Abstract
The general stress response of Bacillus subtilis can be activated by stimuli such as the addition of salt or ethanol and with blue light. In the latter response, YtvA activates σB through a cascade of Rsb proteins, organized in stressosomes. YtvA is composed of an N-terminal LOV (light, oxygen, and voltage) domain and a C-terminal STAS (sulfate transporter and anti-sigma factor) domain and shows light-modulated GTP binding in vitro. Here, we examine the mechanism of YtvA-mediated activation of σB in vivo with site-directed mutagenesis. Constitutive off and constitutive on mutations have been identified. Disruption of GTP binding in the STAS domain eliminates light activation of σB. In contrast, modification of sites relevant for phosphorylation of STAS domains does not affect the stress response significantly. The data obtained are integrated into a model for the structure of full-length YtvA, which presumably functions as a dimer.
LOV2 domains (1), members of the superfamily of PAS domains (2, 3), are abundant in all domains of life and were first identified in plant phototropins (4). These photoreceptors regulate stomatal opening, phototropism, etc. and contain two N-terminal LOV domains that confer light regulation on the C-terminal Ser/Thr kinase domain (4). They also occur in bacteria, in which YtvA from Bacillus subtilis has been best characterized (for a review, see e.g. Ref. 5). Its N-terminal LOV domain binds FMN and shows the typical LOV photochemistry (6, 7): covalent adduct formation between a cysteine and the FMN chromophore. A linker helix, denoted Jα (7), connects the LOV domain to a STAS domain. The latter domain is present in many regulators of the general stress response of B. subtilis (8, 9). Stress via the addition of salt or ethanol (for a review, see Ref. 10) and blue light (11, 12) activates the general stress response via the environmental pathway, which integrates various signals via a large multiprotein complex, called the stressosome (13, 14). YtvA, which mediates light activation of σB (11, 12, 15), co-purifies with other STAS domain proteins in the stressosomes (16).
When cells are stressed, STAS domains of several stressosome proteins (e.g. RsbS and RsbR) are phosphorylated by another intrinsic stressosome component, the serine/threonine kinase RsbT (9, 14, 17, 18). Next, RsbT is released from the complex to trigger RsbU, a protein phosphatase, thus (indirectly) activating σB (19). Phosphorylation of YtvA, however, has never been detected. Rather, it has been demonstrated in vitro that YtvA shows light-dependent GTP binding, presumably at its NTP-binding site in the STAS domain (20).
Little is known about the mechanism of signal transmission in and by YtvA, except that in the C62A mutant, photochemistry in vitro (12) and light activation of σB in vivo (12, 15) are abolished. More detailed information is available for LOV domains of phototropins. A conserved glutamine, which is in hydrogen-bonding contact with the isoalloxazine ring of FMN, rotates its side chain by 180° upon covalent adduct formation (21). Replacement of this residue by leucine in the LOV2 domain of Phy3 from Adiantum results in a considerable reduction of the light-induced structural change (22). The corresponding mutation in phototropin 1 from Arabidopsis impairs autophosphorylation activity (23). The signal generated in the LOV2 domain is transmitted to the downstream kinase domain of phototropin 1 of Avena sativa through disruption of the interaction between its central β-sheet and the C-terminal linker region, the Jα-helix (24).
Here, we study the mechanism of activation of YtvA in vivo, i.e. light-induced activation of the σB response, with site-directed mutagenesis. We focus on three regions of the protein, the flavin-binding pocket, the β-sheet of the LOV domain, and the GTP-binding site, and on potential phosphorylation sites of the STAS domain. We demonstrate that light-activated GTP binding is crucial for functional YtvA. A computational approach was used to model the structure of full-length YtvA. The model suggests that light modulates accessibility of the GTP-binding site of the STAS domain of YtvA.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Genetic Manipulation
Strains and plasmids used in this study are listed in supplemental Table 1. Molecular genetic techniques and DNA manipulation were carried out using standard procedures. The ytvA knock-out strain PB565 was generously provided by Dr. C. Price (University of California, Davis, CA). Transformation of B. subtilis was carried out as described (25, 26) with transformant selection on 30 g/liter tryptic soy broth plates containing 10 μg/ml kanamycin at 37 °C.
To quantify the functionality of the YtvA, derivatives each of them were cloned in pDG148-Stu under control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible spac promoter and transformed to the ytvA knock-out strain PB565. The latter expresses β-galactosidase under control of the ctc promoter (Pctc-lacZ), which is regulated by σB (27).
Site-directed Mutagenesis
Mutants were constructed with pYtvA (see supplemental materials) and the QuikChange II site-directed mutagenesis kit (Stratagene) and verified by DNA sequencing. PCR reactions were carried out with Pfu DNA polymerase (Metabion or Stratagene) and the primers listed in supplemental materials.
Growth Conditions and β-Galactosidase Assays
Stress response experiments were carried out in tryptic soy broth, supplemented with 0.5% glucose, 5 μg/ml chloramphenicol, and 10 μg/ml kanamycin. For each strain, a single colony was inoculated and incubated at 37 °C overnight in a shaking water bath (250 rpm). Overnight cultures were diluted 100-fold and grown exponentially. Then, they were diluted 10-fold and distributed over a series of Erlenmeyer flasks, and incubation was continued in light and dark (flasks wrapped tightly in tinfoil). At around A600 = 0.3, IPTG (final concentration: 1 mm) was added to induce YtvA expression. Culture samples were taken in the light and with minimal amounts of red light as a dark control. A600 of each sample was measured, and part of each sample was flash-frozen in liquid N2 for storage at −80 °C. β-Galactosidase activity was measured and expressed in Miller units as described previously (28).
Western Analysis
Western blots of B. subtilis cell extracts were made using a standard protocol and a dilution of 1:1000 of a polyclonal antiserum (Eurogentec) elicited against polyhistidine-tagged YtvA.
YtvA Purification from Escherichia coli
The coding region of ytvA (gene ID: 937254) was cloned in the expression vector pQE30 (Qiagen) using pQE30YtvA-RV and -FW (see supplemental materials). E. coli M15/pQE30YtvA was grown in production broth (20 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter glucose, 5 g/liter NaCl, 8.7 g/liter K2HPO4, pH 7) at 37 °C in a water bath shaker in the dark, with agitation at 250 rpm. IPTG (1 mm final concentration) was added to exponentially growing cells (A600 = 0.6), which were harvested 3 h later. Polyhistidine-tagged YtvA was purified from cell-free extracts with a HisTrap HP column (Amersham Biosciences) and an ÄKTATM liquid chromatography system using a linear imidazole gradient (from 25 to 500 mm) in 500 mm NaCl plus 20 mm Tris, pH 8.0, to elute the protein from the HisTrap column. After overnight dialysis in 20 mm Tris, pH 8.0, in the dark, the purity of the protein was checked by overloading a Phast SDS gel system.
Illumination
Light refers to white light, from a compact fluorescent lamp. Moderate white light intensities were selected ranging from 25 to 35 microEinsteins · m−2.s−1.
Modeling the Three-dimensional Structure of Full-length YtvA
The crystal structure of the LOV domain of YtvA, plus its helical extension (residues 21–147), has recently been resolved (Protein Data Bank (PDB) code 2PR5 (7)). For the STAS domain, we performed homology modeling using MODELLER version 8.2 (29). The crystal structure of the anti-sigma factor antagonist RsbS from Moorella thermoacetica (PDB code 2VY9 (16)) served as a template, from which the positions of the main chain atoms were retained. Side chains were grown back using sequence alignment (see supplemental Fig. 1) and were subsequently optimized. The STAS domain (residues 148–261) was then docked onto the LOV domain using the Gramm protein docking software (30). Gramm minimizes the intermolecular energy between two protein domains by changing the range of the interatomic potentials. To further optimize the docking of the two protein domains regarding their mutual orientation, we used lowest energy conformation from the Gramm docking procedure as input for the Rosetta server (31). From the 10 lowest energy conformations, we selected the structure with the smallest distance between the LOV C terminus (Ser147) and the STAS N terminus (Thr148). We then removed the coordinates of four amino acids located in the linker (Ala145 to Thr148) and used this structure as a template in another homology modeling procedure to link the two domains. This resulted in five models, from which we selected the conformation in which no atoms would overlap with the FMN co-factor (which the program does not preclude). To further relax the structure of full-length YtvA, we performed energy minimization and molecular dynamics simulations using the GROMACS software package, version 3.3.3 (32). Using the G43a1 force field (33, 34), which includes parameters for FMN, polar and aromatic hydrogens were added to the protein, after which it was energy-minimized. The resulting structure was then solvated in a cubic periodic box of 8092 simple point charge (SPC) water molecules (35) followed by 20 ps of equilibration of water and hydrogen atoms. We then performed 200 ps of molecular dynamics to equilibrate the full system. This was enough to optimize the interaction energy between the protein and the water. The conformation at 187 ps had the lowest protein-water interaction energy and was used as a model for the structure of YtvA to facilitate interpretation of experimental results.
Sequence Analysis
Sequences were aligned with ClustalW (36). Secondary structure predictions and hydrophobic profile analyses were made with Jpred (37) and pepwindowall, respectively. Prediction of coiled-coil conformation was done with PCOIL (38).
RESULTS
Optimal Assay Conditions for the YtvA Mutants
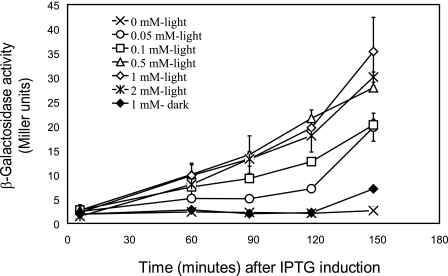
To assay the in vivo activity of YtvA derivatives, each mutant protein was overproduced in a ΔytvA strain containing a genome-encoded σB-reporter (see “Experimental Procedures”). First the optimal conditions for the in vivo analysis of YtvA activity were determined by variation of IPTG concentration and the induction period. Fig. 1 shows that σB activation in a strain that overproduces wild type YtvA occurs only in the presence of light and IPTG. YtvA- and light-dependent σB activation increase linearly in time, up to 120 min after IPTG addition, after which cells, because of entry into stationary phase, also start to activate σB in the dark. Furthermore, optimal YtvA expression and σB activation were observed at concentrations higher than 0.5 mm IPTG. Thus, all YtvA-mutants were then tested for σB light activation at 120 min after the addition of 1 mm of IPTG (final concentration). A positive (overproduction of wild type YtvA), a negative (use of plasmid pDG148-Stu without insert), and a dark control (overproduction of a YtvA variant in the dark) were always included.
FIGURE 1.
Optimal conditions for in vivo analysis of YtvA function. Exponentially growing cells of the σB reporter strain that contains Pctc-lacZ, as well as pYtvA, were incubated in the light (open symbols) or dark (closed symbols) with increasing concentrations of IPTG. The x axis shows the time after the addition of 0 (×), 0.05 (○), 0.1 (□), 0.5 (▵), 1 (♢), (♦), and 2 mm (∗) IPTG. The y axis shows the β-galactosidase activity (in Miller units) at the different IPTG concentrations. Error bars indicate S.E. from two duplicate samples.
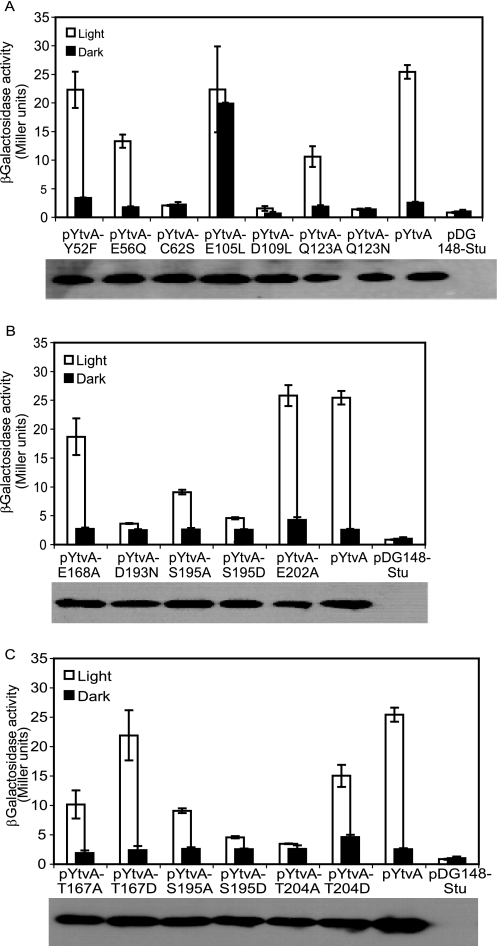
A Western blot was made for each sample to assay the level of YtvA production for each mutant. This revealed that the expression level of each mutant was comparable with that of the wild type (Fig. 2, bottom of each panel), which allows one to conclude that differences found in activities are due to the intrinsic characteristics of each mutant protein.
FIGURE 2.
Effect of mutations in LOV and STAS domains of YtvA on the expression of the σB reporter gene, expressed as Miller units, of the ytvA-overexpressing strains measured 120 min after IPTG addition. In each panel, wild type YtvA and an empty vector (pDG148-Stu) control were also included. Inset of each panel: Western blot analysis of the level of overexpression of each mutant protein. A, mutations in LOV domain as follows: flavin-binding pocket, C62S, Q123A, and Q123N; central β-sheet, E105L and D109L; conserved salt bridge and its surroundings, E56Q and Y52F. B, mutations in STAS domain as follows: GTP binding, D193N, S195A, and S195D; negative charged residues, E168A and E202A. C, mutations in potential phosphorylation sites of STAS domain as follows: T167A, T167D, S195A, S195D, T204A, and T204D.
Primary Photochemistry
It has been proposed that after adduct formation between the sulfur atom of Cys62 and the C4(a) of FMN, a conserved glutamine from the flavin-binding pocket (Gln123 in YtvA) rotates its side chain by 180°, thus altering the hydrogen bonding network around the FMN (7). However, due to limited resolution in the crystallography data of YtvA, this rotation could not be resolved unequivocally.
To confirm and investigate the role of Cys62 and Gln123, respectively, in the function of YtvA, the activity of the C62S, Q123A, and Q123N variants was measured in vivo. As reported earlier (15), mutant C62S is unable to activate the σB response (Fig. 2A). Q123A, which lacks one hydrogen bond with FMN, reduces the activity of YtvA in the light by around 50% (Fig. 2A). Moreover, its replacement by asparagine, which has a similar but shorter side chain, fully abolishes light-induced σB activation (Fig. 2A).
A Conserved Salt Bridge in the LOV Domain Is Dispensable for Biological Activity of YtvA
The PAS domains of YtvA, FixL, and the human eag-related gene (hERG) potassium channel all contain a conserved salt bridge (2) at their surface between Glu56 and Lys97 (Fig. 3). This salt bridge is located in the region of YtvA-LOV undergoing the most pronounced light-induced structural change (7), although the two residues themselves show only minor rearrangements. Disruption of the salt bridge in the E56Q mutant slows down the dark state recovery rate in vitro by a factor of 2 (39). However, in vivo, this mutant shows a similar extent of light activation of σB as the wild type (Fig. 2A). Furthermore, changes of nearby residues such as in the Y52F mutant do not change light activation of σB either.
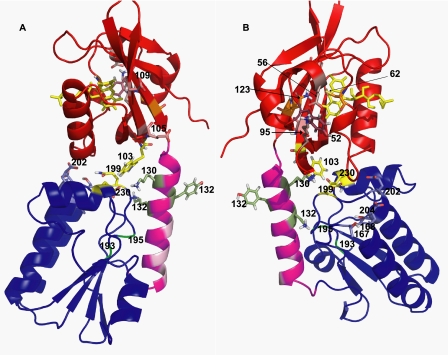
FIGURE 3.
Ribbon representation of the dark state of the full-length YtvA. Parts A and B represent two views of YtvA, before and after 120° rotation around its longest axis. Numbers indicate the position of the relevant residues. Red, LOV21–125. Magenta, linker region 126–147. Dark blue, STAS148–261. Light red, charged amino acids in the central β-sheet of the LOV domain. Light magenta, hydrophobic residues in the linker region. Bright green, potential GTP-binding site (DSLG). Green, residues Tyr132 and Lys134. Yellow, Trp130 and FMN in the LOV domain, Lys130 in the linker region, and Gln199 and Glu230 in the STAS domain.
Role of the Central β-Sheet of the LOV Domain
YtvA-LOV has the common PAS fold, with five antiparallel β-strands and four α-helices forming its core and a C-terminal Jα-helix that extends outwards from the domain (7). Disruption of the interaction between the central β-strand (Hβ) and Jα of phot1 from A. sativa is known to be a key event in signal transduction (24). Glu105 and Asp109 are located in the Hβ of the YtvA-LOV and form intermolecular contacts in the dimeric form of YtvA-LOV (7). Exchange of Glu105 by leucine results in light-independent σB activation (Fig. 2A). Replacement of yet another charged residue from the same β-strand, i.e. Asp109, by leucine completely abolished the light-induced σB response (Fig. 2A).
The GTP-binding Site of the STAS Domain
STAS domains of several proteins, including YtvA and SpoIIAA from B. subtilis, bind ATP and/or GTP (40). Based on sequence analysis, a putative GTP-binding site (i.e. 193DLSG196) has been proposed for YtvA (20). This sequence is localized at the end of the central β-strand and the beginning of a coil of the STAS domain (Fig. 3, bright green). Replacement of Asp193 by an asparagine completely abolishes σB activation, just as the S195D mutation (Fig. 2B). In addition, light activation of σB in the S195A mutant is reduced by about 70% (Fig. 2B).
STAS Domain Phosphorylation
Many STAS domains are phosphorylated upon environmental stress (18). Until now, however, no evidence has been provided that this holds for the STAS domain of YtvA, too. This may be because the equivalent residues from e.g. RsbS and RsbRA are not present in YtvA. YtvA has a negatively charged residue (i.e. Glu168 and Glu202; Fig. 3) at these positions, which might mimic a phosphoryl group (9). Significantly, replacing either of these glutamate residues by an alanine does not abolish light activation of σB (Fig. 2B), although E168A reduces it by 15%. Moreover, by careful inspection of the alignment of STAS domains, two threonines, Thr167 and Thr204, can be identified in YtvA (Fig. 3) that are one or two positions away from conserved phosphorylation sites (supplemental Fig. 2). Substitution of Thr167 by alanine decreases σB activation in response to light with ∼50%, and in the T204A mutant, the light response is abolished (Fig. 2C). Replacement of either of these two residues by the aspartic acid, which may mimic the phosphorylated (i.e. active) state, does not change the activation of σB by YtvA neither in the light nor in darkness (Fig. 2C). Furthermore, SpoIIAA, a STAS domain-containing protein that is involved in sporulation of B. subtilis, is also phosphorylated upon activation (41). This phosphorylation, however, occurs at the serine corresponding to Ser195 of YtvA, which is part of the potential GTP-binding site of YtvA. The S195A mutation reduces the light activation of σB with ∼70% (Fig. 2C).
Modeling the Structure of Full-length YtvA
To facilitate molecular interpretation of our in vivo results, we made a three-dimensional model of a monomer of YtvA using a combination of homology modeling, protein docking, and molecular dynamics simulations (Fig. 3). The STAS domain contains a central four-stranded parallel β-sheet with three helices connecting the strands on one side of the sheet and only one, the C-terminal helix, on the other side. The linker helix flanks the central β-sheet of the STAS domain, which contains the GTP-binding site (Fig. 3, bright green), and it is close to the STAS C-terminal helix. The loops connecting the C-terminal ends of the β-strands to the connecting helices interact with strands 3 and 4 and with helix 4 of the LOV domain. The interactions between the two domains are mostly electrostatic. Several side chains in the linker helix form hydrogen bonds with the backbone atoms of the STAS β-sheet, whereas also several interdomain salt bridges and hydrogen bonds are formed. The side chain of Trp103 interacts with CH2 groups of Lys130, Gln199, and Glu230 (Fig. 3, yellow and green). During the molecular dynamics equilibration, it is possible to observe that hydrogen bonds in the LOV-STAS interface transiently interact with water and form water-mediated hydrogen bonds.
DISCUSSION
YtvA Structure in Vivo
STAS domain proteins often are dimeric (7, 16, 42, 43). Based on electron microscopy data of reconstituted stressosomes, Marles-Wright et al. (16) recently postulated that the RsbR paralogs from B. subtilis, including YtvA, are also incorporated into stressosomes as dimers, such that STAS domain dimers form the core of the stressosome, to which the dimeric N-terminal domain is linked via a helical region. Although the model of YtvA shown in Fig. 3 is that of a monomer, the functionally active form of YtvA in vivo presumably is that of a dimer. Our mutagenesis data are consistent with this; modification in Hβ of YtvA-LOV, which presumably alters the dimer interface, interferes with light activation of the σB response.
In our model, the association of the two domains of YtvA is not primarily driven by shielding of hydrophobic groups from contact with water in the interdomain region. This is because the LOV and STAS domain mostly interact through hydrogen bonds. This suggests that the conformation of the linker helix that connects the two domains plays a more important role in the folding of full-length YtvA than the interactions at the interface between the two separate domains.
Signal Generation in YtvA-LOV
The covalent-adduct state in YtvA is formed from the triplet excited state and relaxes back to the dark state in approximately 1 h (6, 7), making YtvA one of the slowest in recovering in the LOV domain family (5). C62S, in contrast to C62A, cannot form the FMN triplet state (Ref. 44 and data not shown). Earlier in vivo studies of YtvA, in which the illumination conditions were not specified, showed that neither C62A nor C62S could activate σB with light, not even upon overproduction of YtvA (15). Here, we show that the C62S mutation also completely abolishes light activation of σB at saturating light intensities (Fig. 2A). Therefore, generation of the biological signal has to be preceded by covalent adduct formation in YtvA.
Although in the adduct state the sulfur atom of Cys62 lifts C(4a) out of the plane of the flavin ring, additional rearrangement of side chains in the chromophore-binding pocket is induced by light absorption (7). Gln123, which hydrogen-bonds its amide group to O-4 of FMN in the dark, rotates its side chain upon illumination so that this amide group in the lit state forms a hydrogen bond with N-5 of FMN. Significantly, our in vivo studies show that the YtvA mutant Q123A does show light activation of σB, although ∼50% less than wild type (Fig. 2A). It also shows a blue shift in its dark state spectrum (data not shown), implying a change in the electronic environment of FMN in that state. Similar results were observed in full-length phototropin 1 from Arabidopsis thaliana. Its Q575L mutant shows reduced autophosphorylation in the light and a blue shift (∼10 nm) of the absorption maximum of the dark state (23). Shortening the side chain of Gln123 (i.e. the Q123N mutant) locks it in a biologically inactive state (Fig. 2A). These results emphasize that, as in phototropins, alteration of the hydrogen bonding between Gln123 and FMN is a key step for light-induced signal generation in YtvA.
Potential Phosphorylation Sites of YtvA-STAS
Several STAS domain proteins that regulate σB are an integral part of the stressosomes and become phosphorylated upon exposure of B. subtilis to stress (18). So far no evidence was provided that the STAS domain of YtvA can be phosphorylated. Conserved phosphorylation sites (i.e. T/S residues), present in RsbS and RsbR, are absent in YtvA. The latter, instead, contains two negatively charged residues (i.e. Glu168 and Glu202; Fig. 3) at nearby positions. It has been proposed that these latter residues may take over the function of the phosphorylatable residues (18). Our results show that exchange of these glutamates by alanine does not significantly affect light activation of σB (Fig. 2B). Furthermore, substitution of a nearby threonine by an aspartic acid (mimicking the negative charge of a phosphyl group) does not lead to constitutive dark activation of σB. We therefore conclude that phosphorylation of YtvA is not required for σB activation. The impaired σB activation in the T204A mutant presumably is due to the inability to form the stabilizing hydrogen bond between Thr204 and Asn201.
GTP Binding of YtvA
GTP binds to YtvA in vitro, such that the affinity of this binding is modulated by light (20). Here, we demonstrate that disruption of GTP binding (i.e. S195A/S195D and D193N) abolishes light activation of σB (Fig. 2B), which reveals the importance of this nucleotide binding for the biological function of YtvA. We therefore postulate that YtvA serves as a light-dependent NTP recruiter for the RsbT kinase. The properties of the E105L mutant of YtvA support this notion. This mutant binds GTP as well as wild type YtvA but does so independent of light (45),3 consistent with the constitutive activation of this mutant in vivo (Fig. 2A). Modification of residues Lys130 and Lys134 from the linker helix (Fig. 3, green) was recently demonstrated to increase the affinity of YtvA for GTP but with a simultaneous loss of its light dependence3. The effect of these mutations on σB activation in vivo still remains to be tested.
Interdomain Signal Transmission in YtvA
A recent study of engineered hybrid proteins, composed of the LOV domain of YtvA and the histidine kinase domain of FixL and connected via Jα-type helices of different lengths, has revealed that signals are transmitted from the sensory LOV domain to the histidine kinase domain via a 40–60° rotational movement within the α-helical coiled-coil linker (47). Therefore, these hybrid proteins act as light-triggered rotary switches (7, 47).
YtvA and FixL both contain an N-terminal PAS domain, connected to a STAS domain and a histidine kinase domain, respectively, via a Jα-type helix (7). Interestingly, many histidine kinases that have a PAS domain as signal input domain, show conservation of the connecting region between the PAS domain and the Jα-helix (47). This conserved sequence signature (125DIT127) is also present in YtvA (Fig. 4) and is stabilized by a salt bridge between Asp125 and Lys96 and by a hydrogen bond between Thr127 and the backbone of Trp103 (Fig. 3). Furthermore, just next to these stabilizing interactions, Lys97 forms a conserved salt bridge with Glu56. Although one would expect that these interactions provide the necessary structural rigidity for transmission of a light signal from the LOV domain, through the linker helix to the STAS domain, disruption of the Glu56–Lys97 salt bridge only slightly impairs light activation of the σB response (Fig. 2A). The stabilizing role of this salt bridge, however, may be complemented by the Lys96–Asp127 salt bridge. It would therefore be of interest to test double mutants in this region in the future.
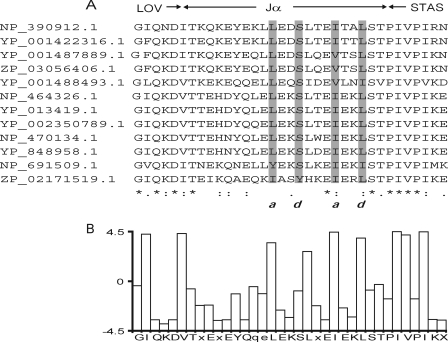
FIGURE 4.
A, alignment of the Jα of LOV-STAS containing proteins. Gray boxes, amino acids with hydrophobic and side chains. Letters a and d indicate hydrophobic side chains and correspond to the repeated heptapeptide motif, characteristic for coiled-coil conformation (a, b, c, d, e, f, and g) (46). B, bar graph that represents the average hydropathy of the residues in the sequence alignment. The legend at the x axis indicates a conserved sequence. GenBankTM numbers are as follows: NP_390912.1, B. subtilis; YP_001422316.1, Bacillus amyloliquefaciens FZB42; YP_001487889.1, Bacillus pumilus SAFR-032, ZP_03056406.1, B. pumilus ATCC 7061; YP_001488493.1, B. pumilus SAFR-032; NP_464326.1, Listeria monocytogenes EGD-e; YP_013419.1, L. monocytogenes str. 4b F2365; YP_002350789.1, L. monocytogenes HCC23; NP_470134.1, Listeria innocua Clip11262; YP_848958.1, Listeria welshimeri serovar 6b str. SLCC5334; NP_691509.1, Oceanobacillus iheyensis HTE831; ZP_02171519.1, Bacillus selenitireducens MLS10.
A closer inspection of the sequence of the YtvA-Jα-region (residues 126–150), using PCOILS (38), indicates that the linker region may form a coiled-coil structure, too. Additionally, by examining an alignment of LOV-STAS proteins, it is possible to recognize the typical hydrophobic pattern (Fig. 4) that defines a coiled-coil structure, just as in the Jα-helix between several PAS domains and their cognate histidine kinase domain (47). Based on the conserved sequences pattern between the above mentioned hybrid protein and YtvA-Jα, we anticipate that the STAS domain is activated via a coiled-coil structure of dimeric YtvA, in a way similar to what has been described for PAS domain containing histidine kinases. The signal is transmitted through the linker helix by a rotary movement, which then promotes GTP binding in YtvA and ultimately activates σB. Such a rotary movement can also explain the reported decrease in tyrosine fluorescence, with concomitant increase in fluorescence from Trp103 (48). We propose that Tyr132 (located in the linker helix) is the residue responsible for the underlying fluorescence energy transfer.
The knowledge gained in this study allows for formulation of exciting new working hypotheses, like that of the functionality of “giraffe” variants of YtvA. This hypothesis is now under investigation.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and supplemental Table 1.
Y. Tang, E. Livoti, Z. Cao, U. Krauss, K.-E. Jaeger, W. Gärtner, and A. Losi, submitted for publication.
- LOV
- light, oxygen, and voltage
- PAS
- Per-ARNT-Sim
- Rsb
- regulator of sigma factor B
- STAS
- sulfate transporter and anti-sigma factor
- FMN
- flavin mononucleotide
- IPTG
- isopropyl-β-d-thiogalactopyranoside.
REFERENCES
- 1.Huala E., Oeller P. W., Liscum E., Han I. S., Larsen E., Briggs W. R. (1997) Science 278, 2120–2123 [DOI] [PubMed] [Google Scholar]
- 2.Taylor B. L., Zhulin I. B. (1999) Microbiol. Mol. Biol. Rev. 63, 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pellequer J. L., Wager-Smith K. A., Kay S. A., Getzoff E. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5884–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie J. M., Reymond P., Powell G. K., Bernasconi P., Raibekas A. A., Liscum E., Briggs W. R. (1998) Science 282, 1698–1701 [DOI] [PubMed] [Google Scholar]
- 5.Losi A. (2007) Photochem. Photobiol. 83, 1283–1300 [DOI] [PubMed] [Google Scholar]
- 6.Losi A., Polverini E., Quest B., Gärtner W. (2002) Biophys. J. 82, 2627–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Möglich A., Moffat K. (2007) J. Mol. Biol. 373, 112–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind L., Koonin E. V. (2000) Curr. Biol. 10, R53–55 [DOI] [PubMed] [Google Scholar]
- 9.Akbar S., Gaidenko T. A., Kang C. M., O'Reilly M., Devine K. M., Price C. W. (2001) J. Bacteriol. 183, 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecker M., Pané-Farré J., Völker U. (2007) Annu. Rev. Microbiol. 61, 215–236 [DOI] [PubMed] [Google Scholar]
- 11.Avila-Pérez M., Hellingwerf K. J., Kort R. (2006) J. Bacteriol. 188, 6411–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki N., Takaya N., Hoshino T., Nakamura A. (2007) J. Gen. Appl. Microbiol. 53, 81–88 [DOI] [PubMed] [Google Scholar]
- 13.Marles-Wright J., Lewis R. J. (2007) Curr. Opin. Struct. Biol. 17, 755–760 [DOI] [PubMed] [Google Scholar]
- 14.Delumeau O., Chen C. C., Murray J. W., Yudkin M. D., Lewis R. J. (2006) J. Bacteriol. 188, 7885–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaidenko T. A., Kim T. J., Weigel A. L., Brody M. S., Price C. W. (2006) J. Bacteriol. 188, 6387–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marles-Wright J., Grant T., Delumeau O., van Duinen G., Firbank S. J., Lewis P. J., Murray J. W., Newman J. A., Quin M. B., Race P. R., Rohou A., Tichelaar W., van Heel M., Lewis R. J. (2008) Science 322, 92–96 [DOI] [PubMed] [Google Scholar]
- 17.Kang C. M., Vijay K., Price C. W. (1998) Mol. Microbiol. 30, 189–196 [DOI] [PubMed] [Google Scholar]
- 18.Kim T. J., Gaidenko T. A., Price C. W. (2004) J. Bacteriol. 186, 6124–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delumeau O., Dutta S., Brigulla M., Kuhnke G., Hardwick S. W., Völker U., Yudkin M. D., Lewis R. J. (2004) J. Biol. Chem. 279, 40927–40937 [DOI] [PubMed] [Google Scholar]
- 20.Buttani V., Losi A., Polverini E., Gärtner W. (2006) FEBS Lett. 580, 3818–3822 [DOI] [PubMed] [Google Scholar]
- 21.Crosson S., Moffat K. (2002) Plant Cell 14, 1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nozaki D., Iwata T., Ishikawa T., Todo T., Tokutomi S., Kandori H. (2004) Biochemistry 43, 8373–8379 [DOI] [PubMed] [Google Scholar]
- 23.Jones M. A., Feeney K. A., Kelly S. M., Christie J. M. (2007) J. Biol. Chem. 282, 6405–6414 [DOI] [PubMed] [Google Scholar]
- 24.Harper S. M., Christie J. M., Gardner K. H. (2004) Biochemistry 43, 16184–16192 [DOI] [PubMed] [Google Scholar]
- 25.Kunst F., Rapoport G. (1995) J. Bacteriol. 177, 2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bongers R. S., Veening J. W., Van Wieringen M., Kuipers O. P., Kleerebezem M. (2005) Appl. Environ. Microbiol. 71, 8818–8824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boylan S. A., Rutherford A., Thomas S. M., Price C. W. (1992) J. Bacteriol. 174, 3695–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenney T. J., Moran C. P., Jr. (1987) J. Bacteriol. 169, 3329–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 30.Tovchigrechko A., Vakser I. A. (2005) Proteins 60, 296–301 [DOI] [PubMed] [Google Scholar]
- 31.Lyskov S., Gray J. J. (2008) Nucleic Acids Res. 36, W233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 33.Daura X., Mark A., Van Gunsteren W. (1998) J. Comput. Chem. 19, 535–547 [Google Scholar]
- 34.van Gunsteren W. F., Billeter S. R., Eising A. A., Hünenberger P. H., Krüger P., Mark A. E., Scott W. R. P., Tironi I. G. (1996) Biomolecular Simulation: The GROMOS96 Manual and User Guide, Hochschuleverlag AG an der ETH Zürich, Zürich, Switzerland [Google Scholar]
- 35.Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Hermans J. (1981) in Intermolecular Forces ( Pullman B. ed) pp. 331–342, Reidel, Dordrecht, The Netherlands [Google Scholar]
- 36.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 37.Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biegert A., Mayer C., Remmert M., Söding J., Lupas A. N. (2006) Nucleic Acids Res. 34, W335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Losi A., Ghiraldelli E., Jansen S., Gärtner W. (2005) Photochem. Photobiol. 81, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 40.Najafi S. M., Harris D. A., Yudkin M. D. (1996) J. Bacteriol. 178, 6632–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Najafi S. M., Willis A. C., Yudkin M. D. (1995) J. Bacteriol. 177, 2912–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quin M., Newman J., Firbank S., Lewis R. J., Marles-Wright J. (2008) Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 64, 196–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray J. W., Delumeau O., Lewis R. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17320–17325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drepper T., Eggert T., Circolone F., Heck A., Krauss U., Guterl J. K., Wendorff M., Losi A., Gärtner W., Jaeger K. E. (2007) Nat. Biotechnol. 25, 443–445 [DOI] [PubMed] [Google Scholar]
- 45.Buttani V., Gärtner W., Losi A. (2007) Eur. Biophys. J. 36, 831–839 [DOI] [PubMed] [Google Scholar]
- 46.McLachlan A. D., Stewart M. (1975) J. Mol. Biol. 98, 293–304 [DOI] [PubMed] [Google Scholar]
- 47.Möglich A., Ayers R. A., Moffat K. (2009) J. Mol. Biol. 385, 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Losi A., Ternelli E., Gärtner W. (2004) Photochem. Photobiol. 80, 150–153 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.