Abstract
Epigenetic reprogramming plays a central role in the development of cloned embryos generated by somatic cell nuclear transfer, and it is believed that aberrant reprogramming leads to the abnormal development of most cloned embryos. Recent studies show that trimethylation of H3K27 (H3K27me3) contributes to the maintenance of embryonic stem cell pluripotency because the differentiation genes are always occupied by nucleosomes trimethylated at H3K27, which represses gene expression. Here, we provide evidence that differential H3K27me3 modification exists between normal fertilization-produced blastocysts and somatic cell nuclear transfer cloned blastocysts; H3K27me3 was specifically found in cells of the inner cell mass (ICM) of normal blastocysts, whereas there was no modification of H3K27me3 in the ICM of cloned blastocysts. Subsequently, we demonstrated that the differentiation-related genes, which are marked by H3K27me3 in embryonic stem cells, were expressed at significantly higher levels in cloned embryos than in normal embryos. The polycomb repressive complex 2 (PRC2) component genes (Eed, Ezh2, and Suz12), which are responsible for the generation of H3K27me3, were expressed at lower levels in the cloned embryos. Our results suggest that reduced expression of PRC2 component genes in cloned embryos results in defective modification of H3K27me3 to the differentiation-related genes in pluripotent ICM cells. This results in premature expression of developmental genes and death of somatic cloned embryos shortly after implantation. Taken together, these studies suggest that H3K27me3 might be an important epigenetic marker with which to evaluate the developmental potential of cloned embryos.
Embryonic stem (ES)3 cells are derived from the inner cell mass (ICM) of a blastocyst. ES cells are pluripotent and can maintain self-renewal under appropriate culture conditions (1–3). Gene expression profiles of ES cells revealed that there is a core pluripotency regulatory circuitry formed by Oct4, Sox2, and Nanog, which coordinated to maintain the transcriptional program required for the maintenance of ES cell pluripotency (4). Results from recent studies have indicated that ES cells possess a bivalent chromatin structure, which has been suggested to be essential for maintaining their self-renewal and pluripotency characteristics. The bivalent chromatin domains of ES cells consisted of large regions of H3K27 methylation and smaller regions of H3K4 methylation (5). Trimethylation of H3K27 is associated with the activity of the polycomb repressive complex 2 (PRC2); and recent studies have indicated that PRC2 specifically targets developmental genes, which are marked by H3K27me3, to maintain ES cell pluripotency by repressing expression of these genes (6–12).
Somatic cell nuclear transfer (SCNT) represents a remarkable process in which the differentiated somatic cell genome can be converted into a totipotent or at least a pluripotent state (13–16). However, the efficiency of cloning is extremely low, as less than 5% of cloned embryos could develop into live animals. Aberrant epigenetic reprogramming has been suggested as the cause of this inefficiency; however, little is known about the chromatin modifications in cloned embryos, except that abnormal DNA methylation has been observed (17, 18). As previously mentioned, histone methylation regulates gene expression and silencing in pluripotent stem cells. It remains unknown if histone methylation could be correctly reprogrammed through SCNT and whether aberrant reprogramming of histone methylation could result in abnormal gene expression that ultimately leads to the death of most SCNT embryos soon after implantation.
Here, we provide evidence demonstrating that loss of the H3K27me3 in ICM cells was widely observed in SCNT-produced blastocysts. Such defective reprogramming of H3K27me3 in somatic clones was caused by inadequate expression of PRC2 component genes, particularly Ezh2, which in turn drove the premature expression of differentiation genes and caused early death of cloned embryos. These results suggest that H3K27me3 may be an important epigenetic marker representing the developmental potential of cloned embryos and that treatments that restore the function of the PRC2 components in clones may increase the survival of cloned embryos.
EXPERIMENTAL PROCEDURES
Collection of Metaphase II-arrested Oocytes and the Preparation of Cumulus Cells
The specific pathogen-free female B6D2F1 (C57BL/6×DBA/2) mice (8–10 weeks old) were superovulated by the sequential injection of 7 IU each of pregnant mare serum gonadotropin and human chorionic gonadotropin (Sigma). Metaphase II oocytes and the surrounding cumulus cells were collected for nuclear transfer. All studies adhered to procedures consistent with the National Institute of Biological Sciences Guide for the care and use of laboratory animals.
SCNT
To generate diploid cloned constructs by SCNT, spindle-chromosome complexes of metaphase II-arrested oocytes were first removed. After spindle-chromosome complex removal, oocytes were kept in CZBG medium until injection (19, 20). Cumulus cell nucleus was separated and injected into the enucleated oocyte using a PiezoDrill micromanipulator. After injection of the cumulus nucleus, the cloned constructs were allowed to recover for several min before they were transferred back to CZBG medium. Reconstructed oocytes were cultured in CZBG medium for ∼1–3 h before activation. Activation was achieved by 6 h of culture of cloned constructs in calcium-free CZBG medium containing 10 mm of strontium chloride and 5 μg/ml of cytochalasin B. Activated constructs were thoroughly washed and cultured in KSOM medium (MR-121-D, Chemicon).
Collection of Fertilized Embryos and Cloned Embryos
For studies requiring zygotes, successfully mated B6D2F1 female mice were sacrificed 20 h post-human chorionic gonadotropin, and zygotes were collected in M2 medium (Sigma) and cultured in KSOM medium. This medium supports efficient development of cumulus cell cloned constructs as well as control fertilized embryos. Finally, the cloned embryos were collected at 26 h (2-cell), 42 h (4-cell), 51 h (8-cell), 69 h (morula), and 91 h (blastocyst) post-strontium chloride treatment as well as control fertilized embryos: 24 h (2-cell), 40 h (4-cell), 45 h (8-cell), 64 h (morula), and 86 h (blastocyst) post-human chorionic gonadotropin injection. To collect late stage blastocysts, the blastocysts were cultured one more day in KSOM medium. To investigate the histone modifications in early stage embryos, the cloned constructs and intracytoplasmic sperm injection-produced embryos used as control were collected as described previously (21).
Immunofluorescent Detection of H3K27me3, H3K4me3, Nanog, Oct4, Cdx2, and Ezh2
All samples were fixed in 4% paraformaldehyde for 30 min followed by permeabilization in phosphate-buffered saline containing 0.5% Triton X-100 for 15 min. After blocking and incubating with rabbit polyclonal antibodies: anti-H3K27me3 (diluted 1:100, Upstate) or anti-H3K4me3 (diluted 1:200, Abcam) for 1 h, samples were incubated with the Alexa Fluor 488 goat anti-rabbit IgG conjugate (diluted 1:1,000, Molecular Probes) for 1 h. To visualize the pluripotent cells and trophectoderm cells, the samples were incubated with anti-Nanog polyclonal antibody (diluted 1:500, Cosmo Bio) or anti-Oct3/4 monoclonal antibody (diluted 1:500, Santa Cruz Biotechnology) and anti-CDX-2 monoclonal antibody (diluted 1:50, BioGenex) for 1 h and secondary antibody conjugated to Alexa 594 goat anti-rabbit IgG conjugate (diluted 1:1,000, Molecular Probes). Mouse monoclonal anti-Ezh2 antibody (diluted 1:100, Cell Signaling Technology) was used to detect Ezh2 in embryos.
Real-time Reverse Transcription (RT)-PCR
RNA was extracted from fertilized embryos and cloned embryos using the PicoPure RNA isolation kit (ARCTURUS) and was reverse-transcribed using the SuperScript III first-strand synthesis system for RT-PCR (18080-051, Invitrogen). Real-time PCR was performed on an ABI Prism 7500 using SYBR Green Premix (Takara). Primer pairs that are specific for PRC2, pluripotent genes, and developmental genes were listed in supplemental Table S2. Gadph was used as control for the quality and amount of the reverse-transcribed RNA.
Western Blot
Whole cell lysates from embryos (260 embryos each: SCNT and fertilized blastocyst) were subjected to immunoblotting with rabbit polyclonal anti-H3K27 antibody (07449, Upstate), mouse monoclonal anti-α-tubulin (T6199, Sigma) and mouse monoclonal anti-Oct3/4 antibody (SC-5279, Santa Cruz Biotechnology). The secondary antibodies were ECL rabbit IgG (NA934, GE Healthcare) and ECL mouse IgG (NA931, GE Healthcare). For detection of Ezh2 (mouse monoclonal anti-EZH2 antibody; AC22, Cell Signaling Technology), lysates from 240 embryos (SCNT and control blastocysts) were subjected to Western blotting.
Statistical Analysis
Values of real-time PCR are expressed as medians or means (with 95% confidence intervals). Correlations between assays were evaluated by linear regression (SPSS version 11.0, SPSS, Inc., Chicago, IL). p values <0.05 were considered significant.
RESULTS
Aberrant Reprogramming of H3K27me3 in Somatic Cell Cloned Blastocysts
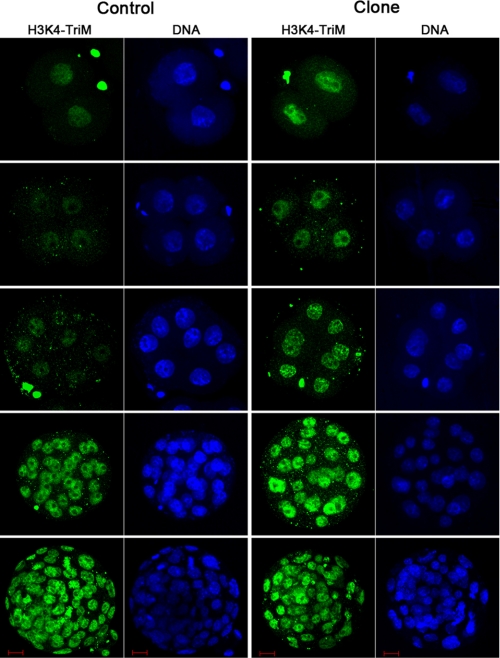
To determine whether the SCNT-produced embryos could recapitulate the ES-like bivalent chromatin structure at blastocyst stage, modification of H3K4me3 and H3K27me3 in both normal fertilized and SCNT cloned embryos during preimplantation development was extensively investigated. As shown in Fig. 1, H3K4me3 was evenly distributed in the nucleus of normal fertilization-produced embryos through the 2-cell to blastocyst stage (Fig. 1, left panel). Compared with normal embryos, the pattern of H3K4me3 modification in somatic cloned embryos showed no significant difference, except the intensity of H3K4me3 fluorescence was much higher in the cloned embryos between the 2- and 8-cell stages (Fig. 1, right panel). However, the intensity exhibited no difference at the morula and blastocyst stage; all of the cells were marked with H3K4me3 modification.
FIGURE 1.
Modification of H3K4me3 in preimplantation embryos. Left panel, fertilization produced embryos at 2-, 4-, 8-cell stages, morula, and blastocyst stages. Scale bar, 10 μm. Right panel, cumulus cell SCNT-produced embryos at 2-, 4-, and 8-cell stages and morula and blastocyst stages. Scale bar, 10 μm.
Next, modification of H3K27me3 was compared in somatic cloned embryos and fertilization-produced embryos. In striking contrast to H3K4me3, there were dramatic changes in H3K27me3 during early development in both normal and cloned embryos (Fig. 2). The nuclei of normal embryos between the 2- and 8-cell stages were positively marked with H3K27me3, whereas the H3K27me3 mark disappeared when the embryos reached morulae stage. These results indicated that global demethylation of H3K27me3 occurred at this specific stage, which is inconsistent with a previous report (22). This result might be caused by the embryos collected at different time point from the previous report. Subsequently, only a small proportion of cells were found positively re-marked with H3K27me3 at the blastocyst stage and asymmetric H3K27me3 marking appeared, which may specify cells with different developmental potentials (Fig. 2, left panel). Compared with normal embryos, there was no marked difference in the pattern of H3K27me3 modification in somatic cell cloned embryos until the morulae stage. As in control embryos, global demethylation of H3K27me3 occurred at morula stage, and no H3K27me3 remained in the cloned embryos. However, there was no evidence of positive marking of H3K27me3 in a small population of cells, as had been observed in the cloned blastocysts (Fig. 2, right panel and Table 1). It became important to determine whether the small population of cells positively marked with H3K27me3 were different from the negative cells in the normal embryos, as this subpopulation may provide insight into the inefficient development of somatic cloned embryos. To exclude the effects of culture, intracytoplasmic sperm injection-produced embryos were cultured in vitro and collected to examine the H3K27me3 modification in parallel with normal embryos. As shown in supplemental Fig. S1, no difference was observed in H3K27me3 marking in intracytoplasmic sperm injection-produced embryos as compared with fertilization-derived embryos cultured in vitro.
FIGURE 2.
Modification of H3K27me3 in preimplantation embryos. Left panel, fertilization produced embryos at 2-, 4-, 8-cell stages, morula, and blastocyst stages. Positive staining of H3K27me3 could be observed in 2- to 8- cell stage. Global demethylation of H3K27 occurred at the morula stage. A small proportion of cells were re-marked with H3K27me3 at the blastocyst stage. Scale bar, 10 μm. Right panel, cumulus cell SCNT-produced embryos at 2-, 4-, and 8-cell, morula, and blastocyst stages, and positive staining of H3K27me3 could be observed in 2- to 8- cell stage. Global demethylation of H3K27 occurred at the morula stage, but no H3K27me3 mark could be detected at the blastocyst stage. Scale bar, 10 μm.
TABLE 1.
H3K27me3-positive blastocysts derived from fertilization and SCNT
| Embryo type | Day 3.5 blastocysts examined | H3K27me3-positive blastocysts (%) |
|---|---|---|
| Fertilization blastocyst | 34 | 34 (100) |
| SCNT blastocyst | 42 | 1 (2) |
Defective Marking of H3K27me3 to the ICM Cells of Cloned Blastocysts
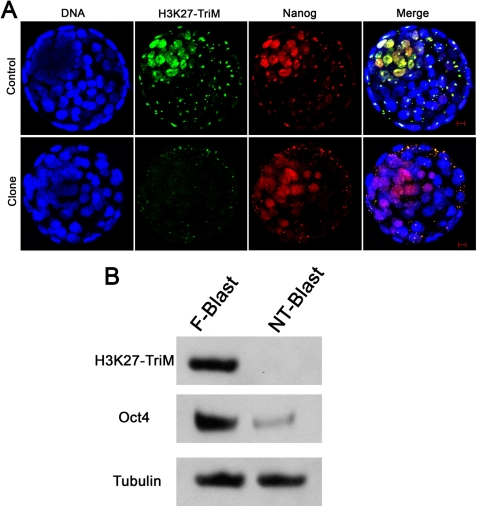
Based on the findings presented above, experiments were performed to define the characteristics of the small population of cells positively marked with H3K27me3 in normal blastocysts. ICM cells are pluripotent cells, which can be maintained in a pluripotent state in vitro as ES cells. Three important transcriptional factors, Oct4, Nanog, and Sox2, are specifically expressed in the ICM cells and are important for maintenance of pluripotency (23–26). In the present study, expression of Nanog and Oct4 was investigated in both types of embryos to distinguish the ICM cells from the trophoblast cells in which Cdx2 is highly expressed (supplemental Fig. S2). As shown in Fig. 3, in the control embryos, the small population of cells that stained positively for H3K27me3 were also stained for Nanog, which indicated that the ICM cells of normal embryos specifically marked with H3K27me3 were pluripotent (Fig. 3, upper panel). Similarly, a small population of cells were found positively stained for Nanog in somatic cloned embryos; however, these Nanog-positive cells were not marked with H3K27me3 (Fig. 3, lower panel). Intracytoplasmic sperm injection-produced blastocysts were collected, and the triple staining results indicated that the Nanog positive cells were specifically marked with H3K27me3, which is similar to fertilization-produced blastocysts (supplemental Fig. S3). Similarly, Oct4, another important transcription factor expressed in pluripotent cells, was examined in both fertilization- and SCNT-produced embryos. As shown in Fig. 4, Oct4-positive cells were detected in both kinds of embryos, but the intensity of Oct4 staining was weaker in clones. In addition, there was a dramatic difference in H3K27me3 staining, which is similar to the observations we described above. Western blotting was further performed to confirm the immunofluorescence staining results. Somatic cloned blastocysts and the normal fertilization-produced blastocysts (260 each) were collected for SDS-PAGE and Western blotting analysis. As shown in Fig. 3B, H3K27me3 could be clearly detected in the normal blastocyst lysates lane, whereas no H3K27me3 could be detected in the somatic cloned blastocysts lane. Similar to a previous report (27, 28), Oct4 expression was much weaker in the somatic clones than the normal embryos. α-Tubulin was used as a control, and there was no difference in its expression between cloned and normal embryos. These results strongly suggested that the ICM cells in somatic cloned embryos are defective in maintaining pluripotency, as modification of H3K27me3 in ES cells has been associated with the repression of differentiation genes.
FIGURE 3.
Co-localization of H3K27me3 with Nanog in the ICM of a fertilization-produced blastocyst. A, upper panel, a fertilization-produced blastocyst, co-localization of H3K27me3 with Nanog staining could be detected in the ICM cells. Scale bar, 10 μm. Lower panel, a cumulus SCNT-produced blastocyst. A small proportion of cells could be stained by Nanog, but the H3K27me3 marker is absent. Scale bar, 10 μm. B, left lane, Western blotting. Fertilization-produced blastocysts showed positive H3K27me3, Oct4. Right lane, cumulus SCNT-produced blastocysts showed weaker Oct4 and non-H3K27me3. α-Tubulin was used as a control and showed no difference between cloned and normal blastocysts.
FIGURE 4.
Co-localization of H3K27me3 with Oct4 in the ICM of a blastocyst. Upper panel, a fertilization-produced blastocyst, co-localization of H3K27me3 with Oct4 staining could be detected in ICM cells. Scale bar, 10 μm. Lower panel, a cumulus SCNT-produced blastocyst. A small proportion of cells could be stained by Oct4, but the H3K27me3 mark is absent. Scale bar, 10 μm.
Defective Marking of H3K27me3 Leads to Up-regulation of Developmental Gene Expression in Cloned Embryos
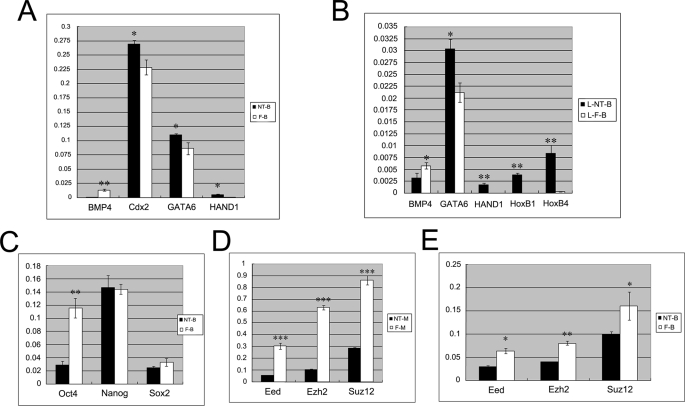
To further investigate if the defective marking of H3K27me3 in the somatic cloned embryos results in aberrant expression of differentiation genes, quantitative real-time RT-PCR was performed to evaluate the expression of key genes whose promoters have been previously shown to be occupied by H3K27me3 in ES cells (12). As shown in Fig. 5, A and B, the expression of the key developmental genes Hind1, HoxB1, and HoxB4, which mediate germ layer differentiation, was significantly elevated in somatic cloned blastocysts. The expression of pluripotent genes Nanog, Sox2 and Oct4 was simultaneously determined (Fig. 5C). There was no difference in the expression of Nanog and Sox2 between somatic cloned blastocysts and fertilization-produced blastocysts. However, Oct4 expression was significantly reduced in somatic clones, which is consistent with previous studies (27, 28). Our results indicated that defective marking of H3K27me3 is associated with the up-regulation of developmental genes, which may promote premature differentiation of the somatic cloned embryos.
FIGURE 5.
Quantitative real-time RT-PCR analysis of PRC2, pluripotent, and developmental genes expression. A and B, the differentiation-related genes (Cdx2, GATA6, HAND1, HoxB1, and HoxB4), were up-regulated significantly in cloned embryos compared with normal embryos at blastocyst and late blastocyst stages. NT-B, cumulus cell SCNT-produced day 3.5 blastocysts; F-B, fertilization-produced day 3.5 blastocysts; L-NT-B, cumulus cell SCNT-produced day 4.5 blastocysts; and L-F-B, fertilization-produced day 4.5 blastocysts. C, expression comparison of pluripotent genes of embryos at the blastocyst stage. SCNT-produced embryos showed similar expression level of Nanog and Sox2 compared with fertilization-produced embryos; however, Oct4 expression was significantly reduced in clones. D and E, the PRC2 components (Eed, Ezh2, and Suz12) responsible for H3K27me3 were down-regulated in cloned embryos, especially at the morula stage. NT-M, SCNT-produced day 2.5 morulae; F-M, fertilization-produced day 2.5 morulae. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Aberrant Expression of PRC2 Genes Causes Defective Marking of H3K27me3 to the ICM Cells of Cloned Blastocysts
Trimethylation of H3K27 is catalyzed by PRC2, which consists of Eed, Ezh2, and Suz12. To investigate whether the defective marking of H3K27me3 in the ICM cells of somatic cloned embryos was caused by aberrant expression of PRC2 components, quantitative RT-PCR was performed to evaluate the expression levels of PRC2 genes in cloned morula and blastocysts. As shown in Fig. 5D, expression of Eed, Ezh2, and Suz12 was significantly lower in the somatic cloned morula than in normal embryos at the same developmental stage. Similarly, expression of these three genes was decreased in cloned blastocysts as compared with normal blastocysts (Fig. 5E). Immunofluorescence staining and Western blotting were performed to investigate the expression of Ezh2 protein in both normal and cloned blastocysts. As shown in Fig. 6A, localization of Ezh2 in the nucleus was detected in the preimplantation control embryos, and the intensity of Ezh2 staining was higher in a small proportion of cells at the blastocyst stage (Fig. 6A, left panel). Similarly, we have found that the small cell population of pluripotent ICM cells is Oct4-positive in normal embryos (supplemental Fig. S4). In contrast, there were dramatic differences between Ezh2 staining in cloned embryos at the blastocyst stage and in control embryos; Ezh2 staining was very weak in the cloned blastocysts (Fig. 6A, right panel). To verify the immunofluorescence staining results, Western blotting was performed. Similar to the H3K27me3 results presented above, there was no obvious Ezh2 protein detected using Western blotting in the lysates generated from 240 cloned blastocysts. In contrast, a very specific band of Ezh2 was detected using the same amount of normal blastocyst lysates (Fig. 6B). These results suggested that defective marking of H3K27me3 in the ICM cells in the somatic cloned blastocysts was directly caused by the down-regulation of PRC2 components in cloned embryos. PRC2 components are essential for embryonic development, and it has been previously demonstrated that knock-out of any of the three genes is embryonically lethal (29–31). It has been shown that knock-out of Ezh2 caused depletion of H3K27me3 modification in ICM cells, which is similar to the phenomenon that we observed in this study. The Ezh2 mutant embryos died soon after implantation, and the dead embryos were found to contain predominantly mesodermic tissue. Our gene expression data indicated that the developmental genes directing mesoderm differentiation were highly expressed in the somatic cloned embryos. Loss of cloned embryos right after implantation has been confirmed in previous studies, and all the data indicated that defective H3K27me3 marking in the ICM cells of somatic cloned embryos may contribute to the premature death of somatic clones.
FIGURE 6.
Detection of Ezh2 expression in preimplantation normal and cloned embryos. A, left panel, expression of Ezh2 in fertilization-produced normal embryos at 2-, 4-, 8-cell, morula, and blastocyst stages detected by immunofluorescence staining. Enhanced Ezh2 staining in a small proportion of cells could be detected at the blastocyst stage. Scale bar, 10 μm. Right panel, expression of Ezh2 in SCNT-produced cloned embryos at 2-, 4-, 8-cell, morula, and blastocyst stages detected by immunofluorescence staining, no obvious Ezh2 staining could be observed in cloned blastocysts. Scale bar, 10 μm. B, Western blotting. Right lane, fertilization-produced blastocysts (F-Blast) showed positive Ezh2 and H3K27me3. Left lane, cumulus SCNT-produced blastocysts (NT-Blast) showed weaker Ezh2 and non-H3K27me3. α-Tubulin was used as control.
DISCUSSION
Overall, these results suggest that inadequate expression of PRC2 components, Eed, Ezh2, and Suz12, particularly Ezh2, leads to inadequate methylation of H3K27 in ICM cells of somatic cloned embryos, and this results in greater expression of developmental genes that induce premature differentiation and death of clones. To our knowledge, this is the first time that these differences have been distinguished between SCNT- and fertilization-produced blastocysts. These studies suggest that H3K27me3 might be an important epigenetic marker with which to evaluate the developmental potential of cloned embryos and that treatments that enhance H3K27me3 in cloned embryos might be an effective strategy to improve the efficiency of cloning.
ICM cells are ES cell counterparts in vivo, and a similar pluripotency regulatory network might be employed to precisely regulate the determination of cell fate during embryonic development. Two master pluripotency regulators, Oct4 and Nanog, are essential for early embryonic survival and mutation of either results in early embryonic lethality (23, 25, 26). In addition, epigenetic modifications also contribute to the maintenance of the pluripotency of ICM cells. It has been previously proven that PRC2 component genes, including Ezh2, Eed, and Suz12, function in early embryonic development (29–31). Mutation of each of these genes during early development will result in loss of H3K27me3 mark in the ICM cells of mutant blastocysts (29). Subsequently, the mutant embryos will differentiate prematurely, which will cause early embryonic death shortly after implantation. These data implicated that the maintenance of pluripotency of ICM cells in the blastocysts requires precise expression of both master pluripotency-regulating genes and PRC2 genes.
The efficiency of cloning has remained extremely low because the first SCNT cloned animal was generated a decade ago, and the cause of this inefficiency is poorly understood (32, 33). Extensive efforts have been undertaken to improve the efficiency of cloning, such as treating the cloned embryos with histone deacetylase inhibitors to alter the global histone acetylation during development. However, even though the cloned embryos developed to the blastocyst stage with higher efficiency, the late stage development was not significantly improved (21, 34–36). Collectively, the data indicated that most somatic clones died soon after implantation, which is similar to the phenomena observed in pluripotency master regulators or PRC2 components mutant embryos.
Overall, these data suggested that the somatic clones are epigenetically defective at the blastocyst stage. Our results further demonstrated that loss of the H3K27me3 mark in the ICM cells of most cloned embryos will cause premature expression of the genes that should be repressed by the activity of PRC2, which results in early embryonic death that phenocopies PRC2 mutants. To date, no non-human primates have ever been successfully cloned by SCNT; moreover, derivation of individual patient-specific autologous nuclear transfer ES cells has not been achieved, and it deserves to be further investigated whether cloned human blastocysts possess the H3K27me3 defects similar to those observed in the mouse clones.
Supplementary Material
Acknowledgments
We thank the lab members for helpful comments on the manuscript.
This work was supported by Grants 2008AA022311 and 2009CB941100 from the Ministry of Science and Technology of China and Grant 30670302 from the National Natural Science Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- ES
- embryonic stem
- RT
- reverse transcription
- SCNT
- somatic cell nuclear transfer
- ICM
- inner cell mass
- PRC
- polycomb repressive complex
- H3K27me3
- H3K27 trimethylation.
REFERENCES
- 1.Evans M. J., Kaufman M. H. (1981) Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 2.Martin G. R. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 4.Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 6.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 7.Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. (2002) Cell 111, 185–196 [DOI] [PubMed] [Google Scholar]
- 8.Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. (2002) Genes Dev. 16, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montgomery N. D., Yee D., Chen A., Kalantry S., Chamberlain S. J., Otte A. P., Magnuson T. (2005) Curr. Biol. 15, 942–947 [DOI] [PubMed] [Google Scholar]
- 10.Pasini D., Bracken A. P., Hansen J. B., Capillo M., Helin K. (2007) Mol. Cell. Biol. 27, 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O'Connor M. B., Kingston R. E., Simon J. A. (2002) Cell 111, 197–208 [DOI] [PubMed] [Google Scholar]
- 12.Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 13.Wilmut I., Schnieke A. E., McWhir J., Kind A. J., Campbell K. H. (1997) Nature 385, 810–813 [DOI] [PubMed] [Google Scholar]
- 14.Wakayama T., Perry A. C., Zuccotti M., Johnson K. R., Yanagimachi R. (1998) Nature 394, 369–374 [DOI] [PubMed] [Google Scholar]
- 15.Hochedlinger K., Jaenisch R. (2002) Nature 415, 1035–1038 [DOI] [PubMed] [Google Scholar]
- 16.Eggan K., Baldwin K., Tackett M., Osborne J., Gogos J., Chess A., Axel R., Jaenisch R. (2004) Nature 428, 44–49 [DOI] [PubMed] [Google Scholar]
- 17.Dean W., Santos F., Stojkovic M., Zakhartchenko V., Walter J., Wolf E., Reik W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13734–13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y. K., Koo D. B., Park J. S., Choi Y. H., Chung A. S., Lee K. K., Han Y. M. (2001) Nat. Genet. 28, 173–177 [DOI] [PubMed] [Google Scholar]
- 19.Chatot C. L., Ziomek C. A., Bavister B. D., Lewis J. L., Torres I. (1989) J. Reprod. Fertil. 86, 679–688 [DOI] [PubMed] [Google Scholar]
- 20.Gao S., Chung Y. G., Williams J. W., Riley J., Moley K., Latham K. E. (2003) Biol. Reprod. 69, 48–56 [DOI] [PubMed] [Google Scholar]
- 21.Wang F., Kou Z., Zhang Y., Gao S. (2007) Biol. Reprod. 77, 1007–1016 [DOI] [PubMed] [Google Scholar]
- 22.Bao S., Miyoshi N., Okamoto I., Jenuwein T., Heard E., Azim Surani M. (2005) EMBO Rep. 6, 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schöler H. R., Balling R., Hatzopoulos A. K., Suzuki N., Gruss P. (1989) EMBO J. 8, 2551–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998) Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 25.Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003) Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- 26.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003) Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 27.Boiani M., Eckardt S., Schöler H. R., McLaughlin K. J. (2002) Genes Dev. 16, 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamazaki Y., Fujita T. C., Low E. W., Alarcón V. B., Yanagimachi R., Marikawa Y. (2006) Mol. Reprod. Dev. 73, 180–188 [DOI] [PubMed] [Google Scholar]
- 29.O'Carroll D., Erhardt S., Pagani M., Barton S. C., Surani M. A., Jenuwein T. (2001) Mol. Cell. Biol. 21, 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faust C., Lawson K. A., Schork N. J., Thiel B., Magnuson T. (1998) Development 125, 4495–4506 [DOI] [PubMed] [Google Scholar]
- 31.Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K. (2004) EMBO J. 23, 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blelloch R., Wang Z., Meissner A., Pollard S., Smith A., Jaenisch R. (2006) Stem Cells 24, 2007–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X., Smith S. L., Tian X. C., Lewin H. A., Renard J. P., Wakayama T. (2007) Nat. Genet. 39, 295–302 [DOI] [PubMed] [Google Scholar]
- 34.Kishigami S., Mizutani E., Ohta H., Hikichi T., Thuan N. V., Wakayama S., Bui H. T., Wakayama T. (2006) Biochem. Biophys. Res. Commun. 340, 183–189 [DOI] [PubMed] [Google Scholar]
- 35.Kishigami S., Bui H. T., Wakayama S., Tokunaga K., Van Thuan N., Hikichi T., Mizutani E., Ohta H., Suetsugu R., Sata T., Wakayama T. (2007) J. Reprod. Dev. 53, 165–170 [DOI] [PubMed] [Google Scholar]
- 36.Enright B. P., Kubota C., Yang X., Tian X. C. (2003) Biol. Reprod. 69, 896–901 [DOI] [PubMed] [Google Scholar]
- 37.Deleted in proof
- 38.Deleted in proof
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.