Abstract
Non muscle myosin II (NMII) is a major motor protein present in all cell types. The three known vertebrate NMII isoforms share high sequence homology but play different cellular roles. The main difference in sequence resides in the C-terminal non-helical tailpiece (tailpiece). In this study we demonstrate that the tailpiece is crucial for proper filament size, overcoming the intrinsic properties of the coiled-coil rod. Furthermore, we show that the tailpiece by itself determines the NMII filament structure in an isoform-specific manner, thus providing a possible mechanism by which each NMII isoform carries out its unique cellular functions. We further show that the tailpiece determines the cellular localization of NMII-A and NMII-B and is important for NMII-C role in focal adhesion complexes. We mapped NMII-C sites phosphorylated by protein kinase C and casein kinase II and showed that these phosphorylations affect its solubility properties and cellular localization. Thus phosphorylation fine-tunes the tailpiece effects on the coiled-coil rod, enabling dynamic regulation of NMII-C assembly. We thus show that the small tailpiece of NMII is a distinct domain playing a role in isoform-specific filament assembly and cellular functions.
Non muscle myosin II (NMII)2 is a major motor protein present in all cell types participating in crucial processes, including cytokinesis, surface attachment, and cell movement (1–3). NMII units are hexamers of two long heavy chains with two pairs of light chains attached. NMII heavy chain is composed of a globular head containing the actin binding and force generating ATPase domains, followed by a large coiled-coil rod that terminates with a short non-helical tailpiece (tailpiece). To carry out its cellular functions, NMII assembles into dimers and higher order filaments by interactions of the coiled-coil rod (4). The assembly process is governed by electrostatic interactions between adjacent coiled-coil rods containing alternating charged regions with specific periodicity (5–9) and is enhanced by activation of the motor domain through regulatory light chain phosphorylation (10–12). The charge periodicity also determines the register and orientation of each NMII hexamer in the filament. Additionally the C-terminal region of the coiled-coil rod contains a distinctive positively charged region and the assembly-competence domains that are crucial for proper filament assembly (5–9, 13).
Three isoforms of NMII (termed NMII-A, NMII-B, and NMII-C) have been identified in mammals (14–16). Although NMII isoforms share somewhat overlapping roles, each isoform has distinctive tissue distribution and specific functions. NMII-A is important for neural growth cone retraction (17, 18) and is distributed to the front of migrating endothelial cells (19). While NMII-B participates in growth cone advancement (20) and was detected in the retracting tails of migrating endothelial cells (19). Furthermore NMII-A and NMII-B have an opposing effect on motility, since depletion of NMII-A leads to increased motility while NMII-B depletion hinders motility (21, 22). NMII-C plays a role in cytokinesis (23) and has distinct distribution in neuronal cells (24). Furthermore one NMII isoform only partly rescue cells in which siRNA was used to reduce the expression of another isoform (23, 25). This functional diversity is achieved despite a significant amino acid sequence identity between the isoforms (overall 64–80%), and the origin of these differential distributions and functions is not completely understood.
Recent studies suggest that the C-terminal portion of NMII-A and NMII-B, particularly the last ∼170 amino acids, is responsible for the differential distribution of these NMII isoforms (26, 27). It was shown that swapping this region between NMII-A and NMII-B resulted in chimeric proteins, which adopted cellular localization according to the C-terminal part (26). This C-terminal ∼170 amino acid coiled-coil region contains the assembly-competence domains and other regions that are critical for filament assembly (5–9, 13) as well as the non-helical tailpiece. As the small tailpiece is also an important regulator of NMII filament assembly (27, 28) capable of changing NMII filament assembly properties; and phosphorylation of NMII tailpiece was shown to interfere with filament assembly (29–33) the tailpiece may be important for allowing NMII to perform its dynamic tasks. Because the coiled-coil regions are highly conserved between NMII isoforms, while the tailpiece is the most divergent, it is therefore a good candidate for mediating NMII isoform-specific functions. However, the exact mechanism by which the tailpiece affects NMII function is not fully understood. Here we show that the tailpiece serves as an isoform-specific control mechanism modulating filament order, assembly, and cellular function.
EXPERIMENTAL PROCEDURES
NMII isoforms used for this study were NMII-A, accession number NP_002464, NMII-B, accession number A59252, and NMII-C accession number AY363100.
Construction of NMII Mutants
GFP-NMII-A–C were kind gifts from Dr. R. S Adelstein, Laboratory of Molecular Cardiology and Laboratory of Molecular Physiology, NIH, Bethesda, MD. To create pET21A-NMII-C-rod fragment (amino acids 1297–2000) NMII-C rod in pBluescript II SK(−) was digested with ClaI followed by fill-in using Klenow fragment, and NotI. The fragment was inserted into pET21A (Clontech Laboratories, Mountain View, CA) that was digested with NheI filled-in and NotI, using Mighty-mix ligation kit (Takara Bio Shiga, Japan). To create the pET21C-NMII-A rod fragment, a fragment encoding amino acids 1482–1960 was removed from pTRE-GFPC3-MHC-A (34) using XmaI filled-in and SalI and transferred to pET21C digested with NheI filled-in and SalI. To correct several problems with the original sequence (25) this subcloning was followed by a series of site-directed mutagenesis reactions using a QuikChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. NdeI site was added to pTRE-GFPC3-MHC-A by site-directed mutagenesis using primer 5′-CAC TCA GGA GCT GCT GCA TAT GGA GAA CCG GCA GAA GC-3′ followed by digestion with NdeI and SacII. The fragment was cloned into pET21C-NMII-A Rod fragment digested with the same enzymes to create pET21C-NMIIA Rod fragment (amino acids 1320–1960) used in this study. To create pQE80-l-NMII-C-193 corresponding to amino acids 1808–2000 of NMII-C, NMII-C rod fragment in pBluescript II SK(−) was digested with Bsp120I filled-in and a combination of SacI and XmaI digestion to receive a detectable blunt-SacI fragment. The plasmid pQE80-l (Qiagen GmbH, Hilden, Germany) was digested with BamHI filled-in followed by SacI digestion. The digested plasmid and fragment were ligated. Tailless NMII constructs were created by converting the coiled-coil ending proline codon on the NMII rod fragments in pET21 (described above) to a terminator codon using site-directed mutagenesis with the following primers: NMII-A, 5′-GGC GCG GGG ACC TGT AAT TTG TCG TGC CCC GCC G-3′; NMII-B, 5′-CGG CTG GAA GAG AAG CTG ATT TAG CCA CCC CGC CTC AGC CGG-3′; and NMII-C, 5′-GGC TCC GGC GTG GCT AAT GAG ACA TTCA CCA CAC GGA C-3′. The tail-swapped constructs were created by adding a KpnI site at the coiled-coil ending the proline codon in all isoforms using site-directed mutagenesis and subcloning the different rod fragment coding regions using KpnI and NdeI. After ligation the KpnI site was restored to the proline codon with a second cycle of site-directed mutagenesis. KpnI inserting primers were as follows: NMII-A, 5′-CAA GCT CAG GCG CGG CGA GGT ACC GTT TGT CGT GCC C-3′; NMII-B, 5′-GAA CCG GCT GAG GCG TGC GGT ACC CAT CAG CTT CTC-3′; and NMII-C, 5′-GAG GAA CAG GCT CCG GCG GGT ACC ACT TAC ATT CAC CAC-3′. Primers used to convert KpnI site to the original proline codon sequence: NMII-A-tailB, 5′-GCT CAG GCG CGG CGA CCT GCC CAT CAG CTT CTC-3′; NMII-A-tailC, 5′-CAA GCT CAG GCG CGG CGA CCT GCC ACT TAC ATT CAC CAC-3′; NMII-B-tailA, 5′-GAA CCG GCT GAG GCG CGG TGG CCC GTT TGT CGT GCCC-3′; NMII-B-tailC, 5′-GAA CCG GCT GAG GCG CGG TGG CCC ACT TAC ATT CAC CAC-3′; NMII-C-tailA, 5′-GAA CAG GCT CCG GCG TGG CCC GTT TGT CGT GCC-3′; and NMII-C-tailB, 5′-GGA ACA GGC TCC GGC GTG GCC CCA TCA GCT TCT CTT C-3′. HA fused to NMII-C was created by removing the GFP tag from GFP-NMII-C using NdeI and SalI. Synthetic primers coding the HA tag 5′-TAT GCT CGA GTA CCC ATA CGA CGT CCC AGA TTA TGC TCT CGA GGC TAG CAC GCG TAC CGG TCT TAA CGG GCC CG-3′, were subsequently ligated into the gap. Tailpiece-swapped full-length NMII were created by transferring the C-terminal region from tail-swapped rod fragments in pET21 (described above) into the GFP- or HA-tagged full-length NMII isoform. Specifically, pET21 containing an NMII-A-tailB rod fragment was digested with SalI and AflI, and the blunt-AflI fragment was ligated into GFP-NMII-A using the same enzymes. GFP-NMII-A-tailC was created by removing a fragment from the NMII-A-tailC rod fragment in pET21 using BamHI digest followed by fill-in reaction and AflI. The fragment was inserted to GFP-NMII-A after digestion with SalI filled-in and AflI. A silent SacII site was created in GFP-NMII-B, NMII-B-tailA rod fragment, and NMII-B-tailC rod fragment by site-directed mutagenesis (as described above) using the following primer: 5′-GCC GAG CTA GCC GCG GAG CGC AGC GCC GC-3′. For GFP-NMII-B-tailA, GFP-NMII-B was digested with SacII and HpaI. NMII-B-tailA rod fragment in pET21 was digested with EcoRI followed by fill-in and SacII digest, the resulting fragment was ligated into GFP-NMII-B. For GFP-NMII-B-tailC, HpaI site was created in the NMII-B-tailC rod fragment in pET21 using 5′-CCA CAC CCC AAT GAT CCA GTT AAC CCT AGA TGC CCC AAG GAC-3′ primer. Next the GFP-NMII-B vector and the NMII-B-tailC rod fragment in pET21 were digested with SacII and HpaI, and the fragments were ligated. Both HA-NMII-C-tailA and HA-NMII-C-tailB were created by digesting HA-NMII-C in pEGFP with BamHI followed by fill-in reaction and SbfI digest. Each of the NMII-C rod fragment chimeras in pET21 were digested using EcoRI and filled-in followed by SbfI digest. Fragments were ligated respectively. Asp mutations were performed on pET21C-NMII-A and pET21A-NMII-C rod fragments described above, using site-directed mutagenesis as described above, using the following primers: NMII-A S1917D-PKC site, 5′-GAA CCG CGA AGT CAG CGA CCT AAA GAA CAA GCT-3′; NMII-A S1944D-CKII site, 5′-GGC GCC GGG GAT GGC GAC GAC GAA GAG GTA G-3′; and NMII-A PKC+CKII double mutant was created using the same primers sequentially. NMII-C rod fragment PKC TT1957–60DD, 5′-GGC CCA CTT ACA TTC GAC ACA CGG GAT GTG CGC CAG GTG TTC-3′; NMII-C rod fragment CKII T1932D, 5′-GAG CTG GAG GAC GTC GAT GAG TCT GCA GAA TC- 3′; NMII-C rod fragment CKII S1973D, 5′-CTG GAA GAG GGC GTG GCT GAT GAC GAG GAA GAG-3′; and NMII-C rod fragment CKII S1985D, 5′-GAG CTG AAC CTG GCG ATG CAC CAG GCC AGG-3′. The NMII-C PKC+CKII double mutant was created using the same primers sequentially. pET21C-NMII-B-642 (amino acids 1336–1975) Asp mutations were described previously (32). All mutations were confirmed by DNA sequencing (Center for Genomic Analysis, The Hebrew University of Jerusalem).
Purification of NMII Fragments from Escherichia coli, Sedimentation Assays, and Negative Staining for Electron Microscopy
Assays were performed as described previously (8, 35). In the electron microscopy experiments, a wash step was added after protein deposition on the grid. Filaments were measured using Image-Pro plus software (Media Cybernetics) on calibrated electron micrographs at ×88000 magnification and subjected to two tailed, two sampled un-equal variance Student's t test using at least 18 measurements. In the sedimentation assays the 50% solubility and area under the graph were calculated from graphs plotted on KaleidaGraph (Synergy Software, Reading, PA) and Excel software (Microsoft Corp.).
Mapping of Phosphorylation Sites
Phosphorylation reactions were performed on NMII-C-193 described above using recombinant CKII (New England Biolabs, Ipswich, MA) or a PKC mix (Sigma-Aldrich) containing isoforms α, βΙ, βΙΙ, γ, δ, ϵ, θ, η, and ζ. The phosphorylation reactions were carried out according to the manufacturer's recommendations and allowed to reach maximal phosphorylation. Phosphorylated NMII-C-193 was separated on SDS-PAGE and analyzed using mass spectrometry as described previously (36) to determine the exact phosphorylation sites.
Creating siRNA-depleted Cell Lines
For NMII depletion, lentivirus particles containing siRNA-expressing plasmids were created. Cells were infected with lentiviruses and examined for NMII expression using specific antibodies. NMII-A-specific siRNA sequence 5′-CGA TTT CCA AAA GCG GAG GCT TCA GAA GGA CTC TCT TGA AGT CCT TCT GAA GCC TCC GCG GGG A-3′ was synthesized and inserted into pLVHTM (Lentiweb) according to instructions. This plasmid has an independent GFP protein transcribed as an infection marker. Because our NMII construct is fused to GFP, we replaced the GFP on pLVHTM with mCherry received from Dr. Michael Brandeis, The Hebrew University. Control sequence 5′-CGC GTC CCC GCC CTT CCG ACA TAT AGG TTT CAA GAG AAC CTA TAT GTC GGA AGG GCT TTT GGA AAT-3′ was created in a similar manner. For NMII-C knockdown 5′-GAT CCC CGC AGC TTC CCA TCT ACA CAT TCA AGA GAT GTG TAG ATG GGA AGC TGC TTT TTG GAA A-3′ was synthesized and ligated into pSUPER digested with BglII and HindIII. A fragment containing the H1 promoter and the siRNA sequence was digested with XhoI and SmaI and ligated into the lentivirus plasmid pSIN18 digested with EcoRV and XhoI. Control siRNA containing the sequence 5′-AAG TGA GAG CAA AGT GAA AGT TCT CTT CAC TTT GCT CTC ACT T-3′ was created as described above. siRNA plasmids were co-transfected with pCMV-dR8.91 and pMD2G (Lentiweb) into HEK293T cells. Lentirirus containing medium was collected 3 days after transfection, filtered through a 0.45-μm filter and centrifuged for 2 h at 23,000 rpm to collect the Lentivirus particles. The particles were resuspended in 500 μl of serum-free medium containing Polybrene (Sigma-Aldrich) and incubated on ice for 30 min. Cells were infected with the Lentivirus particles for 4 h with constant rocking. The NMII levels were determined with isoform-specific NMII antibodies.
Antibodies and Reagents
C-terminal isoform-specific NMII antibodies for Western blotting were generated in rabbits. N-terminal isoform-specific antibodies were received from Dr. R. S. Adelstein. Rat monoclonal HA antibody was from Roche Diagnostics Corporation (Basel Switzerland), Vinculin mouse monoclonal antibodies were from Sigma-Aldrich, and rabbit phosphorylated Paxillin Y118 antibodies were from Invitrogen. Horseradish peroxidase-conjugated secondary antibodies, Rhodamine Red-X Donkey anti-rat and Cy5 goat anti-rabbit secondary antibodies were from Jackson ImmunoResearch Laboratories (Pike West Grove, PA). Rhodamine-Phalloidin was obtained from Molecular Probes (Eugene, OR).
Cell Line, Culture Conditions, and Transient Transfection
B−/B− and C−/C− MEFs were maintained in high glucose Dulbecco's modified Eagle's medium supplemented with 2 mm l-glutamine, 10% fetal calf serum, and antibiotics (100 units/ml penicillin, 100 μg/ml streptomycin, and 1:100 Biomyc3 anti-mycoplasma antibiotic solution (Biological Industries, Beit HaEmek, Israel). A549 lung carcinoma cell line was grown in RPMI 1640 supplemented with 2 mm l-glutamine, 10% fetal calf serum, and antibiotics (100 units/ml penicillin, 100 μg/ml streptomycin, and 1:100 Biomyc3 anti-mycoplasma antibiotic solution. Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. For B−/B− and C−/C− MEFs, 16 h before transfection, 2.5 × 105 cells were plated on 30-mm tissue culture dishes containing coverslips coated with 27 μg/ml collagen I (Sigma). Transfections were carried out with 1 μg of plasmid DNA per 30-mm dish, using JetPEI Transfection Reagent (Polyplus-transfection Inc., Strasbourg, France) with a nitrogen/phosphate ratio = 7, according to the manufacturer's instructions or with 2 μg of DNA in transfection reagent FuGENE HD (Roche Diagnostics). Cells were incubated at 37 °C in a humidified atmosphere for 4–6 h after which the medium was replaced. For A549 cells 5 × 105 cells were plated on 30-mm tissue culture dishes containing coverslips coated with 10 μg/ml fibronectin (Sigma-Aldrich). Transfections were carried out with 2 μg of plasmid DNA per 30-mm dish, using JetPEI Transfection Reagent (Polyplus-transfection Inc., Strasbourg, France) with nitrogen/phosphate ratio = 7, according to the manufacturer's instructions.
Wound Scratch Assay and Confocal Microscopy
For B−/B− and C−/C− MEFs, 16 h after transfection with NMII constructs, the cells were washed twice with 1 ml of starvation medium (high glucose Dulbecco's modified Eagle's medium supplemented with 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.1% fatty acid free bovine serum albumin (Sigma)) and serum-starved for 24 h in 2 ml of starvation medium. After starvation, three parallel scratches were performed using a small pipette tip. Cells were washed once with 1 ml of starvation medium to remove cell debris, and 2 ml of starvation medium containing 25 ng/ml platelet-derived growth factor-BB (Sigma-Aldrich) was added. After 8 h cells were washed three times with phosphate-buffered saline and fixed for 10 min in 1.5 ml of 3.7% formaldehyde in phosphate-buffered saline and subjected to immunostaining as described previously (37). For A549 cells 24-h post transfection cells were washed three times with phosphate-buffered saline and fixed for 10 min in 1.5 ml of 3.7% formaldehyde in phosphate-buffered saline and subjected to immunostaining. Coverslips were mounted on slides using Vectashield mounting medium (Vector Laboratories Inc., Burlingame, CA). Cells were visualized using a 60×, numerical aperture 1.4 objective under a TE2000 inverted confocal laser scanning system (Nikon Corp., Tokyo, Japan). Consecutive Z stakes were taken using a small aperture and converged to create one image using EZ-C1 software (Nikon). Focal adhesion area and number were automatically analyzed using Image-Pro plus software (Media Cybernetics) with calibrated background-reduced final image and subjected to two-tailed, two-sampled un-equal variance Student's t test using at least 16 cells.
RESULTS
The Tailpiece Is Important for Proper NMII Filament Assembly
To study the role of NMII tailpiece in the assembly processes we created different mutant rod fragments of the three NMII isoforms. These rod fragments are composed of ∼2/3 of the complete coiled-coil region and contain all of the known assembly important domains in the coiled-coil (Fig. 1).
FIGURE 1.
Schematic diagram depicting the NMII proteins used in this study. Full-length NMII and coiled-coil rod fragments used for in vivo and in vitro experiments, respectively. Numbers represent amino acid positions in the full-length protein.
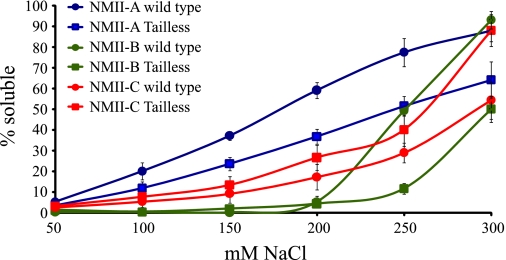
The assembly properties of NMII rod fragments lacking the tailpiece (tailless rod fragment) (Fig. 1 and supplemental Fig. 1A) were examined by sedimentation assay. This assay is considered a good indication of NMII filament assembly capabilities (28, 29, 38, 39). The absence of the tailpiece caused NMII-A and NMII-B tailless rod fragments to create stronger assemblies that became soluble only at higher salt concentrations than intact NMII-A and NMII-B rod fragments (Fig. 2 and Table 1). In contrast, the tailless NMII-C rod fragment created weaker assemblies that became soluble at lower salt concentrations than the intact NMII-C rod fragment. These results indicate that NMII-A and NMII-B tailpieces weakened assembly and NMII-C tailpiece strengthened it.
FIGURE 2.
In vitro self-assembly of tailless NMII rod fragments. 0.1 mg/ml tailless rod fragments were expressed and purified from E. coli as described under “Experimental Procedures.” Proteins were dialyzed against different NaCl concentrations, and the extent of assembly was calculated as the percentage of rod fragment remaining in the supernatant after high speed centrifugation. Results are averages ± S.D. of at least three independent experiments.
TABLE 1.
In vitro self-assembly properties of NMII rod fragment
Biophysical characteristics of the different NMII mutants. Paracrystal width and length were measured using ImagePro software from negatively stained electron micrographs as described in the legend to Fig. 3. Sedimentation assays were carried out as described in the legend to Fig. 2. NaCl concentration resulting in 50% solubility was calculated from supplemental Fig. 4. Area under the graph was calculated from the same graphs as an additional indicator of solubility properties. Values were normalized to area of 100% solubility. Results are averages ± S.D. of at least three independent experiments.
| NMII rod fragment | Filament width | Filament morphology | Filament length | 50% solubility | Solubility, area under the graph |
|---|---|---|---|---|---|
| nm ± S.D. | nm ± S.D. | mm NaCl ± S.D. | % ± S.D. | ||
| NMII-A wild type | 1.13 ± 0.4 | Wide | N/Aa | 180 ± 3.3 | 48.2 ± 4 |
| NMII-B-tailA | 1.77 ± 0.5 | Wide | N/A | 222 ± 5.0 | 32.6 ± 2 |
| NMII-C-tailA | 1.05 ± 0.3 | Wide | N/A | 223 ± 6.3 | 34.5 ± 4 |
| NMII-B wild type | 1.22 ± 0.3 | Wide | N/A | 250 ± 2.1 | 20.5 ± 1 |
| NMII-A-tailB | 1.51 ± 0.3 | Wide | N/A | 235 ± 1.2 | 31.6 ± 2 |
| NMII-C-tailB | 0.91 ± 0.5 | Wide | N/A | 280 ± 4.9 | 22.6 ± 2 |
| NMII-C wild type | 0.39 ± 0.1 | Thin | N/A | 318 ± 4.9 | 14.5 ± 4 |
| NMII-A-tailC | 0.39 ± 0.1 | Thin | N/A | 230 ± 1.3 | 33.3 ± 2 |
| NMII-B-tailC | 0.23 ± 0.05 | Thin | N/A | 232 ± 2.9 | 25.9 ± 2 |
| NMII-A tailless | 0.31 ± 0.06 | Cigar-shaped | 2.85 ± 1.0 | 179 ± 4.6 | 31.6 ± 4 |
| NMII-B tailless | 0.32 ± 0.08 | Cigar-shaped | 3.30 ± 1.0 | 300 ± 2.4 | 8.9 ± 1 |
| NMII-C tailless | 0.35 ± 0.05 | Cigar-shaped | 2.57 ± 0.6 | 261 ± 2.8 | 26.8 ± 5 |
a N/A, not applicable.
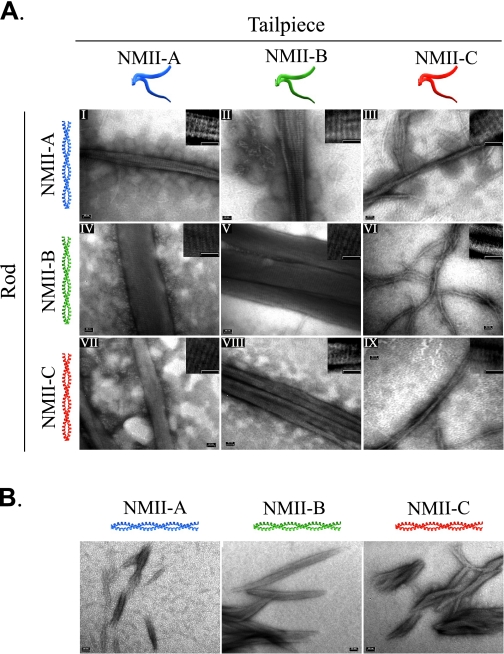
NMII forms ordered filamentous structures called paracrystals, which are salt-dependent and show a distinct pattern of 14.3 nm striations similar to bipolar filaments seen in vivo (40). To further explore the role of the tailpiece in assembly of NMII, we inspected the morphology of paracrystals formed by tailless rod fragments. Wild-type NMII-A and NMII-B formed wide large paracrystals with distinct striations (41). In contrast, the newly discovered NMII-C isoform formed narrow paracrystals (Fig. 3, Ai, Av, and Aix, and Table 1). Removing the tailpiece created very short cigar-like structures with little detectable striation in all NMII isoforms (Fig. 3B). As NMII-C was recently discovered, we confirmed that the tailless and wild-type full-length rod formed paracrystals identical to NMII-C rod fragment counterparts (supplemental Fig. 2). The differences in sedimentation and the formation of paracrystals of a similar size and shape by all three tailless isoforms confirm that the tailpiece is a global modulator of filament assembly and is essential for the proper formation of large filamentous structures (Figs. 2 and 3B). Additionally, peptides corresponding to the tailpieces had no effect on solubility or paracrystal formation when added to the tailless rod fragments (data not shown), indicating the tailpiece must be attached to the rod fragment to carry out its functions.
FIGURE 3.
Tailpiece effect on rod fragment paracrystal formation. Rod fragments and tail-swapped rod fragments were expressed and purified from E. coli, dialyzed against low NaCl buffer, and stained with uranyl acetate prior to viewing by electron microscope as described under “Experimental Procedures.” A, electron micrographs of paracrystals formed by rod fragments with swapped tailpieces. On the left is depicted the coiled-coil part of the chimera protein, and on the top is the tailpiece. B, electron micrograph of paracrystals formed by tailless rod fragments. Insets are enlarged, contrast-enhanced electron micrographs showing filament striations. Tailpiece, the non-helical tailpiece; Rod, the coiled-coil portion as depicted in Fig. 1. Bar, 50 nm.
To examine whether the tailpiece or the coiled-coil region of the rods are important for proper filament assembly, we created rod fragments in which the tailpieces were swapped between the NMII isoforms (Fig. 1 and supplemental Fig. 1, A and B) and examined their paracrystal morphology. To our surprise, placing the tailpiece of NMII-A or NMII-B on the NMII-C rod fragment (NMII-C-tailA or NMII-C-tailB) resulted in the formation of large and wide paracrystals having striations very similar to those of intact NMII-A and NMII-B rod fragments, respectively (Fig. 3A, vii and viii). These differ greatly from NMII-C rod fragment paracrystals, which were very thin (Fig. 3A, ix). NMII-B-tailC and NMII-A-tailC formed very thin filamentous paracrystals, thus resembling the NMII-C rod fragment paracrystals (Fig. 3A, iii and vi). All paracrystals tested showed typical striations (Fig. 3A, inset, and supplemental Table S1). Remarkably these results indicate that the small tailpiece of ∼40 amino acids (see Fig. 6) forces the ∼640 amino acid coiled-coil NMII to form the unique morphology of NMII filaments.
FIGURE 6.
Sequence of NMII isoform tailpieces showing the different phosphorylation sites. Red, phosphorylation by PKC; blue, phosphorylation by CKII; green, novel CKII site; and P, proline ending the coiled-coil. The positively and negatively charged regions are also indicated.
To determine whether the assembly properties of NMII are governed by the tailpiece, we tested the solubility properties of the swapped NMII mutants. Despite the different paracrystal morphology (Fig. 3A) the chimeric proteins NMII-B-tailC and NMII-C-tailB had solubility properties similar to wild type NMII-B and NMII-C rod fragments, respectively (Table 1 and supplemental Fig. 4).
The assembly properties of both NMII-A-tailB and NMII-A-tailC were similar to tailless NMII-A, indicating that NMII-B and NMII-C tailpieces cannot replace NMII-A tailpiece (Fig. 2, Table 1, and supplemental Fig. 4). Additionally, placing NMII-A tailpiece on either NMII-B or NMII-C rod fragments caused a marked increase in their solubility (Table 1 and supplemental Fig. 4), further demonstrating its apparent difference. Hence, it seems that, although paracrystal morphology is dominated solely by the tailpiece, filament assembly properties of each isoform are determined by both the tailpiece and the intrinsic properties of the coiled-coil region.
The Tailpiece Is Important for NMII Functions in Vivo
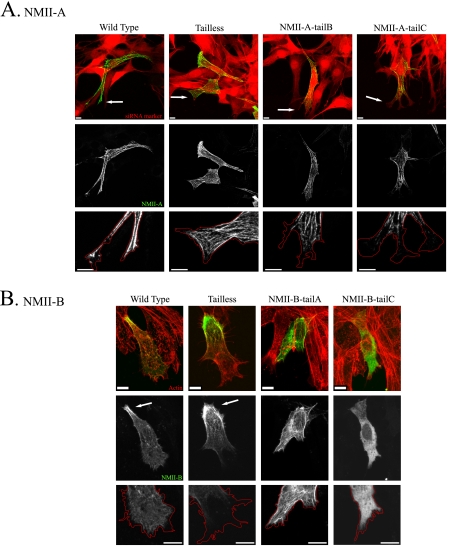
Because the three NMII isoforms have specific cellular distributions and functions, we postulated that the tailpiece with its ability to affect the filament assembly properties of the much larger coiled-coil rod may be important for the different functions of NMII in vivo. To test this we created tailpiece-swapped full-length NMII mutants (supplemental Fig. 1C). NMII-A and NMII-B were fused to GFP while NMII-C was tagged with hemagglutinin (HA), because fusion of GFP to the N-terminal region of NMII-C disrupted its activity (25). The full-length chimeras were transfected into cells lacking the specific NMII isoform and viewed by confocal microscopy. NMII-A chimeras were transiently transfected into mouse embryonic fibroblasts obtained from B−/B− mice (B−/B− MEF) (42). These cells lack NMII-B and NMII-C, and the level of NMII-A was reduced by 84% using specific NMII-A siRNA (supplemental Fig. 1D). The cells were subjected to wound scratch assay to achieve polarized migrating cells. NMII-A wild type was distributed throughout the cell forming large distinct fibers that reached the anterior edge of the cell (in 7 of 9 cells observed) (Fig. 4A).
FIGURE 4.
NMII-A and NMII-B tailpiece effect on isoform localization in vivo. Tailpiece-swapped full-length NMII-A and NMII-B fused to GFP were transfected into cells and subjected to wound scratch assay as described under “Experimental Procedures.” A, NMII-A tail-swapped chimeras expressed in NMII-A siRNA-treated B−/B− MEF cells. Arrows point at cells leading edge. Red, NMII-A siRNA marker; green, GFP-NMII-A. B, NMII-B tail-swapped chimeras expressed in B−/B− MEF cells, actin was detected by Rhodamine-Phalloidin staining. Arrows indicate posterior accumulation of NMII-B. Red, actin; green, NMII-B. Red lines are tracing of cells leading edge. Bar = 10 μm.
NMII-A-tailB caused the exclusion of this chimera protein from the leading edge (Fig. 4A). Furthermore, NMII-A-tailB had posterior localization that is characteristic to wild-type NMII-B but not to wild-type NMII-A (in 5 of 10 cells observed) (Fig. 4A). Surprisingly removing NMII-A tailpiece had little effect on distribution (in 10 of 12 cells observed) (Fig. 4A).
The NMII-B chimeras were expressed in B−/B− MEFs, which express only endogenous NMII-A (supplemental Fig. 1D). Unlike endogenous NMII-A, NMII-B is arranged in more delicate fibrillar structures localized preferentially to the posterior part of the cell and is usually excluded from the leading edge (in 11 of 15 cells observed) (Fig. 4B). Tailless NMII-B accumulated at the cell posterior region, although it was still capable of entering cytoskeletal structures and the cell cortex (in 15 of 17 cells observed) (Fig. 4B). NMII-B-tailA was distributed throughout the cell creating fibers that reached the leading edge similar to endogenous NMII-A (in all 30 cells observed) (Fig. 4). Interestingly NMII-B-tailA still accumulated at the posterior of the cell similar to NMII-B wild-type but to a lesser extent (18 of 30 observed), indicating that some of the cellular localization properties of NMII-B also reside in the head and coiled-coil rod regions. These results indicate that the tailpiece plays a role in the distinctive cellular distribution of NMII-B and can affect NMII-A in vivo.
When examining the effect of placing NMII-C tailpiece on either NMII-A or NMII-B, NMII-B-tailC was almost completely diffuse throughout the cell (in all 19 cells observed). In contrast, NMIIA-tailC formed fibers mainly in the cell body that did not reach the leading edge or the rear of the cell (in 9 of 10 cells observed) (Fig. 4). These results indicate that NMII-C tailpiece may play a role in functions other than cellular localization and support the notion that NMII head and the coiled-coil rod also contain information important for cellular localization.
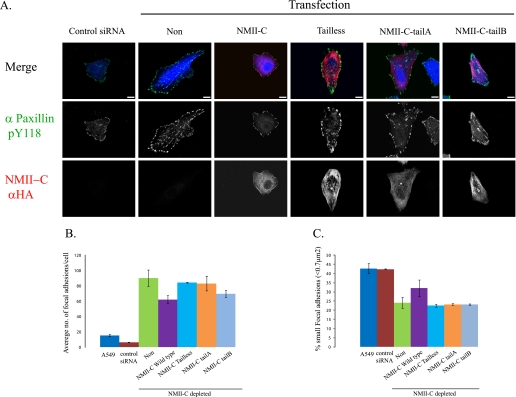
Next we tested the effects of swapping the tailpiece of NMII-C, using A549 lung small cell carcinoma cells in which the level of NMII-C was reduced by ∼92% using specific NMII-C siRNA (supplemental Fig. 1E). Swapping the tailpiece had little effect on NMII-C chimeras cellular distribution and fibrillar morphology (Fig. 5). NMII-C missing the tailpiece as well as the different NMII-C chimeras were distributed evenly throughout the cells, forming very delicate fibers similar to NMII-C wild type (Fig. 5).
FIGURE 5.
NMII-C tailpiece affects focal adhesion. A, HA-tagged tailpiece-swapped NMII-C chimeras were transfected into A549 cells treated with NMII-C specific siRNA (blue). Cells were fixed and immunostained with paxillin pY118 phosphotyrosine antibody (green) and HA antibody (red) and viewed by confocal microscope. Note all NMII-C mutants create delicate fibers throughout the cell. Bar = 10 μm. B, average number of focal adhesion sites per cell in A549 and NMII-C depleted cells transfected with the various NMII-C chimeras. C, percentage of focal adhesion sites smaller than 0.7 μm2 in A549- and NMII-C-depleted cells transfected with the various NMII-C chimeras. Values are an average ± S.D. of at least 16 cells from 2 independent experiments subjected to two-tailed, two-sampled unequal variance Student t test. PA549 versus control = 0.28, PA549 versus NMII-C depleted = 8.29e-29, PNMII-C-depleted versus NMII-C wild-type = 6.77e-7, PNMII-C-depleted versus NMII-C tailless = 0.8, PNMII-C-depleted versus NMII-C-tailA = 0.195, and PNMII-C-depleted versus NMII-C-tailB = 0.3.
However, in the course of characterizing the A549 cell line depleted of NMII-C, we noticed that these cells adhered more firmly than control cells (Fig. 5, B and C). Similar observations were recently reported following the depletion of NMII-C from neuronal cells (24). These results suggest that NMII-C plays a role in the regulation of cell adhesion, and we therefore studied the effect of NMII-C tailpiece swapping on focal adhesion complexes. NMII-C knockdown A549 cells showed a marked increase in both the number and size of focal adhesions compared with untreated cells as viewed by Tyr-118-phosphorylated paxillin (Fig. 5) and vinculin (supplemental Fig. 3). Transfection of NMII-C-depleted cells with wild-type NMII-C was the only construct that reduced the number and size of focal adhesions, whereas cells expressing all other NMII-C mutants had focal adhesion similar to those in NMII-C-depleted cells (Fig. 5). Together these results reveal that the effects the tailpiece exerts on NMII can account for part of the cellular differences observed between the isoforms.
NMII-C Phosphorylation Modulates Its Assembly Properties
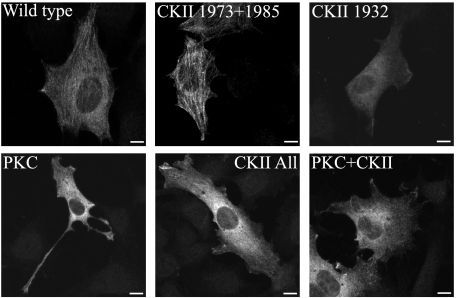
Because phosphorylation of NMII-A and NMII-B tailpieces affects the assembly properties of the rods (29–31), we examined whether phosphorylation of NMII-C also affected its assembly properties. NMII-A and NMII-B tailpieces are known to be phosphorylated by PKC and CKII (29–31). Sequence comparison of the C terminus of the three NMII isoforms and prediction algorithms indicated that NMII-C has putative PKC and CKII phosphorylation sites (Fig. 6). NMII-C was phosphorylated using recombinant PKC isoform mix or CKII, and their specific sites were mapped by mass spectrometry (supplemental Table 2). We found that NMII-C tailpiece has two PKC phosphorylation sites at threonine 1957 and 1960 and two CKII phosphorylation sites at serine 1973 and 1985 (Fig. 6). A third novel CKII phosphorylation site was mapped to threonine 1932, located in the coiled-coil region of NMII-C rod (Fig. 6). Threonine 1957 and 1960 sites were previously found to be phosphorylated by TRPM6 and TRPM7 (43). To study the effect of NMII-C phosphorylation on its solubility properties, we created phosphomimetic NMII-C rod fragments by converting the phosphorylation sites to aspartate. PKC phosphomimetic NMII-C (T1957D/T1960D) increased its solubility compared with unphosphorylated NMII-C (Table 2). However, classic CKII phosphomimetic NMII-C rod fragments (S1973D/S1985D) had no apparent effect on its solubility (Table 2). PKC and CKII phosphomimetic NMII-C (T1957D/T1960D/S1973D/S1985D) showed the same effect as PKC phosphomimetic NMII-C (Table 2), indicating that PKC phosphorylation is solely responsible for the effect on NMII-C rod fragment solubility. Phosphomimicking the novel CKII site (T1932D) had an intermediate effect on NMII-C rod fragment solubility, which did not change after further converting the two classic CKII sites to aspartate (Table 2). To confirm the relevance of the newly discovered phosphorylation sites in vivo, we transfected C−/C− MEF with full-length phosphomimetic forms of NMII-C and examined their cellular distribution. NMII-C wild-type and classic CKII phosphomimetic mutant entered into stress fibers creating delicate networks typical of NMII-C (Fig. 7). However, NMII-C mimicking PKC or PKC plus CKII phosphorylation were completely diffuse. Similar behavior was detected for NMII-C mimicking the novel CKII phosphorylation alone or together with the classic sites (Fig. 7). Hence the in vivo behavior of the phosphomimetic forms of NMII-C resembles the in vitro solubility properties, indicating that the mapped phosphorylation sites are relevant in vivo.
TABLE 2.
In vitro self-assembly of phosphomimetic forms of NMII-C rod fragment
Sedimentation assays were carried out as described in the legend to Fig. 2. NaCl concentration resulting in 50% solubility was calculated from graphs similar to those in Fig. 2. Results are averages ± S.D. of at least three independent experiments.
| NMII-C Asp mutants | 50% solubility |
|---|---|
| mm NaCl ± S.D. | |
| WT | 318 ± 4.9 |
| CKII 1973 + 1985 | 308 ± 3.2 |
| CKII 1932 + 1973 + 1985 | 245 ± 2.3 |
| CKII 1932 only | 222 ± 8.5 |
| PKC 1957 + 1960 | 185 ± 4.8 |
| All | 180 ± 3.7 |
FIGURE 7.
In vivo localization of phosphomimetic NMII-C. HA-tagged NMII-C constructs were transiently transfected into C−/C− MEFs subjected to wound scratch assay and viewed by indirect immunofluorescence with anti-HA antibody as described under “Experimental Procedures.” Bar = 10 μm.
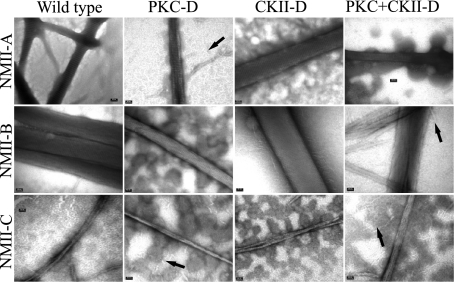
After mapping the NMII-C phosphorylation sites, we went on to test the effect of tailpiece phosphorylation on all NMII isoform paracrystals. All phosphomimetic forms of NMII rod fragments created paracrystals similar to their unphosphorylated counterparts (Fig. 8). Yet, we observed increased amounts of thin filaments formed by rod fragments with PKC only or PKC and CKII sites converted to aspartate (Fig. 8, arrows). Hence, phosphorylation of the tailpiece may affect paracrystal stability and/or formation kinetics, but once formed, the paracrystals resemble those of unphosphorylated rod fragments.
FIGURE 8.
Electron micrographs of paracrystals formed by phosphomimetic NMII rod fragment mutants. Phosphomimicking mutants of the different phosphorylation sites in all three NMII isoforms were created as described under “Experimental Procedures.” Paracrystal morphology was detected by electron microscope as in Fig. 3. PKC-D, CKII-D, PKC+CKII-D, PKC, CKII, and PKC plus CKII phosphorylation sites were converted to aspartate, respectively. Arrows indicate the thin filaments in phosphomimetic NMII. Bar = 50 nm.
DISCUSSION
NMII is revealing itself as a modular molecule with regions of specific properties. The N-terminal head domain carries the actin binding site and force-generating ATPase. The neck region contains the IQ motifs, which bind the regulatory light chain that regulates head activity (4). Important domains effecting filament assembly reside within the long coiled-coil rod. Two assembly-competence domains at the C-terminal are crucial for proper filament assembly (7). Recently we discovered that correct distribution of positive and negative charges along NMII rod is also necessary for proper filament assembly and intracellular localization of NM-II (8). Here we show that the tailpiece is a distinct domain of NMII affecting filament assembly and in vivo properties in an isoform-specific manner.
Our results show that removing the tailpiece of NMII rod fragments changes both their solubility properties and paracrystal morphology. The similar effect on filament assembly caused by omitting the tailpiece from NMII-A and NMII-B rod fragments may result from the sequence similarity of their coiled-coil region (70% identity and 84% similarity), whereas the sequence of NMII-C is more divergent (∼55% identity and ∼75% similarity) explaining its different behavior. This indicates that rods with different sequences have different solubility properties and is in agreement with studies using other myosin II missing the tailpiece (28, 44, 45).
Removing the tailpiece causes all isoforms to create very small paracrystals, and it is plausible that the tailless rods have a limited ability to enter the growing filament due to their relatively negative charge (8, 9). We postulate that the positively charged section of the tailpiece (Fig. 6) overcomes the negatively charged rod, and/or the tailpiece has a structural role allowing more rods to enter the growing filament. The similar morphology of all tailless paracrystals indicates that the tailpiece affects the way NMII filaments are assembled conferring the unique filament morphologies of each NMII isoform. This is further supported by our experiments swapping the tailpiece between the NMII isoforms. Sato et al. (27) proposed that the tailpiece may be necessary for self-recognition and creating homodimers in vivo. Our findings that each tailpiece confers its individual morphology on filaments of the coiled-coil rods may be the underlying mechanism.
Despite major alterations in tailpiece- swapped paracrystal morphology, the tailpiece has a more complex effect on NMII filament assembly properties. Swapping the tailpieces between NMII-B and NMII-C has little effect on their solubility. Since the sedimentation assay challenges electrostatic interactions, as long as the overlap of the individual rod units is similar to that of wild-type NMII, it will result in similar electrostatic interactions and similar solubility. The similar charge distribution between NMII-B and NMII-C tailpieces may allow comparable overlap to occur, resulting in similar filament assembly properties. This suggests that the tailpiece has a distinctive effect on solubility, independent of its effect on paracrystal morphology. Indeed, NMII-A chimeras solubility is similar to tailless NMII-A. NMII-A has a shorter tailpiece compared with NMII-B and NMII-C, with fewer negatively charged amino acids and phosphorylation sites (Fig. 6). Also, it is the only isoform known to interact with MTS1, a protein promoting NMII-A disassembly (29). These differences may also prevent the NMII-A tailpiece from replacing NMII-B and NMII-C tailpieces, so that placing it on either rod fragment interferes with their assembly. Hence, it seems that, whereas paracrystal morphology is dominated solely by the tailpiece, filament assembly properties are determined by the combined properties of the tailpiece and the coiled-coil region.
Expressing swapped tailpiece full-length NMII in live cells indicates that the tailpiece is responsible for part of the isoform-specific cellular functions. Furthermore, the effects of each tailpiece differ from isoform to isoform. The characteristic distribution of NMII-A is transferred to NMII-B when its tailpiece is swapped with that of NMII-A, and vice versa, indicating that the tailpiece is important for the differential distribution of these two isoforms. Sandquist and Means (26) have recently shown that swapping the C-terminal ∼170 amino acids between NMII-A and NMII-B causes the chimeric NMII to act according to its C-terminal part, independent of the head and coiled-coil regions. The region swapped by Sandquist and Means contains the two assembly-competence domains and other regions crucial for filament assembly (5–9, 13) as well as the tailpiece. It is therefore difficult to determine which of these functional regions is responsible for the behavior of the different NMII isoforms in vivo. Our data indicate that the small non-helical tailpiece of ∼40 amino acids is sufficient in determining the cellular localization of NMII-A and NMII-B.
Swapping the tailpiece of NMII-C had little effect on its cellular distribution. However NMII-C tailpiece is important for proper focal adhesion complexes. Only expression of NMII-C wild type could restore focal adhesion size and number in NMII-C-depleted cells. This is further emphasized by the different manner the NMII-C tailpiece effects NMII-A or NMII-B. Whereas NMII-A-tailC formed distinct fibers, NMII-B-tailC was completely diffuse with no detectable fiber structures.
The results presented here suggest that the tailpiece by itself is crucial for the isoform-specific functions in vivo, with the effects of each tailpiece differing from isoform to isoform. This may be expected if the tailpiece plays a differentiating role between the NMII isoform functions. For NMII-A and NMII-B, the tailpiece determines cellular localization, whereas for NMII-C, it is important for focal adhesion structure. These functional differences are further emphasized by the different behavior of the tailless NMII mutants. While removing the tailpiece from NMII-B affects its cellular distribution, the lack of the tailpiece from NMII-A has little effect on its distribution, in agreement with previous studies (46, 47). Although tailless NMII-C presents similar cellular distribution as NMII-C wild-type, it was unable to restores normal focal adhesions.
The large aggregation of tailless NMII-B at the posterior region of moving cells was also seen in cells expressing NMII-B mutants with internal deletions. These deletions also caused a decrease in TX-100 solubility in vivo (8). It is possible that the deleted regions interact with the tailpiece, and therefore both NMII-B mutants have decreased solubility and show similar cellular localization. It is tempting to propose that the differences in cellular distribution and function of each isoform are mediated through the tailpiece effect on filament assembly.
Finally we show that phosphorylation of the newly discovered NMII-C by PKC and CKII is an important mechanism for controlling its cellular distribution. This effect is probably mediated by impairing NMII-C filament assembly properties as seen in the increased solubility of the phosphomimetic mutants and cellular localization. The effect of PKC phosphorylation on the solubility of NMII-C is in agreement with the effect of PKC phosphorylation on NMII-A and NMII-B solubility (29, 32, 33). However other laboratories have shown that phosphorylation of NMII-A and NMII-B by CKII has some effect on their solubility (29, 39). This apparent discrepancy may be due to a different role of CKII phosphorylation in NMII-C filament assembly. The effect of phosphorylation on all NMII isoforms is not reflected in their ability to create paracrystals, because no change in morphology was seen in any of the phosphomimetic NMII isoforms.
Taken together we show that the tailpiece is a distinct region of NMII affecting NMII filament morphology, assembly, and function. We identify three different assembly regulating properties of the non-helical tailpiece. 1) The tailpiece allows the formation of large filaments, overcoming the intrinsic tendencies of the coiled-coil rod to create short cigar-like structures. This property may be due to the overall charge or some other structural effect and is conferred only when the tailpiece is attached to the rod. 2) The tailpiece determines how NMII units will be added to a growing filament overriding any ordering information residing in the long coiled-coil rod. 3) Phosphorylation allows fine-tuning the effect of the tailpiece on the rod, thereby enabling dynamic regulation of NMII rods.
Because each tailpiece has different properties it is well suited to be one of the determining factors ensuring the different spacio-temporal distribution of each isoform in vivo, allowing their different roles. These findings emphasize the importance of this small part of the NMII molecule in assembly and function. Further investigation is needed to elucidate whether the tailpiece achieves its effects by binding, electrostatic interactions, or some structural mechanism.
Supplementary Material
Acknowledgments
We thank Dr. Robert S. Adelstein for B−/B− and C−/C− MEFs, NMII constructs, and NMII-B antibodies and Dr. Ofra Moshel for technical assistance with the mass spectrometry. We are grateful to Michael Rosenberg for discussions throughout the course of these studies. We thank Liron Even-Faitelson, Ravid Straussman, and Ami Ben-Ya'cov for the initial studies and Ahuvi Yearim for his technical assistance.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Tables 1 and 2.
- NMII
- non-muscle myosin II
- PKC
- protein kinase C
- CKII
- casein kinase II
- tailpiece
- non-helical tailpiece
- GFP
- green fluorescent protein
- siRNA
- small interference RNA
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1.Conti M. A., Adelstein R. S. (2008) J. Cell Sci. 121, 11–18 [DOI] [PubMed] [Google Scholar]
- 2.Matsumura F. (2005) Trends Cell Biol. 15, 371–377 [DOI] [PubMed] [Google Scholar]
- 3.Lauffenburger D. A., Horwitz A. F. (1996) Cell 84, 359–369 [DOI] [PubMed] [Google Scholar]
- 4.Sellers J. R. (1999) Myosins, 2nd ed, Oxford, UK: Oxford University Press [Google Scholar]
- 5.Atkinson S. J., Stewart M. (1992) J. Mol. Biol. 226, 7–13 [DOI] [PubMed] [Google Scholar]
- 6.McLachlan A. D., Karn J. (1982) Nature 299, 226–231 [DOI] [PubMed] [Google Scholar]
- 7.Nakasawa T., Takahashi M., Matsuzawa F., Aikawa S., Togashi Y., Saitoh T., Yamagishi A., Yazawa M. (2005) Biochemistry 44, 174–183 [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg M., Straussman R., Ben-Ya'acov A., Ronen D., Ravid S. (2008) PloS ONE 3, e1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Straussman R., Squire J. M., Ben-Ya'acov A., Ravid S. (2005) J. Mol. Biol. 353, 613–628 [DOI] [PubMed] [Google Scholar]
- 10.Bresnick A. R. (1999) Curr. Opin. Cell Biol. 11, 26–33 [DOI] [PubMed] [Google Scholar]
- 11.Tan J. L., Ravid S., Spudich J. A. (1992) Annu. Rev. Biochem. 61, 721–759 [DOI] [PubMed] [Google Scholar]
- 12.Kolega J., Kumar S. (1999) Cell Motil. Cytoskeleton 43, 255–268 [DOI] [PubMed] [Google Scholar]
- 13.Sohn R. L., Vikstrom K. L., Strauss M., Cohen C., Szent-Gyorgyi A. G., Leinwand L. A. (1997) J. Mol. Biol. 266, 317–330 [DOI] [PubMed] [Google Scholar]
- 14.Golomb E., Ma X., Jana S. S., Preston Y. A., Kawamoto S., Shoham N. G., Goldin E., Conti M. A., Sellers J. R., Adelstein R. S. (2004) J. Biol. Chem. 279, 2800–2808 [DOI] [PubMed] [Google Scholar]
- 15.Shohet R. V., Conti M. A., Kawamoto S., Preston Y. A., Brill D. A., Adelstein R. S. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 7726–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simons M., Wang M., McBride O. W., Kawamoto S., Yamakawa K., Gdula D., Adelstein R. S., Weir L. (1991) Circ. Res. 69, 530–539 [DOI] [PubMed] [Google Scholar]
- 17.Kubo T., Endo M., Hata K., Taniguchi J., Kitajo K., Tomura S., Yamaguchi A., Mueller B. K., Yamashita T. (2008) J. Neurochem. 105, 113–126 [DOI] [PubMed] [Google Scholar]
- 18.Wylie S. R., Chantler P. D. (2003) Mol. Biol. Cell 14, 4654–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolega J. (2006) Mol. Biol. Cell 17, 4435–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wylie S. R., Wu P. J., Patel H., Chantler P. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12967–12972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Even-Ram S., Doyle A. D., Conti M. A., Matsumoto K., Adelstein R. S., Yamada K. M. (2007) Nat. Cell Biol. 9, 299–309 [DOI] [PubMed] [Google Scholar]
- 22.Sandquist J. C., Swenson K. I., Demali K. A., Burridge K., Means A. R. (2006) J. Biol. Chem. 281, 35873–35883 [DOI] [PubMed] [Google Scholar]
- 23.Jana S. S., Kawamoto S., Adelstein R. S. (2006) J. Biol. Chem. 281, 24662–24670 [DOI] [PubMed] [Google Scholar]
- 24.Wylie S. R., Chantler P. D. (2008) Mol. Biol. Cell 19, 3956–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bao J., Jana S. S., Adelstein R. S. (2005) J. Biol. Chem. 280, 19594–19599 [DOI] [PubMed] [Google Scholar]
- 26.Sandquist J. C., Means A. R. (2008) Mol. Biol. Cell 19, 5156–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato M. K., Takahashi M., Yazawa M. (2007) Mol. Biol. Cell 18, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodge T. P., Cross R., Kendrick-Jones J. (1992) J. Cell Biol. 118, 1085–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dulyaninova N. G., Malashkevich V. N., Almo S. C., Bresnick A. R. (2005) Biochemistry 44, 6867–6876 [DOI] [PubMed] [Google Scholar]
- 30.Kelley C. A., Adelstein R. S. (1990) J. Biol. Chem. 265, 17876–17882 [PubMed] [Google Scholar]
- 31.Murakami N., Chauhan V. P., Elzinga M. (1998) Biochemistry 37, 1989–2003 [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg M., Ravid S. (2006) Mol. Biol. Cell 17, 1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straussman R., Even L., Ravid S. (2001) J. Cell Sci. 114, 3047–3057 [DOI] [PubMed] [Google Scholar]
- 34.Wei Q., Adelstein R. S. (2000) Mol. Biol. Cell 11, 3617–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Straussman R., Ben-Ya'acov A., Woolfson D. N., Ravid S. (2007) J. Mol. Biol. 366, 1232–1242 [DOI] [PubMed] [Google Scholar]
- 36.Even-Faitelson L., Ravid S. (2006) Mol. Biol. Cell 17, 2869–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Ya'acov A., Ravid S. (2003) J. Biol. Chem. 278, 40032–40040 [DOI] [PubMed] [Google Scholar]
- 38.Hostetter D., Rice S., Dean S., Altman D., McMahon P. M., Sutton S., Tripathy A., Spudich J. A. (2004) PLoS Biol. 2, e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami N., Kotula L., Hwang Y. W. (2000) Biochemistry 39, 11441–11451 [DOI] [PubMed] [Google Scholar]
- 40.Rovner A. S., Fagnant P. M., Lowey S., Trybus K. M. (2002) J. Cell Biol. 156, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kendrick-Jones J., Szent-Gyorgyi A. S., Cohen C. (1971) J. Mol. Biol. 59, 527–529 [DOI] [PubMed] [Google Scholar]
- 42.Lo C. M., Buxton D. B., Chua G. C., Dembo M., Adelstein R. S., Wang Y. L. (2004) Mol. Biol. Cell 15, 982–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark K., Middelbeek J., Dorovkov M. V., Figdor C. G., Ryazanov A. G., Lasonder E., van Leeuwen F. N. (2008) FEBS Lett. 582, 2993–2997 [DOI] [PubMed] [Google Scholar]
- 44.Martin A. F., Bhatti S., Pyne-Geithman G. J., Farjah M., Manaves V., Walker L., Franks R., Strauch A. R., Paul R. J. (2007) Am. J. Physiol. Cell Physiol. 293, C238–C245 [DOI] [PubMed] [Google Scholar]
- 45.Franke J. D., Dong F., Rickoll W. L., Kelley M. J., Kiehart D. P. (2005) Blood 105, 161–169 [DOI] [PubMed] [Google Scholar]
- 46.Breckenridge M. T., Dulyaninova N. G., Egelhoff T. T. (2009) Mol. Biol. Cell 20, 338–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikebe M., Komatsu S., Woodhead J. L., Mabuchi K., Ikebe R., Saito J., Craig R., Higashihara M. (2001) J. Biol. Chem. 276, 30293–30300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.