Abstract
The formation of new blood vessels from pre-existing ones requires highly coordinated restructuring of endothelial cells (EC) and the surrounding extracellular matrix. Directed EC migration is a central step in this process and depends on cellular signaling cascades that initiate and control the structural rearrangements. On the basis of earlier findings that ERK5 deficiency in mouse EC results in massive defects in vessel architecture, we focused on the impact of the MEK5/ERK5 signaling pathway on EC migration. Using a retroviral gene transfer approach, we found that constitutive activation of MEK5/ERK5 signaling strongly inhibits EC migration and results in massive morphological changes. The area covered by spread EC was dramatically enlarged, accompanied by an increase in focal contacts and altered organization of actin filaments. Consequently, cells were more rigid and show reduced motility. This phenotype was most likely based on decreased focal contact turnover caused by reduced expression of p130Cas, a key player in directed cell migration. We demonstrate for the first time that ERK5 signaling not only is involved in EC survival and stress response but also controls migration and morphology of EC.
Directed migration of EC2 is a prerequisite for angiogenic processes in embryonic development and wound healing (1). This process is divided into six phases (for review, see Lamalice et al. (2)): (i) sensing of a chemoattractant, (ii) extension and protrusion at the leading edge, (iii) attachment by assembly of focal contacts, (iv) contraction of the cell body by formation of stress fibers, (v) release of the rear edge by disassembly of focal contacts, and (vi) recycling of adhesive and signaling components. All six stages are highly regulated by a variety of signaling cascades.
MAPK pathways play a crucial role in converting extracellular signals into a variety of intracellular changes, including alteration of cell structure, metabolism, and gene expression. The most recently identified member of the MAPK family is ERK5, also known as big MAPK1 (BMK1) (3). Similar to ERK1/2, ERK5 contains a TEY consensus MEK phosphorylation motif and is activated by mitogens and growth factors. However, although ERK5 is also important for the regulation of cell proliferation, cell survival, and cell differentiation, it differs from ERK1/2 in several aspects (4). First, ERK5 is the only MAPK that is activated not only by mitogens but also by stress conditions such as hyperosmolarity. Furthermore, ERK5 exhibits a long C-terminal regulatory domain exerting transactivating transcriptional properties (5) that distinguish this kinase from any other MAPK. Finally, the TEY motif of ERK5 is not phosphorylated by the MEK1/2 MAPK kinases but by a pathway-specific kinase named MEK5. One of the major downstream targets of ERK5 is MEF2C (myocyte enhancer factor 2C) (6), a transcription factor that has been shown to be important for EC survival (5, 7). ERK5 knock-out mice die at day 10.5 during embryonic development due to severe cardiovascular defects and angiogenic failure in embryonic and extra-embryonic tissues (4, 8, 9). It was shown recently that ERK5 inhibits apoptosis of EC in vitro (7) in a MEF2C-dependent manner, and endothelium-specific knock-out studies showed that ERK5 plays an important role in vascular integrity in vivo (10, 11). Nevertheless, the detailed mechanisms that underlie these processes still remain unknown.
In this study, we address the question of how selective modulation of the MEK5/ERK5 pathway in EC influences the cellular processes involved in EC migration.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following antibodies were used throughout this study: rabbit polyclonal anti-phospho-FAK Tyr925 and mouse monoclonal anti-phospho-ERK (Cell Signaling Technologies); monoclonal anti-phospho-FAK Tyr397 and rabbit polyclonal anti-phospho-FAK Tyr861 (BD Biosciences); mouse monoclonal anti-FAK (Transduction Laboratories); rabbit polyclonal anti-ERK1 and anti-ERK2 (Santa Cruz Biotechnology, Inc.); rabbit anti-human p130Cas, rabbit anti-human p38, rabbit anti-human phospho-p38 Thr180/Tyr182, and rabbit anti-hemagglutinin (Sigma); and rabbit polyclonal anti-ERK5 (Upstate Biotechnology). Mouse monoclonal anti-human integrin α5, β1, and αvβ3 and mouse anti-human phospho-JNK Thr183/Tyr185 antibodies were from BD Biosciences.
Cell Culture
Primary human EC derived from umbilical veins were obtained from Clonetics (via Cell Systems, St. Katharinen, Germany) and PromoCell (Heidelberg, Germany). Cells were grown in HUVEC growth medium and used between passages 3 and 5 as described previously (12). Amphotropic retrovirus producer cells (ϕNXampho) were a gift from G. Nolan (Stanford School of Medicine, Stanford, CA). The culturing conditions used in this work were described previously (13). NIH3T3 mouse fibroblasts were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum.
Transfection of EC with siRNAs
HUVEC (1.2 × 105/well) were seeded in 6-well plates and cultured for 24 h. Cells were transfected with siRNA (ERK5 sense (AGCTGCCCTGCTCAAGTCT), p130Cas sense (GGTCGACAGTGGTGTGTAT), or fluorescein isothiocyanate-labeled control siRNA (Qiagen)) using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Knockdown efficiencies were determined by Western blot analysis 24–72 h post-transfection.
Viral Gene Transfer
Retroviral expression plasmids for constitutively active MEK5DD or dominant-negative MEK5AA mutants were generated by cloning rat MEK5 cDNA with site-directed mutations (S311D/T315D or S311A/T315A, respectively) into the pCFG5-IEGZ retroviral vector (14). Amphotropic ϕNX retrovirus producer cells were transfected and selected with 250 μg/ml Zeocin for 12 days to gain stable producer cells. Virus-containing supernatants were harvested from 5 × 106 ϕNX cells 48 and 72 h after seeding, complemented with 4 μg/ml Polybrene, and added to 2 × 105 HUVEC plated 24 h before on a 100-mm dish. Transduction efficacy was measured by flow cytometric analysis of EGFP expression. The EF.p130Cas.CMV.RFP plasmid was generated by inserting human p130Cas cDNA into the EcoRV-restricted EF.CMV.RFP vector (Addgene) (15–17). For production of recombinant lentiviral particles, 5 × 106 HEK293T cells were cotransfected with 10 μg of expression plasmid EF.CMV.RFP along with 7.5 μg of packaging plasmid psPAX2 (Addgene; coding for human immunodeficiency virus gag-pol) and 2.5 μg of envelope plasmid pMD2.G (Addgene; coding for vesicular stomatitis virus G) (18) using Lipofectamine 2000 (Invitrogen). 24 h after transfection, the medium was replaced with 6 ml of HUVEC medium. HUVEC were infected as described for retroviral transduction.
Lentiviral Transfer of shRNA
Lentiviral shRNA constructs were created using pLVTHM (Addgene) (19). The shRNA hairpin was generated by annealing the sense and antisense oligonucleotides for ERK5-specific shRNA (sense, CGCGTCCCCAGCTGCCTCGCTCAAGTCTTTCAAGAGAAGACTTGAGCAGGGCAGCTTTTTTGGAAAT; and antisense, CGATTTCCAAAAAAGCTGCCCTGCTCAAGTCTTCTCTTGAAAGACTTGAGCAGGGCAGCTGGGGA) and for scrambled control shRNA (sense, CGCGTCCCCGCGAGCTGCCCATTCATCTTTCAAGAGAAGATGAATGGGCAGCTCGCTTTTTGGAAAT; and antisense, CGATTTCCAAAAAGCGAGCTGCCCATTCATCTTCTCTTGAAAGATGAATGGGCAGCTCGCGGGGA). The efficiency of lentiviral shRNA transfer was measured by flow cytometric analysis of coexpressed EGFP.
Proliferation Assay
HUVEC (5 × 104) were seeded and cultured in 6-well plates. EC were trypsinized, stained with trypan blue, and counted with a Neubauer chamber at the times indicated.
Attachment Assay
Attachment assays were performed as described previously (20). Briefly, 96-well plates were coated overnight at 4 °C with different concentrations of fibronectin and blocked thereafter with 1% BSA for 1 h at room temperature. Wells coated with only BSA served as negative controls. HUVEC were trypsinized and washed once in Dulbecco's modified Eagle's medium containing 0.5% BSA, and 5 × 104 cells were added in 50 μl of Dulbecco's modified Eagle's medium with 0.5% BSA to each well. After incubation for 60 min at 37 °C, cells were washed three times with phosphate-buffered saline using an automatic washer, and the amount of attached cells was measured by acidic phosphatase activity.
Cell Migration Assay
Cell migration studies were performed mainly as described (20). Briefly, 48-well plates were coated with fibronectin (20 μg/ml) and blocked with 1% BSA. HUVEC (2 × 105) that were starved in MCDB131 (PAA) with 0.5% BSA without fetal calf serum for 5 h were plated in 0.5 ml of starvation medium in each well. To produce a cell-free space, 1.0-mm thick steel plates were inserted into the wells before seeding the cells and were removed after the cells had been attached to the bottom. This method has the advantage over the frequently used “scratch window” assay in that the substrate in the cell-free space is not destroyed. The migration was monitored by inverted microscopy, and images of cell-free spaces were taken at time points 0 and 16 h after incubation at 37 °C and were used for calculation of the migration velocity.
Automated Wound Healing/Cell Migration Assay with ECIS
An automated wound healing/cell migration assay was applied that uses electric fields to introduce a well defined lesion into a confluent cell monolayer and monitors the repopulation of this wounded area by noninvasive impedance measurements. All measurements were performed with the ECIS 1600R device purchased from Applied BioPhysics Inc. (Troy, NY) together with disposable electrode arrays (type 8W1E) from the same source. Cells (5 × 105/cm2) were seeded in wells that had been precoated with 20 μg/ml fibronectin. Impedance readings to monitor electrode coverage and cell motility were recorded at sensing frequencies of 40 and 4 kHz, respectively, using noninvasive AC voltages. AC voltage pulses of 5.0-V amplitude (root mean square), 40-kHz frequency, and 30-s duration were applied to introduce lesions into the cell layer.
Immunofluorescence
Cells were plated onto 20 μg/ml fibronectin-coated glass coverslips and cultivated for 2 days. Immunofluorescence staining was described previously (20). Paxillin was detected with a murine monoclonal antibody (Signal Transduction). Secondary goat anti-mouse IgG labeled with Cy3 was from BioSource. F-actin was visualized with TRITC-labeled phalloidin (Sigma). Micrographs were acquired using an Axiovert 2000 ApoTome fluorescence microscope (Zeiss, Jena, Germany). Cell sizes were determined using AxioVision software (Zeiss).
Cell Lysis, Immunoprecipitation, and Kinase Assay
Cells were washed twice with ice-cold phosphate-buffered saline and lysed in radioimmune precipitation assay buffer supplemented with 1 mm sodium vanadate, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, and 5 μg/ml aprotinin for 30 min. Lysates were cleared by centrifugation at 10,000 × g for 15 min at 4 °C. Supernatants were resolved by 7.5% SDS-PAGE, and after electroblotting onto a nitrocellulose membrane, proteins were detected with the appropriate antibodies using the ECL detection system (Bio-Rad). For immunoprecipitation, 300 μg of protein from total cell lysates were incubated with the appropriate antibodies and 30 μl of protein A- or G-agarose beads overnight at 4 °C. Beads were washed twice with radioimmune precipitation assay buffer, and immunocomplexes were resolved by SDS-PAGE and analyzed by Western blotting as described (21). Kinase assays was performed as described previously (21) with 10 μg of recombinant glutathione S-transferase-MEF2C.
Real-time Reverse Transcription-PCR
Total RNA was isolated using an RNeasy kit (Qiagen), and subsequently, 1 μg of total RNA was reverse-transcribed with RevertAID polymerase (Fermentas) according to the manufacturer's instructions. cDNA was analyzed by real-time PCR using 2× SYBR Green Brilliant Mastermix (Stratagene) according to the manufacturer's instructions. n-fold values were calculated as described (22). The following primers were purchased from Sigma-Genosys: p130CASfw, 5′-GGTGGGCATGTATGATAAGAAGC-3′; p130CASrev, 5′-GCACCAGGTAGACGCTGTC-3′; GAPDHfw, 5′-GCAAATTTCCATGGCACCGT-3′; and GAPDHrev, 5′-GCCCCACTTGATTTTGGAGG-3′.
RESULTS
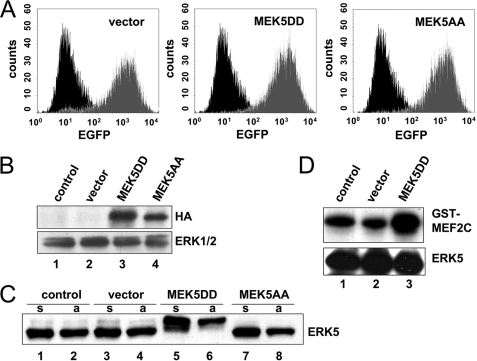
The MEK5/ERK5 signaling module is activated by several growth factors such as vascular endothelial growth factor, basic fibroblast growth factor, and epidermal growth factor, as well as by osmotic stress and UV light (23). All of these stimuli additionally affect other signaling pathways that contribute to the cellular response. Hence, to selectively study the impact of the activated MEK5/ERK5 pathway in EC, we introduced constitutively active or inactive mutants of the ERK5 upstream kinase MEK5 into primary HUVEC. By highly efficient retroviral gene transfer, we achieved ∼90% transgene-positive HUVEC as shown by FACS analysis of EGFP that was coexpressed with the MEK5 mutants from a bicistronic mRNA (Fig. 1A). Expression of hemagglutinin-tagged MEK5 mutants was verified by Western blot analysis (Fig. 1B, upper panel). To test the functional effect of the transdominant mutants, we analyzed the phosphorylation status of ERK5 by its mobility retardation in Western blots (Fig. 1C) and its kinase activity using the specific substrate glutathione S-transferase-MEF2C (Fig. 1D) in an in vitro kinase assay. Constitutive activation of ERK5 by expression of MEK5DD resulted in a pronounced retention of the ERK5 band in Western blots (Fig. 1C, lanes 5 and 6), indicating its increased phosphorylation. Concordantly, MEK5DD expression led to enhanced ERK5 activity as revealed by the kinase assay (Fig. 1D, lane 3). In turn, expression of the dominant-negative MEK5 mutant MEK5AA resulted in repressed basal phosphorylation of endogenous ERK5 (Fig. 1C, lanes 7 and 8).
FIGURE 1.
Retroviral gene transfer of constitutively active and constitutively inactive mutants of MEK5 into HUVEC alters ERK5 activity. A, the efficiency of retroviral gene transfer indicated by EGFP reporter expression (gray traces) was measured by cytometric analysis 96 h after transduction in comparison with control cells (black traces). B, expression of MEK5 mutants was studied by Western blot analysis 96 h after transduction. Recombinant proteins were detected by a hemagglutinin (HA) tag-specific antibody. Immunoblotting of ERK1/2 served as a loading control. C, endogenous ERK5 expression in cells with MEK5 mutants was analyzed. Note the retarded migration of ERK5 after expression of the constitutively active MEK5 mutant. Cells were starved for 5 h and either kept in suspension (s) or attached to fibronectin for 40 min (a). D, the enzymatic activity of ERK5 was measured in an in vitro immunocomplex kinase assay with recombinant glutathione S-transferase (GST)-MEF2C as a specific substrate. The efficiency of MEF2C phosphorylation was determined by PhosphorImager scanning, and the efficiency of ERK5 immunoprecipitation was measured by Western blotting with a specific antibody.
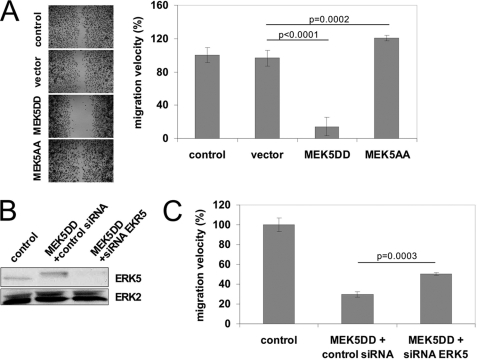
Formation of new blood vessels requires direct migration of activated EC toward the angiogenic stimulus. We were interested in whether ERK5 signaling interferes with motility and migration of EC. Thus, we performed lateral migration assays on fibronectin-coated cell culture plates. Surprisingly, constitutive activation of ERK5 signaling did not enhance the migration velocity of EC, as would have been expected for a pathway triggered by growth factors (6). Instead, migration was almost completely blocked by expression of MEK5DD (Fig. 2A). Accordingly, cell migration was slightly but significantly increased in the presence of the dominant-negative mutant MEK5AA. Similar results were obtained in EC in which endogenous ERK5 was partially knocked down via lentiviral expression of ERK5-specific shRNA (supplemental Fig. 1). To further show that reduction in MEK5DD EC migration was indeed mediated by ERK5 activation, we selectively knocked down ERK5 by synthetic siRNAs in these cells. Unfortunately, a complete knockdown of ERK5 was lethal for cells. However, in cells in which ERK5 was partially knocked down to a level still tolerated by HUVEC (Fig. 2B), the MEK5DD-induced migration block was also partially reverted (Fig. 2C), indicating that the retarded migration of MEK5DD cells was in fact mediated by increased activity of ERK5. The migratory block was EC-specific because murine fibroblasts expressing MEK5DD showed no reduced cell migration (supplemental Fig. 2A), although MEK5DD was expressed and endogenous ERK5 was activated (supplemental Fig. 2B). Interestingly, expression of the dominant-inactive mutant MEK5AA in fibroblasts lead to a reduced rather than an increased migration velocity (supplemental Fig. 2A). Although MEK5 was described to be highly specific for activation of ERK5, we tested the transduced EC for possible cross-activation of other MAPK pathways. As shown in supplemental Fig. 3, only a mild activation of ERK1/2 and no changes in p38 or JNK activation were observed after expression of MEK5DD in EC. However, this ERK activation should support rather than inhibit cell migration.
FIGURE 2.
Constitutive activation of MEK5/ERK5 signaling blocks lateral migration of HUVEC. A, lateral migration was measured as described previously (20). Left, the images show representative bright-field micrographs of EC stained with crystal violet after 16 h of migration (magnification ×5). Right, the relative mean migration velocity ± S.D. from three independent experiments normalized to control cells is depicted. B, the knockdown efficiency of ERK5 was analyzed by Western blotting. Immunoblotting of ERK1/2 served as a loading control. C, knockdown of ERK5 MAPK in HUVEC by specific siRNAs (third bar) partially restored MEK5DD-mediated inhibition of EC migration (middle bar). p values were obtained by Student's t test.
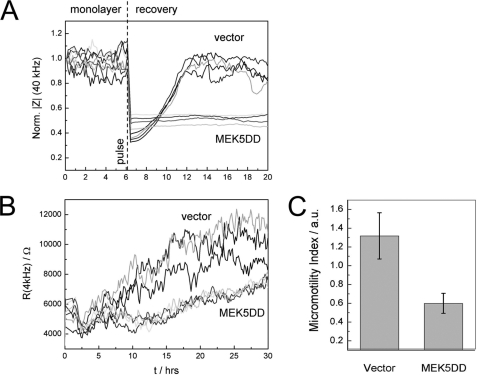
The MEK5DD-dependent inhibition of EC migration was independently confirmed in ECIS (24, 25)-based wound healing assays (Fig. 3A). When cells attach and spread on the electrode surface, the electrode impedance increases as the presence of the dielectric cell bodies force the current to flow either around or through the cells. After a confluent cell layer has been established, an invasive voltage pulse of several volts can be applied to locally kill all cells that are adhered to the electrode. The repopulation of the cell-free electrode space due to migration of surrounding cells can be monitored by noninvasive impedance measurements in real time.
FIGURE 3.
Constitutive activation of ERK5 inhibits wound healing and cell motility. A, MEK5DD-expressing HUVEC do not migrate into the artificially induced wounded area on a gold electrode as indicated by the missing recovery impedance values after electric wounding of the monolayer. Normalized impedance at a sampling frequency of 40 kHz of three independent experiments is depicted. B, cell micromotion, detected by fluctuation of resistance (4 kHz) over time, is reduced in MEK5DD-expressing HUVEC. C, quantitative analysis of EC micromotion. The bars show the average S.D. of increments for consecutive data frames of 64 s. a.u., arbitrary units.
Vector-transduced cells rapidly closed the artificial wound within 6 h, whereas MEK5DD-transduced cells completely failed to re-establish impedance levels of the monolayer (normalized to 1) (Fig. 3A). A quite high level of non-directed movement (micromotion (26)) of vector-transduced cells on the electrode was observed, as measured by the fluctuation of impedance with time. In MEK5DD-expressing HUVEC, we observed a significant reduction in micromotion as indicated by drastically decreased resistance fluctuations in these cells (Fig. 3B). To quantify the observed changes in micromotion, we calculated the average S.D. of the resistance increments for consecutive data windows of 64 s. This parameter (referred to as the micromotion index) reflects the non-directional movement of cells and the turnover of cell-substrate contacts (Fig. 3C) (26).
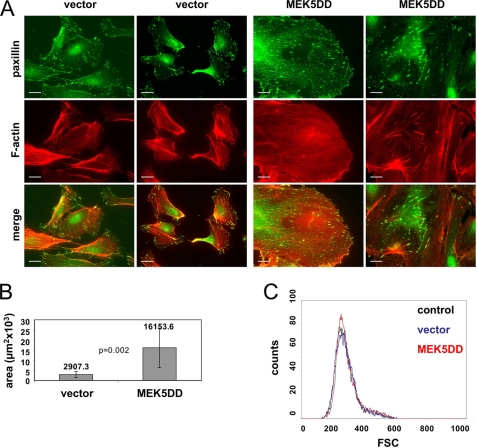
Cell migration requires a coordinated structural change of the cytoskeleton and highly dynamic reorganization of focal contacts. To determine whether the modified migration and motility of MEK5DD-expressing cells are based on altered cytoskeleton architecture, we performed immunofluorescence stainings of these cells and vector-transduced EC (Fig. 4A). Actin fibers were detected by TRITC-labeled phalloidin, and focal contacts were visualized with an antibody that recognizes paxillin. Although vector-transduced cells exhibited a highly organized microfilament network with long actin fibers running through the whole cell body connecting distal cell protrusions (Fig. 4A, left panels), the MEK5DD-overexpressing cells were characterized by shorter and partially thicker actin fibers that also were distributed over the whole cell body but often randomly orientated (Fig. 4A, right panels). In vector-transduced cells, clusters of focal contacts were located preferentially at the cell periphery, whereas expression of active MEK5DD led to an increased number of focal contacts that were distributed over the entire basolateral membrane. These additional focal contacts were connected by short and thick actin fibers (Fig. 4A, lower panels). Spread MEK5DD-expressing cells always showed a rounded cell shape with a minimum of protrusions but with a dramatic increase in the area covered by the cell body. Quantification of these areas using AxioVision software revealed that the mean cell area covered by randomly chosen MEK5DD-expressing cells was about five times larger compared with vector-transduced cells (Fig. 4B). However, the increase in lateral cell dimensions of MEK5DD cells was not due to changes in cell volume. FACS analysis of detached HUVEC revealed no change in cell volume (Fig. 4C), indicating that MEK5DD expression induced enhanced spreading or flattening rather then a general increase in cell size. HUVEC expressing dominant-inactive MEK5AA showed no detectable morphological changes compared with vector-transduced cells (data not shown).
FIGURE 4.
Expression of MEK5DD induces drastic morphological changes. A, cells were plated onto uncoated glass coverslips for 96 h, fixed with paraformaldehyde, and stained for paxillin (green) and F-actin (red). As the morphology of HUVEC vary significantly, two different cell images for each staining are presented. Scale bars = 20 μm. B, the mean covered area ± S.D. of 70 randomly chosen spread cells is depicted. Covered areas were measured with AxioVision software. p values were obtained by Student's t test. C, shown are the results from flow cytometric study of control, vector-transduced, and MEK5DD-transduced EC in a forward scatter light (FSC) histogram analysis. 104 cells were analyzed.
The increased number of focal contacts and their equal distribution over the whole basolateral membrane in MEK5DD-expressing cells, as well as the enlarged spreading of these cells, might be due to a changed function of integrin receptors on their surface, which are responsible for formation of focal contacts after binding to extracellular matrix proteins. First, we quantified the amount of integrin α5, β1, or αvβ3 on the surface of these cells. No differences in integrin levels were observed on MEK5DD cells compared with vector-transduced cells in our FACS analyses (supplemental Fig. 4A), thus excluding that the observed effects might be due to increased amounts of integrins on MEK5DD cells. Consistent with similar integrin levels on vector-transduced versus MEK5DD-expressing EC, also no differences were observed in the initial attachment to fibronectin between control cells, vector-transduced cells, MEK5DD-expressing cells, and MEK5AA-transduced cells (supplemental Fig. 4B), indicating that the initial activation of integrin receptors was not altered in MEK5DD cells. Hence, the delayed migration of MEK5DD cells and the enhanced number of focal contacts, as well as changes in the F-actin organization, cannot be explained by altered expression of integrins or their initial activation during attachment of cells to matrix proteins. Nevertheless, it should be noted that spread MEK5DD cells attached more tightly to their substrates than did MEK5AA- or vector-transduced cells. Indeed, they always showed a delayed detachment during routine subculturing by trypsin/EDTA (supplemental Fig. 5). Taken together, the enhancing effect of MEK5DD on focal contact formation and cell-substrate interaction seems to occur after initial attachment of cells by reorganization of focal contacts and the affiliated integrins during spreading of the cells.
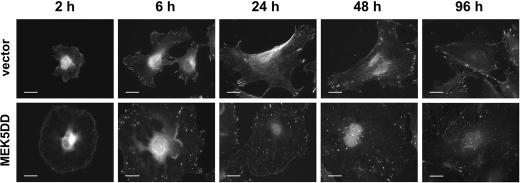
Thus, we analyzed by immunofluorescence the spreading of EC and focal contact and stress fiber formation during this process. As data in Fig. 5 show, the initiation of spreading of both cell types did not differ, and the cell morphology and focal contact formation were also similar in vector-transduced (upper panels) and MEK5DD-transduced (lower panels) cells during the first 2 h of attachment. The major focal contacts were localized at the cell periphery in both cell types, and the microfilaments were still less developed, but MEK5DD cells already showed an enlarged cell surface. However, 6 h post-adhesion, the number of focal contacts and their distribution in MEK5DD-expressing cells differed strikingly from those in control cells. Changes in cell shape and in cytoskeleton organization were also clearly seen from 6 h onward. This correlates with the previously observed flat cell body and increased number of focal contacts distributed over the lateral membrane (Fig. 4) and the distinct organized actin cytoskeleton in MEK5DD cells (supplemental Fig. 6).
FIGURE 5.
Expression of constitutively active MEK5 in HUVEC leads to changes in cell spreading. Vector- or MEK5DD-transduced HUVEC were plated on glass coverslips, fixed at the indicated times, and stained for paxillin with a specific antibody. Scale bars = 20 μm.
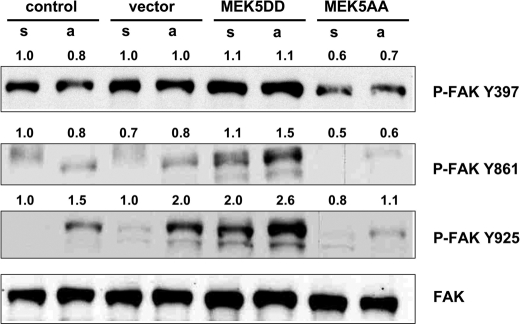
Keeping in mind that the intensity of cell-substrate interaction was reduced in MEK5DD-expressing cells (as shown by ECIS) (Fig. 3, B and C), we hypothesized that focal contact assembly and disassembly might be differentially regulated in cells with constitutive ERK5 signaling. Such a difference would most likely also influence the dynamics of the cytoskeleton organization. A major regulator of focal contact architecture is FAK. Integrin engagement upon attachment of cells leads to autophosphorylation of FAK Tyr397, resulting in conformational changes of the kinase and subsequent binding to Src. The latter phosphorylates FAK at additional tyrosine residues, including Tyr861 and Tyr925, which then allows binding of downstream targets such as p130Cas (for review, see Ref. 27). We analyzed the attachment-induced phosphorylation status of FAK in the presence or absence of MEK5DD with phospho-specific antibodies by Western blotting (Fig. 6). Constitutive activation of MEK5 only mildly increased phosphorylation of the initial autoactivation site Tyr397 but enhanced phosphorylation of FAK at Tyr861 and Tyr925 in comparison with vector-transduced or control cells. Interestingly, phosphorylation of these sites in MEK5AA cells was reduced in comparison with control or vector-transduced cells. The increased phosphorylation pattern of FAK in cells expressing MEK5DD suggests an enhanced dynamic rearrangement of focal contacts and activation of promigratory regulators such as p130Cas. Consequently, cell motility and migration of these cells should be enhanced (27). Thus, the altered FAK phosphorylation is in disagreement with the observed effects on cell migration and the cytoskeletal changes induced by constitutively active MEK5, and the key event leading to the phenotype of the MEK5DD cells has to occur downstream of FAK. Therefore, we examined the expression levels of p130Cas, a downstream target of FAK known to be essential for cell migration (28–30).
FIGURE 6.
Constitutive activation of ERK5 results in hyperphosphorylation of FAK. Cells were starved for 5 h and either kept in suspension (s) or attached to fibronectin (a). Phosphorylation of FAK was analyzed by Western blotting using phospho-specific antibodies as indicated to the right of the first three panels. Immunoblotting with a FAK-specific antibody served as a loading control (lower panel). Phosphorylation intensity was estimated densitometrically as the relative intensity of the p-bands to the appropriate loading controls. Values for control cells kept in suspension were taken as unity.
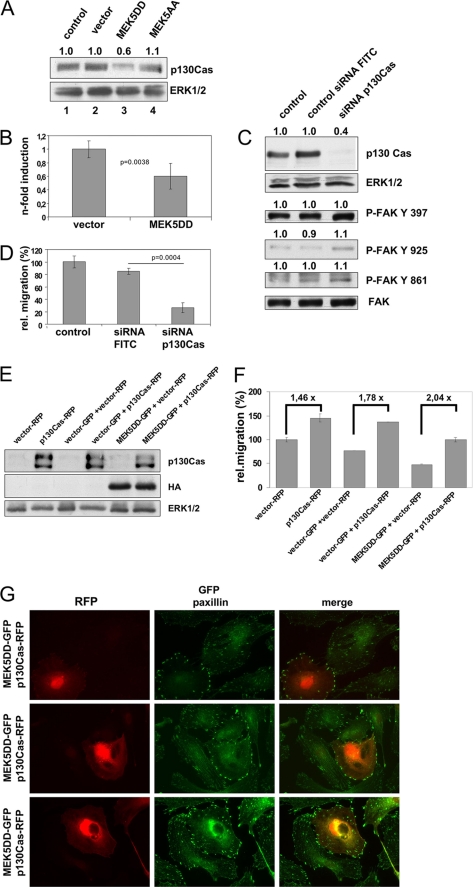
By Western blot analysis with specific antibodies, we detected reduced amounts of p130Cas in MEK5DD-expressing HUVEC (Fig. 7A). Real-time reverse-transcription PCR confirmed this finding and revealed reduced p130Cas mRNA levels (Fig. 7B). We could not observe this effect in fibroblasts expressing MEK5DD (supplemental Fig. 2B), which concordantly also showed no block in cell migration. This points again to an EC-specific effect of ERK5 signaling in cell migration. To test whether reduced levels of the promigratory factor p130Cas might be responsible at least partially for the motility alterations induced by constitutive MEK5 activation, we used an RNA interference approach to knock down p130Cas. Transfection of synthetic siRNAs resulted in massive down-regulation of p130Cas 48 h after transfection (Fig. 7C, upper panel). Interestingly, phosphorylation of FAK at Tyr861, Tyr925, and Ser910 in these cells was increased (Fig. 7C, lower panels), similar to MEK5DD cells (Fig. 6 and supplemental Fig. 8), whereas phosphorylation at Tyr397 was not changed in cells with down-regulated p130Cas. In a lateral migration assay, these cells showed diminished migration (Fig. 7D), indicating that reduced expression levels of p130Cas indeed result in reduced EC migration. Reduced expression of p130Cas was also observed in MEK5DD-expressing cells. On the contrary, overexpression of recombinant p130Cas (Fig. 7E) rescued, albeit not completely, the reduced migration capacity of MEK5DD cells and, as expected, further enhanced the motility of vector-transduced cells (Fig. 7F). Additionally, coexpression of p130Cas and MEK5DD also re-established the disturbed lateral EC migration and resulted in redistribution of focal contacts from the cell body to the cell periphery (Fig. 7G), indicating that the impaired migration phenotype of MEK5DD cells is, at least in part, due to reduced expression of p130Cas in these cells.
FIGURE 7.
MEK5DD expression forces down-regulation of p130Cas. A, p130Cas expression (upper panel) in HUVEC expressing different MEK5 mutants was analyzed by Western blotting. ERK1/2 served as a loading control (lower panel). B, expression of p130Cas was estimated by real-time PCR with p130Cas-specific primers, calibrated to glyceraldehyde-3-phosphate dehydrogenase mRNA levels, and normalized to vector-transduced cells (left bar, arbitrary set to 1). C, reduction in p130Cas expression resulted in enhanced FAK phosphorylation. Knockdown of p130Cas with specific siRNAs was analyzed by Western blotting (first panel). The loading control was validated by ERK1/2 levels. Transfection efficiency was >90% as validated by FACS analysis of fluorescein isothiocyanate (FITC)-labeled control siRNA (not shown). Phosphorylation of FAK at different tyrosine residues was studied as described in the legend to Fig. 6. D, reduction in p130Cas expression resulted in decreased lateral migration. The same cells described for C were analyzed for their migration properties. Relative (rel.) mean migration velocity ± S.D. of three independently repeated experiments is shown. E and F, overexpression of p130Cas in MEK5DD cells rescued the migration phenotype. E, HUVEC were subsequently transduced with MEK5DD- and p130Cas-containing retroviruses. According to cytometric analysis, 78–99% of these cells were positive for both reporters (not shown). Expression of recombinant p130Cas was verified with anti-Cas antibody, whereas MEK5DD was verified with anti-hemagglutinin (HA) tag antibody. ERK1/2 served as a loading control (upper panels). F, lateral migration of the double-transduced cells is shown. Relative mean migration velocity ± S.D. from three independent experiments normalized to control cells is depicted. Numbers over bars show the acceleration -fold of cell motility. Statistical analysis was performed by Student's t test and revealed the following values: p = 0.0013 for vector-RFP versus p130Cas-RFP; p < 0.0001 for vector-GFP + vector-RFP versus vector-GFP + p130Cas-RFP; p < 0.0001 for MEK5DD-GFP + vector-RFP versus MEK5DD- GFP + p130Cas-RFP; p < 0.0001 for vector-GFP + vector-RFP versus MEK5DD-GFP + vector-RFP; and p < 0.0001 for vector-GFP + vector-RFP versus MEK5DD-GFP + p130Cas-RFP. G, overexpression of p130Cas in MEK5DD cells reverted the altered distribution of focal contacts. Double-infected cells were plated for 96 h and stained for paxillin to reveal focal contacts (green). Paxillin was detected by an Alexa 488-labeled secondary antibody. The additional intense green color of cell bodies reflects the GFP fluorescence caused by MEK5DD-GFP expression. Cells containing recombinant p130Cas (red) were visualized by RFP fluorescence. Because of the high variation between separate HUVEC, three different images are shown to better underline the observed effect. p values were obtained by Student's t test analysis.
DISCUSSION
EC migration is a central step in the formation of new blood vessels. It is coordinated by a variety of cellular signaling cascades that control the structural rearrangements of cytoskeleton, extracellular matrix, and focal contacts.
In this study, we have demonstrated that ERK5 MAPK can be added to the signaling molecules that regulate EC migration processes. Selective activation of ERK5 signaling by a constitutively active MEK5 mutant blocked basic cell migration and resulted in increased flattening of cells, altered actin fiber architecture, and enhanced number of focal contacts. In accordance to this, EC migration is slightly enhanced in cells expressing the dominant-inactive mutant MEK5AA or an ERK5-specific shRNA (supplemental Fig. 1). Previous studies in different cell and tissue types have shown the involvement of ERK5 in cytoskeletal architecture and cell migration. Transgenic mice expressing MEK5DD in heart tissue demonstrate severe morphological changes in cardiomyocytes (31). However, in keratinocytes, ERK5 signaling acts in a promigratory fashion (32). Schramp et al. (33) also demonstrated reduced podocyte formation in ERK5-deficient fibroblasts activated by constitutively active Src kinase. This finding is in accordance with our data obtained in murine fibroblasts expressing dominant-inactive MEK5AA. In summary, ERK5 signaling regulates cell migration and cytoskeleton remodeling in a cell type-specific manner.
We could greatly rule out that proliferative effects might influence the observed result because MEK5DD-expressing cells proliferated faster than vector- or MEK5AA-transduced EC (supplemental Fig. 5), which is in good accordance with the already described promitotic effect of the ERK5 signaling pathway (7). Additionally, our migration experiments never lasted >20 h, which is greatly below the reduplication time of all EC analyzed. Reduced migration of EC expressing MEK5DD was confirmed by ECIS wound healing assay. Moreover, we observed massively reduced micromotion of these cells, pointing again to decreased cytoskeletal dynamics and reduced fluctuation of cell-substrate contacts. In a time-resolved cell spreading experiment, we demonstrated that EC expressing MEK5DD developed their typical morphological phenotype not during initial attachment but during subsequent spreading or flattening. Accumulation of focal contacts occurred in a time-dependent manner in MEK5DD cells, whereas the number of focal contacts remained constant after spreading in vector-transduced cells. Moreover, MEK5DD-expressing EC showed an equal distribution of focal contacts over the whole circle-shaped lateral membrane, whereas focal contacts remained concentrated at the tips of cell protrusions in vector-transduced cells.
Taken together, these findings point to an altered balance of focal contact assembly and disassembly in MEK5DD cells. This results in enlarged flattening of cells with less protrusions but an increased number of extracellular matrix-cell contact points distributed over the large and flat cell bodies, finally leading to an impaired motility. Indeed, we have shown differences in FAK phosphorylation dependent on the activation state of the expressed MEK5. FAK is a central signaling molecule in focal contact turnover. Although expression of MEK5DD in EC resulted in hyperphosphorylation of FAK, the presence of the kinase-dead MEK5AA led to FAK hypophosphorylation. Dynamic phosphorylation of FAK is a prerequisite for focal contact turnover (for review, see Mitra et al. (27)). However, the phosphorylation pattern of FAK in MEK5DD-expressing cells should rather support than repress cell migration. Constant hyperactivation of FAK and its downstream target p130Cas is a hallmark for transformed tumor cells, resulting in enhanced migration and motility and reduced matrix-cell interaction and stress fiber formation (34, 35). Presently, we can only speculate about the molecular link between ERK5 activation and FAK phosphorylation. A recent work by Villa-Moruzzi (36) demonstrated direct phosphorylation of FAK at Ser910 by ERK5 in fibroblasts, suggesting the involvement of ERK5 in regulation of FAK phosphorylation and turnover of focal contacts in a way similar to ERK1/2. We verified this observation in EC and showed that phosphorylation of FAK at Ser910 was enhanced in cells expressing MEK5DD but decreased in cells expressing MEK5AA (supplemental Fig. 8A). Moreover, phosphorylation of this serine residue was also enhanced in cells with down-regulated p130Cas (supplemental Fig. 8B), which, similar to MEK5DD-expressing cells, showed a reduced motility. The physiological relevance of FAK phosphorylation at this position in the turnover of focal adhesions and in cell migration is, however, only poorly understood and needs further investigations.
Analysis of p130Cas, a downstream target of FAK, revealed that ERK5 activation resulted in decreased p130Cas levels. Mechanistically, the reduced mRNA levels point either to a reduced transcription rate or to a reduction in mRNA stability. ERK5 activates a panel of transcription factors, can itself directly interact with DNA, and force transcriptional activity (5). Ongoing studies address the molecular mechanism of ERK5-mediated down-regulation of p130Cas and attend to the question of whether hyperphosphorylation of FAK is a compensatory consequence of the MEK5DD-induced reduction of p130Cas. Our analysis of FAK phosphorylation in cells with down-regulated p130Cas (Fig. 7C) is in line with this assumption. In addition, constitutive overexpression of p130Cas rescued, although only partially, the lateral migration capacity and focal contact distribution of EC that were altered by ERK5 hyperactivation. Honda et al. (37) showed already 10 years ago that p130Cas deficiency in mice leads to abnormal vessel development and severe changes in cell morphology. Murine p130Cas−/− fibroblasts are also characterized by a disturbed actin cytoskeleton and a flat round-shaped cell body (37). Although there is only a partial reduction of p130Cas protein levels in MEK5DD-expressing cells, we observed a very similar phenotype in EC. In summary, we have demonstrated that ERK5 MAPK signaling plays a pivotal role in EC migration and cytoskeleton organization in addition to the already described function of ERK5 in EC survival.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–8.
- EC
- endothelial cell(s)
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- MAPK/ERK kinase
- FAK
- focal adhesion kinase
- JNK
- c-Jun N-terminal kinase
- HUVEC
- human umbilical vein endothelial cell(s)
- siRNA
- small interfering RNA
- EGFP
- enhanced green fluorescent protein
- shRNA
- short hairpin RNA
- BSA
- bovine serum albumin
- ECIS
- electric cell-substrate impedance sensing
- TRITC
- tetramethylrhodamine isothiocyanate
- FACS
- fluorescence-activated cell sorter
- RFP
- red fluorescent protein.
REFERENCES
- 1.Mauriz J. L., González-Gallego J. (2008) J. Pharm. Sci. 97, 4129–4154 [DOI] [PubMed] [Google Scholar]
- 2.Lamalice L., Le Boeuf F., Huot J. (2007) Circ. Res. 100, 782–794 [DOI] [PubMed] [Google Scholar]
- 3.Zhou G., Bao Z. Q., Dixon J. E. (1995) J. Biol. Chem. 270, 12665–12669 [DOI] [PubMed] [Google Scholar]
- 4.Nishimoto S., Nishida E. (2006) EMBO Rep. 7, 782–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasler H. G., Victoria J., Duramad O., Winoto A. (2000) Mol. Cell. Biol. 20, 8382–8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato Y., Kravchenko V. V., Tapping R. I., Han J., Ulevitch R. J., Lee J. D. (1997) EMBO J. 16, 7054–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pi X., Yan C., Berk B. C. (2004) Circ. Res. 94, 362–369 [DOI] [PubMed] [Google Scholar]
- 8.Regan C. P., Li W., Boucher D. M., Spatz S., Su M. S., Kuida K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9248–9253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sohn S. J., Sarvis B. K., Cado D., Winoto A. (2002) J. Biol. Chem. 277, 43344–43351 [DOI] [PubMed] [Google Scholar]
- 10.Hayashi M., Kim S. W., Imanaka-Yoshida K., Yoshida T., Abel E. D., Eliceiri B., Yang Y., Ulevitch R. J., Lee J. D. (2004) J. Clin. Invest. 113, 1138–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi M., Lee J. D. (2004) J. Mol. Med. 82, 800–808 [DOI] [PubMed] [Google Scholar]
- 12.Goebeler M., Gillitzer R., Kilian K., Utzel K., Bröcker E. B., Rapp U. R., Ludwig S. (2001) Blood 97, 46–55 [DOI] [PubMed] [Google Scholar]
- 13.Denk A., Goebeler M., Schmid S., Berberich I., Ritz O., Lindemann D., Ludwig S., Wirth T. (2001) J. Biol. Chem. 276, 28451–28458 [DOI] [PubMed] [Google Scholar]
- 14.Lindemann D., Patriquin E., Feng S., Mulligan R. C. (1997) Mol. Med. 3, 466–476 [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X., Zhan X., D'Costa J., Tanavde V. M., Ye Z., Peng T., Malehorn M. T., Yang X., Civin C. I., Cheng L. (2003) Mol. Ther. 7, 827–838 [DOI] [PubMed] [Google Scholar]
- 16.Riedl J., Crevenna A. H., Kessenbrock K., Yu J. H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T. A., Werb Z., Sixt M., Wedlich-Soldner R. (2008) Nat. Methods 5, 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb D. J., Donais K., Whitmore L. A., Thomas S. M., Turner C. E., Parsons J. T., Horwitz A. F. (2004) Nat. Cell Biol. 6, 154–161 [DOI] [PubMed] [Google Scholar]
- 18.Szulc J., Wiznerowicz M., Sauvain M. O., Trono D., Aebischer P. (2006) Nat. Methods 3, 109–116 [DOI] [PubMed] [Google Scholar]
- 19.Wiznerowicz M., Trono D. (2003) J. Virol. 77, 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wixler V., Hirner S., Müller J. M., Gullotti L., Will C., Kirfel J., Günther T., Schneider H., Bosserhoff A., Schorle H., Park J., Schüle R., Buettner R. (2007) J. Cell Biol. 177, 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinev D., Jordan B. W., Neufeld B., Lee J. D., Lindemann D., Rapp U. R., Ludwig S. (2001) EMBO Rep. 2, 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 23.Abe J., Kusuhara M., Ulevitch R. J., Berk B. C., Lee J. D. (1996) J. Biol. Chem. 271, 16586–16590 [DOI] [PubMed] [Google Scholar]
- 24.Giaever I., Keese C. R. (1986) IEEE Trans. Biomed. Eng. 33, 242–247 [DOI] [PubMed] [Google Scholar]
- 25.Wegener J., Keese C. R., Giaever I. (2000) Exp. Cell Res. 259, 158–166 [DOI] [PubMed] [Google Scholar]
- 26.Giaever I., Keese C. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7896–7900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitra S. K., Hanson D. A., Schlaepfer D. D. (2005) Nat. Rev. Mol. Cell Biol. 6, 56–68 [DOI] [PubMed] [Google Scholar]
- 28.Abassi Y. A., Rehn M., Ekman N., Alitalo K., Vuori K. (2003) J. Biol. Chem. 278, 35636–35643 [DOI] [PubMed] [Google Scholar]
- 29.Cary L. A., Han D. C., Polte T. R., Hanks S. K., Guan J. L. (1998) J. Cell Biol. 140, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klemke R. L., Leng J., Molander R., Brooks P. C., Vuori K., Cheresh D. A. (1998) J. Cell Biol. 140, 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicol R. L., Frey N., Pearson G., Cobb M., Richardson J., Olson E. N. (2001) EMBO J. 20, 2757–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnoux V., Nassour M., L'Helgoualc'h A., Hipskind R. A., Savagner P. (2008) Mol. Biol. Cell 19, 4738–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schramp M., Ying O., Kim T. Y., Martin G. S. (2008) J. Cell Biol. 181, 1195–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson T. R., Khandrika L., Kumar B., Venezia S., Koul S., Chandhoke R., Maroni P., Donohue R., Meacham R. B., Koul H. K. (2008) Mol. Cancer Res. 6, 1639–1648 [DOI] [PubMed] [Google Scholar]
- 35.Kaneda T., Sonoda Y., Ando K., Suzuki T., Sasaki Y., Oshio T., Tago M., Kasahara T. (2008) Cancer Lett. 270, 354–361 [DOI] [PubMed] [Google Scholar]
- 36.Villa-Moruzzi E. (2007) Biochem. J. 408, 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honda H., Oda H., Nakamoto T., Honda Z., Sakai R., Suzuki T., Saito T., Nakamura K., Nakao K., Ishikawa T., Katsuki M., Yazaki Y., Hirai H. (1998) Nat. Genet. 19, 361–365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.