Abstract
The Arabidopsis thaliana locus At5g06580 encodes an ortholog to Saccharomyces cerevisiae d-lactate dehydrogenase (AtD-LDH). The recombinant protein is a homodimer of 59-kDa subunits with one FAD per monomer. A substrate screen indicated that AtD-LDH catalyzes the oxidation of d- and l-lactate, d-2-hydroxybutyrate, glycerate, and glycolate using cytochrome c as an electron acceptor. AtD-LDH shows a clear preference for d-lactate, with a catalytic efficiency 200- and 2000-fold higher than that for l-lactate and glycolate, respectively, and a Km value for d-lactate of ∼160 μm. Knock-out mutants showed impaired growth in the presence of d-lactate or methylglyoxal. Collectively, the data indicated that the protein is a d-LDH that participates in planta in the methylglyoxal pathway. Web-based bioinformatic tools revealed the existence of a paralogous protein encoded by locus At4g36400. The recombinant protein is a homodimer of 61-kDa subunits with one FAD per monomer. A substrate screening revealed highly specific d-2-hydroxyglutarate (d-2HG) conversion in the presence of an organic cofactor with a Km value of ∼580 μm. Thus, the enzyme was characterized as a d-2HG dehydrogenase (AtD-2HGDH). Analysis of knock-out mutants demonstrated that AtD-2HGDH is responsible for the total d-2HGDH activity present in A. thaliana. Gene coexpression analysis indicated that AtD-2HGDH is in the same network as several genes involved in β-oxidation and degradation of branched-chain amino acids and chlorophyll. It is proposed that AtD-2HGDH participates in the catabolism of d-2HG most probably during the mobilization of alternative substrates from proteolysis and/or lipid degradation.
l- and d-lactate dehydrogenases belong to evolutionarily unrelated enzyme families (1). l-Lactate is oxidized by l-lactate:NAD oxidoreductase (EC 1.1.1.27), which catalyzes the reaction l-lactate + NAD → pyruvate + NADH, and by l-lactate cytochrome c oxidoreductase (l-lactate cytochrome c oxidoreductase, EC 1.1.2.3), which catalyzes the reaction l-lactate + 2 cytochrome c (oxidized) → pyruvate + 2 cytochrome c (reduced). Both groups are found in eubacteria, archebacteria, and eukaryotes. All known plant sequences belong to the EC 1.1.1.27 group (1). On the other hand, d-lactate is oxidized by d-lactate:NAD oxidoreductase (d-lactate:NAD oxidoreductase, EC 1.1.1.28), which catalyzes the reaction d-lactate + NAD → pyruvate + NADH, and by d-lactate cytochrome c oxidoreductase (d-lactate cytochrome c oxidoreductase, EC 1.1.2.4), which catalyzes the reaction d-lactate + 2 cytochrome c (oxidized) → pyruvate + 2 cytochrome c (reduced).
Although l-lactate dehydrogenase belongs to the most intensely studied enzyme families (2, 3), our knowledge about the structure, kinetics, and biological function of d-LDH3 is limited. d-LDHs have mainly been identified in prokaryotes and fungi where they play an important role in anaerobic energy metabolism (4–10). In Saccharomyces cerevisiae and Kluyveromyces lactis, a mitochondrial flavoprotein d-lactate ferricytochrome c oxidoreductase (d-lactate cytochrome c oxidoreductase), catalyzing the oxidation of d-lactate to pyruvate, is required for the utilization of d-lactate (8, 11). In S. cerevisiae it was suggested that d-LDH is involved in the metabolism of methylglyoxal (MG) (12).
In eukaryotic cells, d-lactate results from the glyoxalase system (13, 14). This system is the main MG catabolic pathway, comprising the enzymes glyoxalase I (lactoylglutathione lyase, EC 4.4.1.5) and glyoxalase II (hydroxyacylglutathione hydrolase, EC 3.1.2.6). MG (CH3-CO-CHO; see structure in Fig. 4) is a cytotoxic compound formed primarily as a by-product of glycolysis through nonenzymatic phosphate elimination from dihydroxyacetone phosphate and glyceraldehyde 3-phosphate (15), and its production in various plants is enhanced under stress conditions such as salt, drought, cold, and heavy metal stress (16, 17). Moreover, the overexpression of glyoxalase I or II was shown to confer resistance to salt stress in tobacco and rice (17, 18). It is assumed that the role of the MG pathway, from MG synthase to d-lactate cytochrome c oxidoreductase in the extant metabolism, is to detoxify MG, whereas in the early state of metabolic development it might function as an anaplerotic route for the tricarboxylic acid cycle (15).
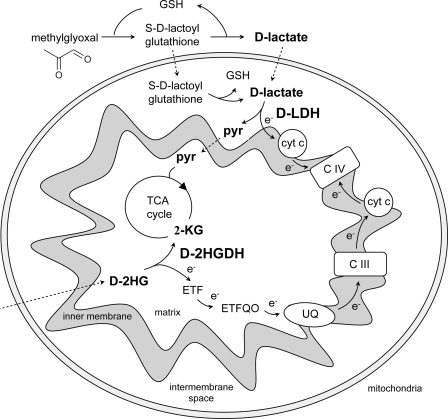
FIGURE 4.
Scheme showing the involvement of AtD-LDH in the methylglyoxal pathway and of AtD-2HGDH in the respiration of substrates from proteolysis and/or lipid degradation. d-Lactate resulting from the glyoxalase system is converted to pyruvate by AtD-LDH. The electrons originated may be transferred to the respiratory chain through cytochrome c in the intermembrane space. d-2-HG produced in the peroxisomes (as shown in supplemental Fig. S3) is transported to the mitochondria and converted to 2-ketoglutarate by AtD-2HGDH. Electrons are donated to the electron transport chain through the ETF/ETFQO system. Dotted files represent possible transport processes. 2-KG, 2-ketoglutarate. CIII, complex III. CIV, complex IV. e−, electron. ETF, electron transfer protein. ETFQO, ETF-ubiquinone oxidoreductase. GSH, glutathione. Pyr, pyruvate. TCA cycle, tricarboxylic acid cycle; UQ, ubiquinone.
Glyoxalase I catalyzes the formation of S-d-lactoylglutathione from the hemithioacetal formed nonenzymatically from MG and glutathione, although glyoxalase II catalyzes the hydrolysis of S-d-lactoylglutathione to regenerate glutathione and liberate d-lactate. Glyoxalase I and II activities are present in all tissues of eukaryotic organisms. Glyoxalase I is found in the cytosol, whereas glyoxalase II localizes to the cytosol and mitochondria (13, 19, 20). Although glyoxalase I and II were extensively characterized, there are only few reports on the characterization of d-LDH. Recently, Atlante et al. (13) showed that externally added d-lactate caused oxygen consumption by mitochondria and that this metabolite was oxidized by a mitochondrial flavoprotein in Helianthus tuberosus.
The complete sequence of Arabidopsis thaliana opened the way to search for genes encoding d-LDHs. Based on similarity with the d-LDH from S. cerevisiae (DLD1), an A. thaliana ortholog was identified. In this study, the isolation and structural and biochemical characterization of the recombinant mature d-LDH from A. thaliana (AtD-LDH) and its paralog, which was found to be a d-2-hydroxyglutarate dehydrogenase (AtD-2HGDH), is described. Whereas AtD-LDH has a narrow substrate specificity and the preferred substrates are d-lactate and d-2-hydroxybutyrate, AtD-2HGDH showed activity exclusively with d-2-hydroxyglutarate. Based on gene coexpression analysis and analysis of corresponding knock-out mutants, the participation of these previously unrecognized mitochondrial activities in plant metabolism is discussed.
EXPERIMENTAL PROCEDURES
Cloning of Mature AtD-LDH and AtD-2HGDH cDNA in an Expression Vector
Total RNA was isolated from 100-mg leaves of 4-week-old plants using the TRIzol reagent (Invitrogen). One microgram of total RNA was reverse-transcribed into first strand cDNA using the SuperScriptII reverse transcriptase (Invitrogen) and used for PCR amplification. To express the mature enzymes, specific primers containing the first codon downstream of the predicted cleavage site were generated. Oligonucleotide primers were designed to introduce a BamHI site both at the 5′ and at the 3′ end of the fragment. The primers used for the amplification were as follows: AtD-LDH-F (5′-GGATCCGGCGATAGCTGCCTCCG-3′) and AtD-LDH-R (5′-GGATCCTTAGAAACATACATGAGGAGGAATTAAC-3) and AtD-2HGDH-F (5′-GGATCCGGTGTCAGGCTTTGTG-3) and AtD-2HGDH-R (5′-GGATCCTTAGTTGGAGAAGAGAGAGTGAGG-3). First-strand synthesis products were amplified by PCR using the PfuTurbo DNA polymerase (Stratagene) and the following conditions: initial denaturation at 95 °C for 1.5 min followed by 37 cycles of denaturation at 95 °C for 30 s, annealing at 64 °C for 40 s, elongation at 72 °C for 2 min, and a final elongation step at 72 °C for 10 min. The amplified products were cloned into pCR-BluntII-TOPO (Invitrogen) and the generated plasmid, called pTopoBluntAtD-LDH and pTopoBluntAtD-2HGDH, was sequenced using the PRISM fluorescent dye-terminator system (Applied Biosystems). The inserts were cut out with BamHI and ligated into a BamHI linearized pET16b vector (Novagen). The resulting expression vectors designated as pET16b-AtD-LDH and pET16b-AtD-2HGDH were transformed into the BLR DE3 pLysS Escherichia coli strain (Novagen).
Expression and Purification of Recombinant AtD-LDH and AtD-2HGDH
The vectors pET16b-AtD-LDH and pET16b-AtD-2HGDH generate fusion proteins with an N-terminal His tag facilitating the purification of the recombinant proteins. The BLR cells transformed with the vectors were grown in LB medium at 37 °C and agitation at 220 rpm in the presence of 50 μg/ml carbenicillin, 50 μg/ml chloramphenicol, and 5 μg/ml tetracycline until the culture reached an A600 nm of 0.6. To induce the expression of AtD-LDH and AtD-2HGDH, 1 mm isopropyl β-d-thiogalactopyranoside was added to the culture, and the bacteria were allowed to grow for another hour. The cells were then harvested by centrifugation at 4,000 × g for 10 min, resuspended in 35 ml of 20 mm Tris-HCl (pH 8.0) containing 0.5 mg/ml phenylmethylsulfonyl fluoride, sonicated, and centrifuged at 10,000 × g for 15 min at 4 °C to remove debris. The supernatant was used for protein purification using immobilized metal ion chromatography on Ni2+-nitrilotriacetic acid-agarose (Ni2+-NTA, Qiagen). The column was equilibrated and washed with 100 mm Na2PO4 buffer (pH 8.0) containing 300 mm NaCl and 10 mm imidazole. The protein was eluted using 3 ml of 100 mm Na2PO4 (pH 7.2) and 250 mm imidazole. The yield of the expressed proteins (greater than 95% pure) was about 2 mg/liter E. coli culture. The purified enzymes were used immediately for the kinetic measurements or, alternatively, concentrated using Centricon YM-30 (Amicon) and incubated with protease factor Xa (1:100 v/v) at 10 °C for 2 h to remove the N terminus encoded by the expression vector (20 amino acid residues). The proteins were further purified using a Ni2+-NTA column to eliminate the cleaved fusion peptide. The eluted proteins were used immediately for kinetic measurements.
Substrate Screening and Enzymatic Activities
The pH dependence of the AtD-LDH and AtD-2HGDH activities was determined spectrophotometrically using a reaction mixture containing 50 mm K2PO4 (pH 6.0–9.5) and 200 μm cytochrome c or, alternatively, 3 mm phenazine methosulfate (PMS) and 200 μm 2,6 dichlorophenolindophenol (DCIP) in a final volume of 0.2 ml. In the case of AtD-LDH, 10 mm of either glycolate or sodium d-lactate and for AtD-2HGDH 10 mm Na-d-2-hydroxyglutarate (d-2HG) were used as substrates. Activities were measured at 25 °C by recording the reduction of DCIP at 600 nm (ϵ, 22 cm−1 mm−1) or, alternatively, cytochrome c at 550 nm (ϵ, 18.6 cm−1 mm−1). When assaying with cytochrome c, the resulting absorption was divided by 2 because 2 mol of cytochrome c are reduced for each mole of substrate oxidized. A base line was determined by monitoring the reaction mix for 4 min before the addition of purified enzyme (0.2–40 μg, depending on the substrate used). The substrate specificity of the enzymes was assayed using 10 mm substrate (see Tables 2 and 3) at pH 8.75 and cytochrome c or PMS and DCIP as electron acceptors as described above. The Km and Vmax values for the best substrates were determined at pH 8.75 with cytochrome c or PMS and DCIP and varying the concentration of the substrate while keeping the concentrations of the other components constant. All constants were calculated with at least two different enzyme batches each containing at least triplicate determinations and adjusted to nonlinear regression (SigmaPlot). One unit of enzyme activity is defined as the amount that oxidizes 1 μmol of substrate/min. This corresponds to the reduction of 2 μmol of cytochrome c or 1 μmol of DCIP.
TABLE 2.
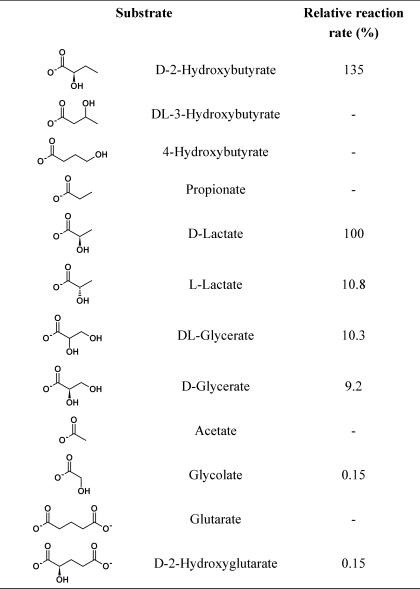
Enzymatic activities of recombinant AtD-LDH using various substrates and cytochrome c as electron acceptor
Activity was measured under the standard assay conditions described under “Experimental Procedures”. 100% activity is defined for the substrate d-lactate and represents 1.1 ± 0.03 μmol min−1 mg−1.
TABLE 3.
Kinetic parameters of recombinant AtD-LDH and AtD-2HGDH
Kinetic data were best fit by nonlinear regression analysis. Parameters represent the mean ± S.E. of two independent enzyme preparations.
| With cytochrome c |
|||
|---|---|---|---|
| Km | kcat | kcat/Km | |
| μm | min−1 | min−1mm−1 | |
| AtD-LDH | |||
| d-2HB | 61 ± 2 | 88 ± 1.2 | 1450 |
| d-lactate | 164 ± 11 | 65 ± 1.8 | 395 |
| l-lactate | 4486 ± 374 | 7.0 ± 0.3 | 2.0 |
| d-glycerate | 8871 ± 1379 | 6.0 ± 0.8 | 1.0 |
| Glycolate | 432 ± 31 | 0.1 ± 0.0 | 0.2 |
| With DCIP |
|||
|---|---|---|---|
| d-lactate | 317 ± 31 | 73 ± 1.7 | 231 |
| l-lactate | 7134 ± 456 | 5 ± 0.1 | 0.7 |
| Glycolate | 596 ± 44 | 0.4 ± 0.0 | 0.7 |
| AtD-2HGDH | |||
| d-2HG | 584 ± 45 | 48 ± 0.9 | 82 |
The oxidation of glycolate, d- and l-lactate, and d-2HG by NAD or NADP (1 mm) as well as the reduction of glyoxylate, pyruvate, and 2-ketoglutarate by NADH or NADPH (1 mm) was measured at 340 nm (ϵ, 6.22 cm−1 mm−1) as described above. The possible oxidase activity on glycolate using O2 was measured as described previously (21).
Native Molecular Mass Estimation
The molecular masses of the recombinant proteins were estimated by gel filtration chromatography on a ÄKTA purifier system (GE Healthcare) using a Superdex 200 10/300 GL column (GE Healthcare). The column was equilibrated with 20 mm Tris-HCl (pH 8.0) and calibrated using molecular mass standards. The samples and the standards were applied separately in a final volume of 10 μl at a constant flow rate of 0.5 ml/min.
Separation and Analysis of the Prosthetic Group
The absorption spectra of AtD-LDH and AtD-2HGDH were measured with a Hach DR 5000 UV-visible spectrophotometer in 2 nm steps. The cofactors were released from the recombinant purified proteins (0.7 mg) by heating at 95 °C for 15 min, and the denatured protein was removed by centrifugation. The resulting supernatants (20 μl), riboflavin (2 μl, 2 mm), FMN (2 μl, 2 mm), and FAD (2 μl, 2 mm) were spotted on cellulose 300 thin layer chromatography plates and run in the dark with buffer A consisting of 5% Na2HPO4 in aqueous solution or, alternatively, with buffer B consisting of 1-butanol:HAc:water (4:1:5). Sample positions were subsequently detected under UV light. The same samples were analyzed on a native polyacrylamide gel (0.75 m Tris (pH 8.45)) consisting of an 8% T, 3% C stacking gel and an 18% T, 6.1% C separation gel. The gel was run at 100 V in darkness using as anode buffer 0.2 m Tris (pH 8.9) and as cathode buffer 0.1 m Tris and 0.1 m Tricine (pH 8.25).
Screening for tDNA Insertion Lines and Plant Growth Conditions
Seeds of tDNA insertion lines Salk_001490 (dldh1-1), Salk_026859 (dldh1-2), Salk_061383 (d2hgdh1-1), and Sail_844_G06 (d2hgdh1-2) were obtained from the Nottingham Arabidopsis Stock Center. Homozygous plants were isolated by two sets of PCRs. The first PCR was performed using the gene-specific primers d-LDH1–1-F (5′-ATAAGAAAAGAGGCGCTGTGG-3′) and d-LDH1–1-R (5′-TTAGAAACATACATGAGGAGGAATTAACTTTCCC-3′) for dldh1-1, or d-LDH1–2-F (5′-ATGGCTTTCGCTTCAAAATTC-3′) and d-LDH1–2-R (5′-GTAGCACACATGCCTCCTATGG-3′) for dldh1-2, or d-2HGDH1–1-F (5′-CGAGACAACAGGGAGTGATG-3′) and d-2HGDH1–1-R (5′-GTTGGAGAAGAGAGAGTGAGGAAG-3′) for d2hgdh1-1, or d-2HGDH1–2-F (CTTTGTTTGGATTGCGATGA) and d-2HGDH1–2-R (CATTGTACAAGCACAAGCCAAT) for d2hgdh1-2. The second PCR was carried out with the tDNA left border primer SALK-LB (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′) in combination with d-LDH1–1-F, d-LDH1–2-R, or d-2HGDH-R using the lines dldh1-1, dldh1-2, and d2hgdh1-1, respectively. In the case of line d2hgdh1-2, the second PCR was carried out using the tDNA left border primer Sail-LB (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′) and d-2HGDH1–2-R. A. thaliana wild type (Columbia-0) and the tDNA insertion lines were grown in pots containing 3 parts soil (Gebr. Patzer KG, Sinntal-Jossa) and 1 part vermiculite (Basalt Feuerfest, Linz) in a growth cabinet at a 16:8 h light:dark cycle at 22 °C day and 18 °C night temperatures and at a photosynthetically active photon flux density of 100 μmol quanta m−2 s−1. Alternatively, plants were grown in liquid medium containing 4.6 g/liter Murashige and Skoog (MS) and 3% sucrose at pH 5.6 in the dark at 28 °C and constant agitation. Growth of plants in the presence of different concentrations of l-lactate, MG, and glycolate was conducted on ½ MS agar plates in a growth cabinet as described above.
Semi-quantitative Reverse Transcription-PCR Analysis
To determine the expression of both homologous genes in the insertion mutants, total RNA was isolated from 100 mg of leaves using the TRIzol reagent (Invitrogen). RNA was converted into first strand cDNA using the SuperScriptII reverse transcriptase (Invitrogen). PCRs were conducted in a final volume of 10 μl using 1 μl of the transcribed product and TaqDNA polymerase (Qiagen). The pairs of primers used were d-LDH1–1-F and d-LDH1–1-R for dldh1-1 and dldh1-2 or d-2HGDH-F and d-2HGDH-R for d2hgdh1-1 and d2hgdh1-2. Amplification conditions were as follows: 1.5 min of denaturation at 95 °C; 35 cycles at 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 2 min, followed by 5 min at 72 °C. As control, the actin2 gene was amplified by 28 cycles using the following primers: Actin2-F (5′-TAACTCTCCCGCTATGTATGTCGC-3′) and Actin2-R (5′-CCACTGAGCACAATGTTACCGTAC-3′).
Protein Crude Extracts and Mitochondrial Preparations
Leaves from 3-week-old soil-grown plants and roots from liquid culture-grown plants (500 mg) from wild type and the tDNA insertion lines were ground in liquid N2 in the presence of 500 μl of 20 mm Tris-HCl (pH 8.0) containing 0.05% Triton X-100 and 1 mg/ml phenylmethylsulfonyl fluoride. The homogenates were clarified by centrifugation at 20,000 × g for 5 min and subjected to native PAGE. Mitochondria isolation and intactness of the preparations were essentially as described by Keech et al. (22). Purity of the mitochondrial preparations was analyzed by Western blot using antibodies raised against the cytosolic malate dehydrogenase from Ananas comosus, which react with the cytosolic and chloroplastic isoforms but not with the mitochondrial one (donated by Florencio Podestá, University of Rosario).
PAGE and Protein Determination
Protein concentration was determined according to the method of Bradford (23). Denaturing SDS-PAGE was performed using 12.5% (w/v) polyacrylamide gels according to Laemmli (24). Proteins were visualized with Coomassie Blue or electroblotted onto a nitrocellulose membrane for immunoblotting. For Western blots, the membrane was incubated with anti-His antibodies (1:10; Qiagen). Bound antibodies were visualized using alkaline phosphatase-conjugated goat anti-rabbit IgG according to the manufacturer's instructions (Sigma). Native PAGE was performed employing an 8% (w/v) polyacrylamide gel. Electrophoresis was run at 100 V and 4 °C. Gels were assayed for activity by incubation in the dark at 30 °C with 50 mm K2PO4 (pH 8.75), 5 mm substrate, 0.05% (w/v) nitro blue tetrazolium, and 150 μm PMS in a final volume of 5 ml.
Gene Coexpression Network Analysis
Genes that are coexpressed with At5g06580 and At4g36400 were identified using the ATTED-II data base (25) and summarized in supplemental Table 1. The Mutual Rank, which is based on weighted Pearson correlation coefficients, is used to assess the extent of gene coexpression. The Mutual Rank is calculated as the geometric mean of the correlation rank of gene A to gene B and of gene B to gene A. The Gene Ontology annotations were manually actualized and corrected.
RESULTS
In Silico Identification of d-LDH Orthologous Proteins in A. thaliana and Phylogenetic Analysis of Homologs
To identify candidate genes encoding d-LDH enzymes, the A. thaliana genome was searched with the S. cerevisiae d-LDH (ScD-LDH or DLD1, UniProtKB/Swiss-Prot accession number P32891) protein sequence, using the BLAST search algorithm. The top hit was a polypeptide of 567 amino acids (62.2 kDa) encoded by locus At5g06580. This protein contains a predicted mitochondrial targeting sequence (26). A subsequent BLAST search in the A. thaliana genome using the At5g06580 translated sequence yielded a second protein with 24% identity. This 559-amino acid protein (61.4 kDa) was also predicted to localize to mitochondria (26) and is encoded by locus At4g36400.
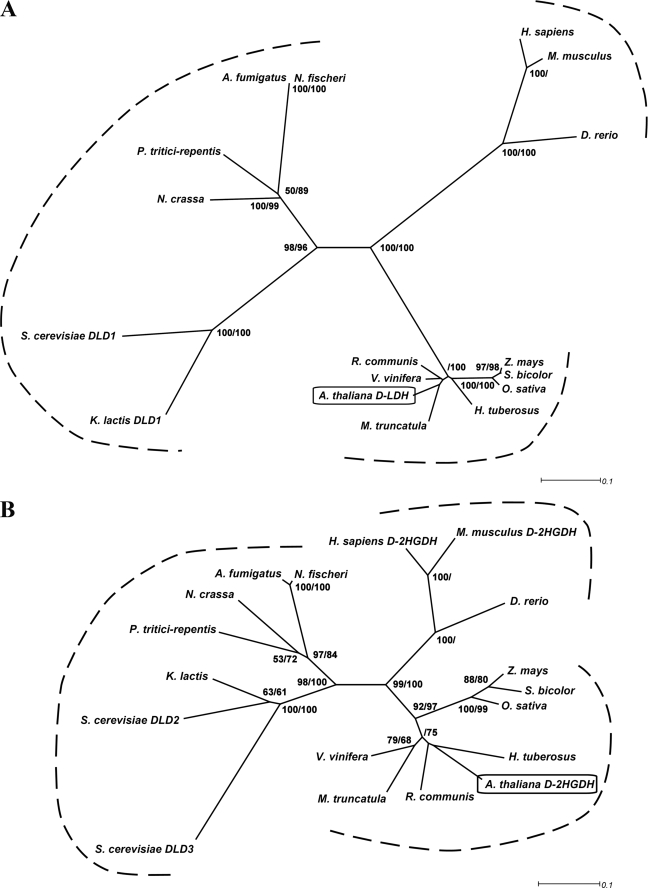
A phylogenetic tree generated using a multiple sequence alignment of orthologous sequences of At5g06580 and At4g36400 showed two well resolved groups, which include At5g06580 (Fig. 1A) or At4g36400 (Fig. 1B), respectively. Interestingly, plants, mammalia, and fungi always possess both sequences, each belonging to one of the two groups (Fig. 1 and supplemental Fig. S1). Furthermore, the phylogenetic analysis clearly shows that plant, mammalian, and fungi proteins group into different clusters (Figs. 1A and 2A). The retention of both enzymes in all higher organisms investigated indicates that these proteins may have universal roles in metabolism.
FIGURE 1.
Phylogenetic relationships of AtD-LDH and AtD-2HGDH. Protein sequences were aligned using ClustalW. The phylogenetic trees were constructed by the neighbor-joining method using BioNJ and visualized using Dendroscope (59). Bootstrap values at the nodes represent the percentage support from 500 replicates with neighbor-joining and maximum likelihood, respectively. Only bootstrap values over 50% are shown. A, phylogenetic tree of AtD-LDH homologous proteins. The following sequences are included: A. thaliana d-LDH (Q94AX4); H. tuberosus (EL444323, EL437620, and EL440912); Zea mays (B4G146); Sorghum bicolor (Sb02g003640); Oryza sativa (B9FVM4); Ricinus communis (B9SDP2); Vitis vinifera (A7Q036); Medicago truncatula (BF636550, BF636365, BF636697, BQ123259, CA858805, CA858793, and DY617162); H. sapiens (Q86WU2); Danio rerio (Q803V9); M. musculus (Q7TNG8); S. cerevisiae DLD1 (P32891); K. lactis (Q12627); Neurospora crassa (Q7SE05); Pyrenophora tritici-repentis (B2WBK4); Aspergillus fulmigatus (Q4WE96); and Neosartorya fischeri (A1D828). B, phylogenetic tree of AtD-2HGDH homologous proteins. The following sequences are included: A. thaliana d-2HGDH (O23240); H. tuberosus (EL452354); Z. mays (B4FWJ7); S. bicolor (e_gw1.2.1014.1); O. sativa (Q7XI14); R. communis (B9S687); V. vinifera (A7QE54); M. truncatula (MtAC136451_19.4); H. sapiens (Q8N465); D. rerio (A1L258); M. musculus (Q8CIM3); S. cerevisiae DLD2 (P46681); S. cerevisiae DLD3 (P39976); K. lactis (Q6CL48); N. crassa (Q7RYX6); P. tritici-repentis (B2WHR3); A. fulmigatus (Q4WE96); and N. fischeri (A1D092).
FIGURE 2.
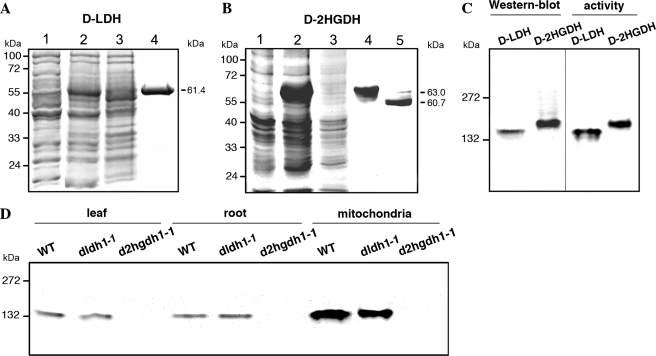
A. thalianad-LDH and d-2HGDH analyzed by gel electrophoresis. Coomassie-stained SDS-PAGE of the progress of recombinant d-LDH (A) and d-2HGDH (B) purification. Lane 1, 20 μg of noninduced cell culture lysate. Lane 2, 20 μg of cell culture lysate after 2 h of induction with isopropyl β-d-thiogalactopyranoside. Lane 3, 20 μg of Ni2+-NTA column flow through. Lane 4, 5 μg of purified recombinant d-LDH or d-2HGDH. Lane 5, d-2HGDH after factor Xa digestion. The molecular masses of the purified proteins are indicated on the right. Molecular weight markers run in parallel are indicated on the left. C, native PAGE of recombinant d-LDH and d-2HGDH analyzed by Western blot using anti-His antibodies (on the left) and stained for activity using d-lactate and d-2HG as substrates in the case of d-LDH and d-2HGDH, respectively (on the right). The reaction was stopped after 30 min of incubation in the respective solutions. Molecular weight markers run in parallel are indicated on the left. D, native PAGE of leaf, root, and leaf mitochondria extracts from wild type (WT), dldh1-1, and d2hgdh1-1 analyzed for d-2HGDH activity using d-2HG as substrate. The reaction was stopped after 30 min of incubation. Molecular weight markers run in parallel are indicated on the left.
The only two characterized proteins orthologous to At5g06580, S. cerevisiae DLD1 (supplemental Fig. S1A) and K. lactis DLD1, show d-LDH activity (8, 10, 27), and thus, At5g06580 was designated as AtD-LDH. Bacterial d-LDHs resemble the proteins of the At5g06580 group, but the sequences are less conserved (9, 28). On the other hand, four proteins orthologous to At4g36400 have been described previously. The enzymes from Homo sapiens (supplemental Fig. S1B) and Mus musculus display d-2-hydroxyglutarate dehydrogenase (d-2HGDH) activity (29), whereas the proteins from S. cerevisiae (ScDLD2 (supplemental Fig. S1B) and ScDLD3) have d-LDH activity (30). However, ScDLD2 and ScDLD3 have not been tested with d-2HG, and thus, previous investigations could not exclude that this could be a substrate for these enzymes. At4g36400 was therefore designated as AtD-2HGDH.
Expression and Purification of the Recombinant A. thaliana d-LDH and d-2HGDH
Analysis of the N-terminal region of AtD-LDH and AtD-2HGDH predicted the existence of mitochondrial targeting sequences of 56 and 33 amino acid residues in length (26). With the aim of determining the catalytic properties of the mature proteins (starting at amino acid residues 57 and 34, respectively, see supplemental Fig. S1, A and B), the truncated AtD-LDH and AtD-2HGDH cDNAs were cloned in the pET-16b expression vector and expressed in E. coli. Polypeptides of ∼61.0 and 63.0 kDa (59.0 and 60.7 kDa corresponding to AtD-LDH and AtD-2HGDH mature protein sequences, respectively, plus a 2.4-kDa N-terminal sequence encoded by the vector, including the His-10 epitope) were present in the extracts of isopropyl β-d-thiogalactopyranoside-induced cells but not in extracts from noninduced cells (Fig. 2, A and B). These proteins were purified to near homogeneity by Ni2+-NTA-agarose chromatography (Fig. 2, A and B) with yields of 0.5–2 mg of protein per liter of E. coli culture. The purified proteins were analyzed by Western blot using antibodies against the His tag. SDS-PAGE indicated unique immunoreactive bands with the same molecular masses as obtained by Coomassie Blue staining (data not shown), and native PAGE revealed unique immunoreactive bands compatible with the formation of dimers (Fig. 2C). The purified enzymes were rather unstable, and 2 days of storage in a buffer containing 25% glycerol at −20 °C resulted in a 90% loss of activity.
Structural Properties of AtD-LDH and AtD-2HGDH
The purified AtD-LDH and AtD-2HGDH were bright yellow, suggesting that a cofactor was associated with the enzymes. The UV-visible absorption spectrum revealed absorption maxima at 375 and 450 nm, which perfectly matches those of the FAD and FMN standards and are well documented as indicators for a flavin cofactor. To determine the identity of the prosthetic group, the enzymes were boiled, and the denatured proteins were removed by centrifugation. The supernatants containing the cofactors were analyzed by thin layer chromatography and by native PAGE using FMN, FAD, and riboflavin as standards. Using both methods, the Rf values of the enzyme cofactors of both enzymes were highly similar to those of FAD (Table 1).
TABLE 1.
RF values of flavins by ascending TLC and PAGE
Solvent systems used are as follows: A, 5% Na2HPO4 in H2O, and B, 1-butanol:HAc:H2O (4:1:5).
| Flavin standard |
AtD-LDH cofactor | AtD-2HGDH cofactor | |||
|---|---|---|---|---|---|
| Riboflavin | FMN | FAD | |||
| TLC A | 0.38 | 0.72 | 0.61 | 0.58 | 0.59 |
| TLC B | 0.45 | 0.22 | 0.07 | 0.07 | 0.09 |
| PAGE | 0.09 | 0.94 | 0.82 | 0.80 | 0.83 |
Additionally, using the cofactor isolated from 120 μg of boiled AtD-LDH and AtD-2HGDH in a final volume of 500 μl, the absorption at 450 nm was 0.05. Taking into account that the extinction coefficient of FAD is 11.3 mm−1 cm−1 and the calculated molecular masses of the recombinant AtD-LDH and AtD-2HGDH are 59.0 and 60.7 kDa, respectively, it can be calculated that each mole of monomeric enzyme binds 1 mol of FAD. In addition, size exclusion chromatography indicated native molecular masses of 135 and 140 kDa for AtD-LDH and AtD-2HGDH, respectively, demonstrating that the enzymes form homodimers.
Catalytic Properties of Recombinant AtD-LDH
The substrate specificity of AtD-LDH was screened using a number of related substances as substrates and DCIP or cytochrome c as acceptor molecules (Tables 2 and 3). The highest activity was found with cytochrome c, for which Km values of 53 and 28 μm were calculated using d- and l-lactate as substrates, respectively. Using a saturating concentration of cytochrome c, the highest catalytic rates were determined with d-2-hydroxybutyrate (d-2HB) and d-lactate, whereas those with l-lactate, d-glycerate, and glycolate were 10.8, 9.2, and 0.15% that with d-lactate, respectively (Table 2). The affinity for l-lactate and d-glycerate was very low with Km values of 8.9 and 4.5 mm, respectively. These values are 27- and 54-fold higher than that with d-lactate, respectively (Table 3). The affinities for d-2HB, d-lactate, and glycolate were high with Km values of 61, 164, and 432 μm, respectively (Table 3). In this way, AtD-LDH had a high catalytic efficiency with d-lactate using cytochrome c as acceptor molecule showing about 200- and 2000-fold higher preference for d-lactate compared with l-lactate and glycolate, respectively. A similar kinetic behavior was obtained using the in vitro electron acceptor DCIP (Table 3). AtD-LDH showed the highest catalytic efficiency toward d-2HB and no activity with d-3HB and d-4HB (Table 2). No activity was detected using NAD or NADP as electron acceptors, and O2 did not function as the acceptor for glycolate electrons. Similar catalytic properties were obtained with the recombinant enzyme after removal of the N terminus encoded by the expression vector. A further evaluation of the kinetic properties showed that AtD-LDH was able to catalyze the reverse reactions of reduction of glyoxylate and pyruvate in the presence of NADH or NADPH, although in both cases the activities were ∼4% of the forward reaction measured with d-lactate and cytochrome c.
The pH optimum for d-lactate oxidation extended over a broad pH range 8.0–9.0 (data not shown). The same results were obtained using l-lactate and glycolate as substrates (data not shown). The enzyme retained at least 25% activity down to pH 7.25, whereas it rapidly lost activity at lower pH values.
Catalytic Properties of Recombinant AtD-2HGDH
A substrate screening similar to that described above for AtD-LDH was also performed with the purified AtD-2HGDH. AtD-2HGDH was highly specific and catalyzed the efficient oxidation of d-2HG using DCIP as electron acceptor with a Km of 584 μm for the substrate (Table 3). The enzyme had a very low activity toward d-lactate, d-2HB, and meso-tartrate with DCIP, but this was so close to the detection limit that kinetic parameters could not be accurately determined. When using cytochrome c as electron acceptor, the activity with d-2HG was only 5% of that using DCIP, and the enzyme also showed low activities toward d-lactate, d-2HB, and meso-tartrate as with DCIP. Based on these results, we assumed that in vivo AtD-2HGDH may donate the electrons to an electron acceptor other than cytochrome c. The enzyme did not use glycolate as a substrate with either of the acceptors. AtD-2HGDH was not able to catalyze the oxidation of d-2HG using NAD or NADP nor the reduction of 2-ketoglutarate using NADH or NADPH. Maximal enzymatic activity, which was investigated with saturating d-2HG and DCIP concentrations, was observed over the pH range of 8.5–9.5. Similar results were obtained with the recombinant enzyme after removal of the His tag (Fig. 2B).
d-LDH and d-2HGDH Activities in Extracts of A. thaliana Wild Type and Knock-out Plants
The purified recombinant AtD-LDH and AtD-2HGDH were analyzed by native PAGE and stained for activity using different substrates and nitro blue tetrazolium as an electron acceptor. In accordance with the in vitro kinetic data, AtD-LDH showed an activity band when the gels were incubated with d-lactate (Fig. 2C) and d-2HB (data not shown) for 30 min. If the incubation time was prolonged for 12 h, a faint active band was also observed with l-lactate and glycolate as substrates (data not shown). On the other hand, AtD-2HGDH showed an active band after 30 min incubation with d-2HG (Fig. 2C), whereas after overnight incubation activity staining was observed with d-lactate and d-2HB. In both cases, the activity bands had mobilities compatible with dimeric oligomeric states as shown by immunoblot analysis (Fig. 2C).
Furthermore, A. thaliana leaf and root crude extracts and extracts of isolated mitochondria were analyzed by native PAGE and stained for activity using the same substrates as above. Interestingly, the activity of AtD-LDH could not be detected by native electrophoresis in the extracts. As the purified recombinant enzyme exhibited enzymatic activity after native PAGE (Fig. 2C), it is possible that the protein lost its activity during the preparation of extracts. It is also feasible that due to the low level of expression of AtD-LDH under normal conditions of growth (31), the specific activity of the extracts is under the detection limit of the assay.
A single active band with similar mobility of that displayed by the recombinant expressed AtD-2HGDH was detected in leaf and root crude extracts and extracts of isolated mitochondria using d-2HG as a substrate after 30 min of incubation (Fig. 2D). As in the case of the isolated protein, after overnight incubation a low level of activity could also be detected with d-lactate and d-2HB as substrates (data not shown). Thus, these results demonstrated that the recombinant enzyme displayed the same substrate preferences and catalytic efficiencies as the enzyme present in the plant extracts. It is worth mentioning that d-2HGDH activity was enriched in the mitochondrial extracts in comparison with the crude extracts. These results together with the observation that the mitochondrial preparation did not show immunoreactive bands with antibodies that recognize chloroplastic and cytosolic malate dehydrogenases point to a mitochondrial localization of AtD-2HGDH.
To correlate the activities and the substrate specificities determined in vitro with those found in mitochondria and crude extracts, two independent tDNA insertion lines for each gene were isolated (supplemental Fig. S2A). Homozygous lines for each mutant were confirmed by genomic PCR and designated dldh1-1 (Salk_001490), dldh1-2 (Salk_026859), d2hgdh1-1 (Salk_061383), and d2hgdh1-2 (Sail_844_G06). Semiquantitative reverse transcription-PCR using primer pairs designed to span the tDNA insertion sites were used to investigate the gene expression in the insertion lines. AtD-LDH mRNAs were detected in the wild type and both d2hgdh lines, but no amplification products were observed in dldh1-1 and dldh1-2 (supplemental Fig. S2B). On the other hand, AtD-2HGDH mRNAs were detected in the wild type and dldh mutants, but no amplification products were observed in the d2hgdh mutant lines (supplemental Fig. S2B). Protein extracts from leaves and roots and mitochondrial preparations of the knock-out d2hgdh1-1 analyzed by native PAGE showed no active band when incubated with d-2HG (Fig. 2D), d-lactate, or d-2HB (data not shown). The same results were obtained with extracts of the allelic knock-out line d2hgdh1-2, unambiguously demonstrating that locus At4g36400 encodes a d-2HGDH, which is responsible for the total d-2HGDH activity detected in A. thaliana.
d-Lactate and Methylglyoxal Affect dldh Seedling Growth
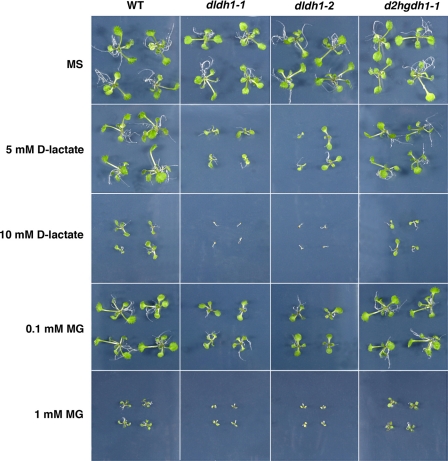
During growth under standard greenhouse conditions or on MS plates (Fig. 3) the knock-out lines showed no evident differences in development or morphology when compared with the wild type. To determine the effect of d-lactate and MG on the growth of A. thaliana seedlings, wild type plants and the knock-out mutants were germinated on MS medium supplemented with different concentrations of the compounds. When the seeds were germinated in the presence of d-lactate concentrations up to 5 mm or MG concentrations up to 0.1 mm developmental retardation was observed in both dldh lines (Fig. 3), whereas wild type, d2hgdh1-1 (Fig. 3), and d2hgdh1-2 (data not shown) plants grew significantly better. At higher d-lactate or MG concentrations both dldh1-1 and dldh1-2 plants were only able to germinate, but seedlings did not develop beyond the cotyledon stage and died (Fig. 3). It is worth mentioning that in the presence of MG and d-lactate wild type, d2hgdh1-1 and d2hgdh1-2 plants remain smaller than in media without supplementation, indicating that both MG and d-lactate exert a dose-dependent toxic effect, which affects general plant development. These results undoubtedly demonstrate that in vivo AtD-LDH is able to metabolize d-lactate. Moreover, MG exerts a higher damaging effect in both allelic dldh lines indicating its impaired detoxification through the loss of d-LDH activity in these plants.
FIGURE 3.
Effect of methylglyoxal and d-lactate on plant growth. Two-week-old wild type (WT), dldh1-1, d-ldh1-2, and d2hgdh1-1 seedlings grown on MS solid media and in the same media supplemented with different concentrations of d-lactate or MG.
DISCUSSION
A. thaliana Locus At5g06580 Encodes a d-LDH
The open reading frame of A. thaliana locus At5g06580 encodes a polypeptide of 62.2 kDa that binds a FAD group as shown by thin layer chromatography and native PAGE (Table 1). Consistent with this, the predicted protein has conserved FAD-binding motifs (a FAD_binding_4 and a FAD-linked oxidase, C-terminal domain) (supplemental Fig. S1A). The enzyme is active as a dimer, in contrast to the isoforms from Rhodopseudomonas palustris, which consists of four subunits of 57 kDa (10). The kinetic data indicate that the A. thaliana protein acts as a d-LDH (Table 3). The enzyme can also act on l-lactate, d-glycerate, and glycolate, although with very low catalytic efficiency due to a low affinity toward l-lactate and d-glycerate and an extremely low catalytic rate toward glycolate (Table 3). From all the substances tested, AtD-LDH showed the highest activity rate with d-2HB and the lowest one with glycolate (Table 2). These properties, i.e. high activity d-lactate and d-2HB and low or no activity with l-lactate and glycolate, resemble those of characterized d-LDHs from S. cerevisiae, R. palustris, and Peptostreptococcus elsdenii (7, 10, 27). In mammalia, d-2HB is produced principally in hepatic tissues as a by-product during l-threonine catabolism (32) or glutathione synthesis (33) and is a metabolic end product in Fusarium species (34). To our knowledge, there are no reports about the biological function of this metabolite in plant tissues. In this way, the high activity of AtD-LDH toward this substrate could be a consequence of a conserved substrate recognition site and thus could represent a side reaction of no physiological importance. Because AtD-LDH showed an extremely low catalytic rate toward glycolate, it can be assumed that it is not involved in glycolate metabolism in planta, as the level of this metabolite in mitochondria may be very low. However, the expression of this open reading frame in E. coli mutants lacking any of the three subunits of glycolate oxidase restored the capacity of glycolate oxidation (35), indicating that the enzyme would be able to metabolize glycolate, at least when high concentrations of this substrate are present as in this complementation experiment. However, the same toxic effect was observed in two independent dldh lines and the wild type grown in agar plates supplemented with glycolate even at concentrations up to 50 mm (data not shown), suggesting that AtD-LDH would not be involved in the metabolization of glycolate in planta.
The screening of related substances as possible substrates allows us to conclude that AtD-LDH is active on d-enantiomers of 2-hydroxy acids (Table 2). Moreover, the results obtained strongly indicate that At5g06580 should be re-classified into the EC 1.1.2.4 group (d-LDH cytochrome c-dependent). The enzyme was previously classified into the EC 1.1.99.14 group (35). It is worth mentioning that studies on activities classified into this last group were performed with tissue extracts or enriched fractions, but to our knowledge the corresponding genes/proteins have never been isolated.
AtD-LDH Is Involved in the Methylglyoxal Pathway and Donates Electrons to the Electron Transport Chain
In plants, fungi, and animals, d-lactate is formed from MG by the consecutive action of glyoxalase I and glyoxalase II. In A. thaliana, glyoxalase I (At1g67280) and glyoxalase II (At1g06130) are ubiquitously expressed. Glyoxalase I is found exclusively in the cytosol (14), and glyoxalase II is localized to the cytosol and mitochondria (13, 19), and AtD-LDH has previously been localized to mitochondria (35). In this way, d-lactate would be produced directly at the site of AtD-LDH or, alternatively, imported from the cytosol. In this respect, H. tuberosus mitochondria were shown to take up externally added d-lactate by means of a d-lactate/H+ symporter or a d-lactate/malate carrier (13, 36). The ubiquitous expression of AtD-LDH (31), its mitochondrial localization (26, 35), and its clear preference for d-lactate indicate that this enzyme is a candidate for the oxidation of d-lactate produced in mitochondria or cytosol through the glyoxalase system in A. thaliana (Fig. 4). Moreover, growth in media supplemented with MG or d-lactate was impaired in independent knock-out lines with disruptions in the d-LDH gene, clearly indicating that in planta this enzyme contributes to the metabolization of MG to d-lactate. In accordance with this, transcriptional co-response pattern analysis (25) showed that AtD-LDH is directly connected with glyoxalase I in the coexpression network.
The in vitro biochemical data obtained with the recombinant purified AtD-LDH showed that cytochrome c is a good acceptor for the electrons obtained from d-lactate through its oxidation to pyruvate. Similar kinetics were previously described for S. cerevisiae and K. lactis, where the oxidation of d-lactate through a mitochondrial d-LDH could be coupled to cytochrome c (8, 11), and for R. palustris d-LDH, where the electrons are transferred to cytochrome c2 (10). Additionally, Atlante et al. (13) showed that a mitochondria-localized enzyme oxidizes d-lactate with concomitant electron transport to complex III in H. tuberosus. Thus, it is conceivable that the oxidation of d-lactate by d-LDH is linked to the mitochondrial respiratory chain, most likely by directly donating electrons to cytochrome c (Fig. 4). Taken together, in A. thaliana electrons originating from d-lactate may be transferred to the respiratory chain, especially under stress conditions when the glyoxalase system is highly active. Work aimed at identifying the physiological electron acceptor is being undertaken.
A. thaliana Locus At4g36400 Encodes a d-2HGDH
The open reading frame of A. thaliana locus At5g36400 encodes a polypeptide of 61.4 kDa that contains FAD as prosthetic group (Table 1). As in the case of AtD-LDH, the predicted protein has predicted FAD_binding_4 and a FAD-linked oxidase, C-terminal domain (supplemental Fig. S1B), and the enzyme is active as a dimer. Analysis of the substrate specificity revealed that the purified recombinant product of A. thaliana locus At4g36400 catalyzes the oxidation of d-2HG with a high preference over all other substrates tested, and thus, it is a d-2HGDH. These results were confirmed by native electrophoresis coupled to activity staining of root and leaf extracts and mitochondrial preparations of A. thaliana wild type and the knock-out mutants (Fig. 4B), which also demonstrated that AtD-2HGDH is responsible for the total d-2HGDH activity detected in the tissues tested.
AtD-2HGDH is related to mitochondrial FAD-dependent enzymes from other organisms that are capable of oxidizing a variety of d-2-hydroxy acids (29, 37, 38). These enzymes act primarily on d-2HG but showed lower activity rates and lower affinities toward d-lactate, d-malate, and meso-tartrate. In contrast to this, the plant enzyme seems to have developed more selectivity as it showed to be extremely specific for d-2HG. On the other hand, AtD-2HGDH does not efficiently use cytochrome c as an electron acceptor and is not able to transfer electrons to NAD or NADP. The nature of the in vivo electron acceptor remains to be determined.
d-2HGDH Is Most Probably Involved in the Catabolism of Propionyl-CoA Derived from β-Oxidation
Although the substrate of AtD-2HGDH, d-2HG, is a known metabolic intermediate in bacteria, yeast, and mammalia, little is known about the biological function of this metabolite in plant tissues. In mammals, d-2HG is formed as a degradation product of aminolevulinate (39) and it was also shown to be formed within the mitochondria from 2-ketoglutarate through a hydroxyacid-oxoacid-transhydrogenase in a reaction in which γ-hydroxybutyrate is converted to succinic semialdehyde (40). However, no ortholog of this enzyme has been found in plants.
d-2HG accumulates in different mammalian tissues during d-2HG aciduria (41). This fatal neurometabolic disease is caused by mutations in either d-2HGDH (42), the electron transfer protein (ETF) (43), or the ETF-ubiquinone oxidoreductase (ETFQO) (43). ETF and ETRQO serve to transfer electrons collected by soluble mitochondrial dehydrogenases, also from d-2HG, to the respiratory chain via coenzyme Q to complex III (43, 44). In addition, the mammalian ETF/ETFQO system plays a key role in β-oxidation of fatty acids and catabolism of amino acids and choline (45, 46). Higher plants also contain ETF/ETFQO homologs, which supply the mitochondrial respiratory chain with electrons originating from substrates other than sucrose (45, 46). The analysis of A. thaliana knock-out mutants in these genes demonstrated that they are involved in the catabolism of Leu and chlorophyll, which are activated during dark-induced senescence and sugar starvation and are essential for viability in sucrose-starved conditions (45, 46).
The question arises as to which compound could be the source of d-2HG in plants tissues. The production of d-2HG could be the result of the condensation of propionyl-CoA and glyoxylate through the action of a 2-hydroxyglutarate synthase (EC 2.3.3.11) in peroxisomes. The existence of this activity was previously reported in E. coli and Aspergillus glaucus grown on propionate (47, 48). Propionyl-CoA is produced in the final step of odd-chain fatty acids, Val and Leu catabolism through β-oxidation. Propionyl-CoA is also a final product of metabolism of the methyl branched acids, phytanic acid, derived from the degradation of phytol, the hydrophobic tail of chlorophyll (49).
Propionyl-CoA is not a common metabolic intermediate in other pathways, so it must be converted into something else to be effectively metabolized. Propionyl-CoA could be converted to succinyl-CoA, which can enter the tricarboxylic acid cycle, but in plants the existence of this catabolic pathway is still not clear as no orthologs corresponding to mammalian or bacterial propionyl-CoA carboxylase were found (49). Recently, it was postulated that propionyl-CoA may be converted into β-hydroxypropionate by a modified peroxisomal β-oxidation pathway (49). The existence of a d-2HGDH activity (this study) allows us to postulate an alternative route for propionyl-CoA degradation in which mitochondrial d-2HGDH and a peroxisomal d-2HG synthase would act in concert to transfer carbon from peroxisomes to the mitochondria during respiration of substrates from proteolysis and/or lipid degradation (supplemental Fig. S3). In this way, d-2HG is converted into 2-ketoglutarate, which is fed into the tricarboxylic acid cycle. These steps parallel those of acetyl-CoA detoxification, in which acetyl-CoA is converted into citrate by peroxisomal citrate synthase and citrate is then transported to the mitochondria to fuel respiration (50).
In accordance with the origins of propionyl-CoA in plants, analysis of transcriptional co-response patterns (25) showed that AtD-2HGDH is coexpressed with the core enzymes of peroxisomal β-oxidation, enzymes of the β-oxidation cycle of unsaturated fatty acids, and the peroxisomal ABC transporter 1 (PXA1) importing long-chain fatty acids for degradation to acetyl-CoA, catalase, and citrate synthase (supplemental Table 1S and supplemental Fig. S3), all activities required for fatty acid respiration (51, 52). Moreover, several enzymes involved in Val and Ile degradation are also included in the same cluster (supplemental Table 1S and supplemental Fig. S3). Peroxisomal β-oxidation in the degradation of Val is supported by the characterization of a peroxisomal β-hydroxyisobutyryl-CoA hydrolase (supplemental Fig. S3) (53), and the synthesis of the polyhydroxyalkanoates in peroxisomes implies that there is a source of propionyl-CoA from Val and/or Ile in the peroxisomes (54).
In addition, several enzymes involved in the chlorophyll degradation and the peroxisomal phytanic acid oxidation pathways are also found in the same coexpression network (supplemental Table 1S and supplemental Fig. 3). Although the exact mechanism of phytol or chlorophyll degradation in plants is still not completely elucidated (55), it was postulated that phytol is oxidized to the respective aldehyde phytenal, which subsequently might be converted to phytanic acid, as was shown for the degradation of phytol in animals (56). Moreover, A. thaliana possesses a phytanoyl-CoA dioxygenase (supplemental Table 1S), and it was recently shown that A. thaliana ETFQO is involved in phytol degradation (46). Interestingly, ETFQO (At2g43400) and ETFbeta (At5g43430) are also in the same coexpressed gene network. This observation and the fact that AtD-2HGDH does not effectively use cytochrome c as electron acceptor indicate that most probably AtD-2HGDH donates electrons to the electron transport chain through the ETF/ETFQO (supplemental Fig. S3).
Examination of the Affymetrix-based expression-profiling data base AtGenExpress (57) indicates that the expression of AtD-2HGDH parallels the basal level of expression of genes encoding enzymes of β-oxidation in a wide variety of tissues. Thus, AtD-2HGDH might be required for the metabolization of propionyl-CoA derived from membrane lipid, chlorophyll, and protein turnover. In silico analysis of the subcellular localization and mitochondrial proteome analysis indicate that AtD-2HGDH is located to mitochondria (26, 58), as it is orthologous from rat and human (29, 40). In this way, the mitochondrial localization of AtD-2HGDH further supports the model proposed above (supplemental Fig. S3). Further studies are being conducted to determine the exact contribution of AtD-2HGDH to plant metabolism.
Supplementary Material
This work was supported in part by a Deutsche Forschungsgemeinschaft grant (to V. G. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- d-LDH
- d-lactate dehydrogenase
- d-2HG,
- d-2-hydroxyglutarate
- d-2HGDH
- d-2-hydroxyglutarate dehydrogenase
- DCIP
- dichlorophenolindophenol
- d-LDH
- d-lactate dehydrogenase
- ETF
- electron transfer protein
- ETFQO
- ETF-ubiquinone oxidoreductase
- MG
- methylglyoxal
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid
- PMS
- phenazine methosulfate
- DCIP
- 2,6-dichlorophenolindophenol
- MS
- Murashige and Skoog
- d-2HB
- d-2-hydroxybutyrate.
REFERENCES
- 1.Cristescu M. E., Innes D. J., Stillman J. H., Crease T. J. (2008) BMC Evol. Biol. 8, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lodi T., Guiard B. (1991) Mol. Cell. Biol. 11, 3762–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Passarella S., de Bari L., Valenti D., Pizzuto R., Paventi G., Atlante A. (2008) FEBS Lett. 582, 3569–3576 [DOI] [PubMed] [Google Scholar]
- 4.Mizushima S., Hiyama T., Kitahara K. (1964) J. Gen. Appl. Microbiol. 10, 33–44 [Google Scholar]
- 5.Garvie E. I. (1980) Microbiol. Rev. 44, 106–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogata M., Arihara K., Yagi T. (1981) J. Biochem. 89, 1423–1431 [DOI] [PubMed] [Google Scholar]
- 7.Brockman H. L., Wood W. A. (1975) J. Bacteriol. 124, 1454–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodi T., Ferrero I. (1993) Mol. Gen. Genet. 238, 315–324 [DOI] [PubMed] [Google Scholar]
- 9.Reed D. W., Hartzell P. L. (1999) J. Bacteriol. 181, 7580–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horikiri S., Aizawa Y., Kai T., Amachi S., Shinoyama H., Fujii T. (2004) Biosci. Biotechnol. Biochem. 68, 516–522 [DOI] [PubMed] [Google Scholar]
- 11.Lodi T., O'Connor D., Goffrini P., Ferrero I. (1994) Mol. Gen. Genet. 244, 622–629 [DOI] [PubMed] [Google Scholar]
- 12.Labeyrie F., Slonimski P. (1964) Bull. Soc. Chim. Biol. 44, 1793–1828 [PubMed] [Google Scholar]
- 13.Atlante A., de Bari L., Valenti D., Pizzuto R., Paventi G., Passarella S. (2005) Biochim. Biophys. Acta 1708, 13–22 [DOI] [PubMed] [Google Scholar]
- 14.Thornalley P. J. (1990) Biochem. J. 269, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalapos M. P. (1999) Toxicol. Lett. 110, 145–175 [DOI] [PubMed] [Google Scholar]
- 16.Xu Y., Chen X. (2006) J. Biol. Chem. 281, 26702–26713 [DOI] [PubMed] [Google Scholar]
- 17.Yadav S. K., Singla-Pareek S. L., Ray M., Reddy M. K., Sopory S. K. (2005) Biochem. Biophys. Res. Commun. 337, 61–67 [DOI] [PubMed] [Google Scholar]
- 18.Singla-Pareek S. L., Yadav S. K., Pareek A., Reddy M. K., Sopory S. K. (2008) Transgenić Res. 17, 171–180 [DOI] [PubMed] [Google Scholar]
- 19.Maiti M. K., Krishnasamy S., Owen H. A., Makaroff C. A. (1997) Plant Mol. Biol. 35, 471–481 [DOI] [PubMed] [Google Scholar]
- 20.Talesa V., Rosi G., Contenti S., Mangiabene C., Lupattelli M., Norton S. J., Giovannini E., Principato G. B. (1990) Biochem. Int. 22, 1115–1120 [PubMed] [Google Scholar]
- 21.Fahnenstich H., Scarpeci T. E., Valle E. M., Flügge U. I., Maurino V. G. (2008) Plant Physiol. 148, 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keech O., Dizengremel P., Gardeström P. (2005) Physiol. Plant. 124, 403–409 [Google Scholar]
- 23.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25.Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009) Nucleic Acids Res. 37, D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W. B., Flügge U. I., Kunze R. (2003) Plant Physiol. 131, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregolin C., Singer T. P. (1963) Biochim. Biophys. Acta 67, 201–218 [DOI] [PubMed] [Google Scholar]
- 28.Sun Z. Y., Dowd S. R., Felix C., Hyde J. S., Ho C. (1995) Biochim. Biophys. Acta 1252, 269–277 [DOI] [PubMed] [Google Scholar]
- 29.Achouri Y., Noël G., Vertommen D., Rider M. H., Veiga-Da-Cunha M., Van Schaftingen E. (2004) Biochem. J. 381, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chelstowska A., Liu Z., Jia Y., Amberg D., Butow R. A. (1999) Yeast 15, 1377–1391 [DOI] [PubMed] [Google Scholar]
- 31.Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007) PLoS ONE 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammer V. A., Rogers Q. R., Freedland R. A. (1996) J. Nutr. 126, 2218–2226 [DOI] [PubMed] [Google Scholar]
- 33.Fernández-Checa J., Lu S., Ookhtens M., DeLeve L., Runnegar M., Yoshida H., Saiki H., Kannan R., Garcia-Ruiz C., Kuhlenkamp J., Kaplowitz N. (1993) in Hepatic Anion Transport and Bile Secretion: Physiology and Pathophysiology ( Tavoloni N., Berk P. D. eds) pp. 363–395, Marcel Dekker, New York [Google Scholar]
- 34.Carlier J. P., Henry C., Lorin V., Rouffignat K. (1997) Lett. Appl. Microbiol. 25, 371–374 [DOI] [PubMed] [Google Scholar]
- 35.Bari R., Kebeish R., Kalamajka R., Rademacher T., Peterhänsel C. (2004) J. Exp. Bot. 55, 623–630 [DOI] [PubMed] [Google Scholar]
- 36.de Bari L., Valenti D., Pizzuto R., Paventi G., Atlante A., Passarella S. (2005) Biochem. Biophys. Res. Commun. 335, 1224–1230 [DOI] [PubMed] [Google Scholar]
- 37.Tubbs P. K., Greville G. D. (1961) Biochem. J. 81, 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cammack R. (1969) Biochem. J. 115, 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindahl G., Lindstedt G., Lindstedt S. (1967) Arch. Biochem. Biophys. 119, 347–352 [DOI] [PubMed] [Google Scholar]
- 40.Struys E. A., Verhoeven N. M., Brunengraber H., Jakobs C. (2004) FEBS Lett. 557, 115–120 [DOI] [PubMed] [Google Scholar]
- 41.Struys E. A. (2006) J. Inherited Metab. Dis. 29, 21–29 [DOI] [PubMed] [Google Scholar]
- 42.Struys E. A., Salomons G. S., Achouri Y., Van Schaftingen E., Grosso S., Craigen W. J., Verhoeven N. M., Jakobs C. (2005) Am. J. Hum. Genet. 76, 358–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frerman F. E., Goodman S. I. (2001) in The Metabolic and Molecular Bases of Inherited Disease ( Beaudet A. L., Scriver C. R., Sly W. S., Valle D. eds) Vol. 2, pp. 2357–2365, McGraw-Hill Medical Publishing Division, New York [Google Scholar]
- 44.Parker A., Engel P. C. (2000) Biochem. J. 345, 429–435 [PMC free article] [PubMed] [Google Scholar]
- 45.Ishizaki K., Schauer N., Larson T. R., Graham I. A., Fernie A. R., Leaver C. J. (2006) Plant J. 47, 751–760 [DOI] [PubMed] [Google Scholar]
- 46.Ishizaki K., Larson T. R., Schauer N., Fernie A. R., Graham I. A., Leaver C. J. (2005) Plant Cell 17, 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bleiweis A. S., Reeves H. C., Ajl S. J. (1967) J. Bacteriol. 94, 1560–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves H. C., Ajl S. J. (1962) J. Bacteriol. 84, 186–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lucas K. A., Filley J. R., Erb J. M., Graybill E. R., Hawes J. W. (2007) J. Biol. Chem. 282, 24980–24989 [DOI] [PubMed] [Google Scholar]
- 50.Raymond P., Spiteri A., Dieuaide M., Gerhardt B., Pradet A. (1992) Plant Physiol. Biochem. 30, 153–161 [Google Scholar]
- 51.Goepfert S., Poirier Y. (2007) Curr. Opin. Plant Biol. 10, 245–251 [DOI] [PubMed] [Google Scholar]
- 52.Pracharoenwattana I., Cornah J. E., Smith S. M. (2005) Plant Cell 17, 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zolman B. K., Monroe-Augustus M., Thompson B., Hawes J. W., Krukenberg K. A., Matsuda S. P., Bartel B. (2001) J. Biol. Chem. 276, 31037–31046 [DOI] [PubMed] [Google Scholar]
- 54.Arai Y., Nakashita H., Suzuki Y., Kobayashi Y., Shimizu T., Yasuda M., Doi Y., Yamaguchi I. (2002) Plant Cell Physiol. 43, 555–562 [DOI] [PubMed] [Google Scholar]
- 55.Hörtensteiner S. (2006) Annu. Rev. Plant Biol. 57, 55–77 [DOI] [PubMed] [Google Scholar]
- 56.van den Brink D. M., van Miert J. N., Dacremont G., Rontani J. F., Jansen G. A., Wanders R. J. (2004) Mol. Genet. Metab. 82, 33–37 [DOI] [PubMed] [Google Scholar]
- 57.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004) Plant Physiol. 136, 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee C. P., Eubel H., O'Toole N., Millar A. H. (2008) Mol. Cell. Proteomics 7, 1297–1316 [DOI] [PubMed] [Google Scholar]
- 59.Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. (2007) Bioinformatics 8, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.