586, 5383–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S. L., Kozak J. A., Jiang W., Yeromin A. V., Chen J., Yu Y., Penna A., Shen W., Chi V., Cahalan M. D. (2008) J. Biol. Chem. 283, 17662–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lis A., Peinelt C., Beck A., Parvez S., Monteilh-Zoller M., Fleig A., Penner R. (2007) Curr. Biol. 17, 794–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scrimgeour N., Litjens T., Ma L., Barritt G. J., Rychkov G. Y. (2009) J. Physiol. 587, 2903–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita M., Navarro-Borelly L., McNally B. A., Prakriya M. (2007) J. Gen. Physiol. 130, 525–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derler I., Fahrner M., Carugo O., Muik M., Bergsmann J., Schindl R., Frischauf I., Eshaghi S., Romanin C. (2009) J. Biol. Chem. 284, 15903–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scrimgeour N., Litjens T., Ma L., Barritt G. J., Rychkov G. Y. (2009) J. Physiol. 587, 2903–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakowski D., Parekh A. B. (2007) Cell Calcium 42, 333–339 [DOI] [PubMed] [Google Scholar]
- 36.Parekh A. B. (2003) J. Physiol. 547, 333–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z., Neher E. (1993) J. Physiol. 469, 245–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romanin C., Gamsjaeger R., Kahr H., Schaufler D., Carlson O., Abernethy D. R., Soldatov N. M. (2000) FEBS Lett. 487, 301–306 [DOI] [PubMed] [Google Scholar]
- 39.Peinelt C., Vig M., Koomoa D. L., Beck A., Nadler M. J., Koblan-Huberson M., Lis A., Fleig A., Penner R., Kinet J. P. (2006) Nat. Cell Biol. 8, 771–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gifford J. L., Walsh M. P., Vogel H. J. (2007) Biochem. J. 405, 199–221 [DOI] [PubMed] [Google Scholar]