Abstract
Protective antigen (PA), the binding subunit of anthrax toxin, is the major component in the current anthrax vaccine, but the fine antigenic structure of PA is not well defined. To identify linear neutralizing epitopes of PA, 145 overlapping peptides covering the entire sequence of the protein were synthesized. Six monoclonal antibodies (mAbs) and antisera from mice specific for PA were tested for their reactivity to the peptides by enzyme-linked immunosorbent assays. Three major linear immunodominant B-cell epitopes were mapped to residues Leu156 to Ser170, Val196 to Ile210, and Ser312 to Asn326 of the PA protein. Two mAbs with toxin-neutralizing activity recognized two different epitopes in close proximity to the furin cleavage site in domain 1. The three-dimensional complex structure of PA and its neutralizing mAbs 7.5G and 19D9 were modeled using the molecular docking method providing models for the interacting epitope and paratope residues. For both mAbs, LeTx neutralization was associated with interference with furin cleavage, but they differed in effectiveness depending on whether they bound on the N- or C-terminal aspect of the cleaved products. The two peptides containing these epitopes that include amino acids Leu156–Ser170 and Val196–Ile210 were immunogenic and elicited neutralizing antibody responses to PA. These results identify the first linear neutralizing epitopes of PA and show that peptides containing epitope sequences can elicit neutralizing antibody responses, a finding that could be exploited for vaccine design.
Bacillus anthracis is a Gram-positive, facultatively anaerobic, rod-shaped bacterium that secretes a variety of toxins, including anthrax toxin. This toxin is made up of three proteins as follows: protective antigen (PA),3 edema factor (EF), and lethal factor (LF). Like other binary toxins, anthrax toxin follows the same pattern where distinct subunits are involved in the different steps at which it can act. The B subunit (PA) is involved in receptor binding and cellular internalization into the cytoplasm, whereas the A subunit (EF and/or LF) bears the enzymatic activity (1). Anthrax can occur naturally in animals by spore transmission via ingestion, inhalation, or an open skin wound, but it can also be a result of bioterrorism and biological warfare (2).
PA is the component of the currently licensed anthrax vaccine that elicits protective antibodies. Recent studies have demonstrated that a strong humoral response to truncated recombinant PA contributes to a protective immune response to anthrax (3, 4). Accordingly, there is considerable interest in ascertaining the location and immunogenicity of B-cell epitopes on the PA molecule. The development of numerous monoclonal antibodies (mAbs) to different epitopes on the PA molecule that influence PA functions, in conjunction with epitope mapping, has provided an opportunity to study the fine antigenic structure of PA. Most of these epitopes have been defined in mice (5–8), in macaques (9), in rabbits (10), as well as in vaccinated humans (11). Consequently, the epitopes described thus far are localized to three discrete regions within the PA. These regions correspond to the 2β2–2β3 loop of domain 2, the domain 3-domain 4 boundary, and the small loop of domain 4. The 2β2–2β3 loop of domain 2 is responsible for mediating membrane insertion, and many neutralizing murine mAbs target this region (5, 8). The boundary between domains 3 and 4, which does not have a known functional activity, has been suggested as a region recognized by polyclonal antibodies from vaccinated humans and rabbits (6, 12). The cellular receptor binding region is localized to the small loop of domain 4, and this region has been described to be recognized by two neutralizing mAbs (7, 9). With the exception of a neutralizing mAb that bound to PA20 (13), no B-cell epitopes have been reported in domain 1. Therefore, identification of further dominant antigenic epitopes is pivotal for understanding the minimal immunogenic region of PA that will allow for precise direction of potent immune responses.
Two mAbs to PA have been reported previously by our laboratory, one known as 7.5G binds to domain 1 and can neutralize the cytotoxic activity of lethal toxin (LeTx) (13). The other, mAb 10F4, binds to domain 4 and has weak neutralizing activity. In addition, we now describe four new anti-PA mAbs, of which only one neutralizes LeTx. The characterization of the B-cell epitopes in PA recognized by protective and nonprotective mAbs is important to better understand the antigenic structure of this toxin, and such information is potentially useful for the design of vaccines and passive immune therapies against B. anthracis. Because PA has been identified as an effective subunit vaccine, we wanted to identify the specific epitopes that provide the protection and use them as immunogens. Using mAbs and 145 overlapping peptides covering the entire sequence of PA, we identify the first linear neutralizing epitopes in domain 1 of PA, and we demonstrate that two peptides containing epitopes in domain 1 were capable of inducing strong LeTx-neutralizing antibody responses.
EXPERIMENTAL PROCEDURES
B. anthracis
B. anthracis Sterne strain 34F2 (pXO1+, pXO2−) was obtained from Dr. Alex Hoffmaster at the Centers for Disease Control and Prevention (Atlanta, GA). Bacterial cultures were grown in brain heart infusion broth (Difco) at 37 °C for 18 h while shaking. Recombinant PA and LF were obtained from Wadsworth Laboratories, New York State Department of Health (Albany, NY). Fragments of PA comprising one or more domains were expressed in Escherichia coli as described previously (13).
Mouse Immunization with GALXM-PA Conjugate
Female BALB/c mice (6–8 weeks old) were obtained from the NCI, National Institutes of Health (Bethesda). Two mice received an initial conjugate dose of 50 μl, whereas the third mouse was immunized with 100 μl of galactoxylomannan-protective antigen (GalXM-PA) conjugate (14) in complete Freund's adjuvant (CFA). All mice were subsequently immunized with 50 μl of the conjugate in incomplete Freund's adjuvant (IFA) at day 14. Serum titers were analyzed to determine antigen response. Mice where boosted daily with 50 μl of the conjugate 3 days prior to fusion.
mAb Production
Hybridomas making mAbs to PA were generated by standard techniques from splenocytes of GalXM-PA-immunized BALB/c mice (15). The GalXM-PA conjugate had been synthesized to generate antibodies to GalXM for an independent study (14), and we took advantage of the availability of mice with high titers to PA to make additional PA-binding mAbs. Briefly, splenocytes from GalXM-PA-immunized mice were fused to NSO myeloma cells at a ratio of 4:1. NSO is the nonproducing mouse myeloma fusion partner. Two weeks later, hybridoma supernatants were screened by ELISA for antibody reactivity to PA. Hybridoma clones were then selected and stabilized by cloning twice in soft agar. The isotypes of the murine mAbs were established by ELISA using isotype-specific reagents (Southern Biotechnology, Birmingham, AL).
Determination of VH and VL Sequences
Total RNA was isolated from hybridoma cell lines producing mAbs to EF using TRIzol reagent (Invitrogen) as per the manufacturer's instructions. Briefly, 1 ml of TRIzol reagent was used per 106 log phase hybridoma cells, and 50 ng of RNA was used for cDNA synthesis with oligo(dT) primer and superscript II reverse transcriptase (Qiagen, Valencia, CA). Universal 5′ (sense) variable region and specific 3′ (antisense) constant region primers were used in a PCR to generate cDNA encoding the variable domains of mAbs. The primers are as follows: 3′MsCγ, AGACCTATGGGGCTGTTGTTTTGGC; 3′MsCμ, GACATTTGGGAAGGACTGACTCTC; 3′MsCκ, TGGATACAGTTGGTGCAGCATCAGC; 5′VHuni, TGAGGTGCAGCTGGAGGAGTC; and 5′Vκuni, GACATTCTGATGACCCAGTCT. The PCR was performed using 1 μg of the cDNA templates, with 2.5 mm each of deoxynucleoside triphosphate and 125 nm each of primer under the following conditions with Taq polymerase (Roche Applied Science): 94 °C for 1 min, 50 °C for 1 min, and 72 °C for 1.5 min for 40 cycles, followed by a final 10-min extension at 72 °C. The amplified cDNAs were gel-purified (Qiagen, Valencia, CA) and then sequenced at the Sequencing Facility of the Cancer Center at the Albert Einstein College of Medicine.
LeTx Neutralization Assay
The LeTx neutralization assay used J774 macrophage-like cells treated with PA and LF, and mAb antitoxin activity was measured using the MTT assay for cell viability. All incubations were done at 37 °C in a 5% CO2 atmosphere, 95% relative humidity. J774 cells were plated in 96-well flat bottom microtiter plates at a density of 3 × 104 cells/well in 200 μl of Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 17–19 h prior to the assay. The following day, 100 μl of the cell supernatant was removed from wells containing J774 cells and then incubated with 100 ng each of PA and LF and/or a range of dilutions of each antibody sample for 4 h. Cell viability was determined by the addition of 25 μl/well of a 5 mg/ml stock solution of MTT, and the incubation continued for 2 h. The assay was terminated by addition of 100 μl/well of the extraction buffer (12% SDS, 45% N,N-dimethylformamide) and incubated overnight. The optical density values were measured at 570 nm (Labsystem Multiskan, Franklin, MA). All antibodies were tested at least three times, and the average was taken for wells receiving LeTx plus antibody or LeTx alone.
Proteolytic Digestion of PA
PA was digested with furin (Sigma) or trypsin (Promega, Madison, WI). For trypsin digestions, 10 μg of rPA in 150 mm NaCl2, 20 mm Tris, pH 8.2, was mixed with trypsin (1 μg/ml) for 30 min at room temperature in a 20-μl volume. For furin digests, 10 μg of PA was incubated in 20 μl of 1 mm CaCl2, 1 mm β-mercaptoethanol, 0.5% Triton X-100, 100 mm Hepes, pH 7.5, and mixed with 0.02–10 units of furin for 30 s to 15 min at 30 °C. PA was cleaved by chymotrypsin (1:1; Sigma) for 30 min at 30 °C. Digested products were separated on a 12% SDS-PAGE (16). The proteins were visualized by staining the gel with GelCode Blue Stain (Pierce).
SDS-PAGE and Western Blot
To determine the domain location of the epitopes recognized by the various mAbs to PA, SDS-PAGE and Western blotting were employed. PA or its proteolytic digests were mixed with Laemmli sample buffer containing β-mercaptoethanol, boiled for 5 min, and then separated on SDS-polyacrylamide gradient gel (10–20%). Proteins were then visualized by staining overnight with GelCode Blue Stain (Pierce). Proteins were transferred to nitrocellulose membrane (0.20 μm pore size) by electrophoretic transfer. The membranes were blocked with 5% dry milk in Tris-buffered saline, 0.1% Tween 20 (TBST) and then incubated with mAbs 2.9H, 16A12, 19D9, or 20G7 overnight at 4 °C. After washing with TBST, the membranes were incubated at room temperature for 1 h with horseradish peroxidase-labeled goat isotype-specific antibody. The ECL chemiluminescence kit (Pierce) was used to reveal horseradish peroxidase activity according to the manufacturer's instructions.
Peptide Synthesis
To map the functional linear epitopes of PA, biotinylated soluble peptides representing the entire length of PA were synthesized as 15-mer, overlapping by 10 residues (total of 145 peptides), at the Proteomics Resource Center, Rockefeller University, New York. All peptides were created using an Intavis MultiPepTM (Intavis, Koln, Germany) Wang resins (p-alkoxy-benzyl alcohol) (Bachem, Torrance, CA) using Fmoc (9-fluorenylmethyloxycarbonyl) nitrogen terminal protected amino acids (Anaspec, San Jose, CA) (17). Coupling reactions were conducted using 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate and 1-hydroxybenzotriazole in N-methylpyrrolidinone as the primary solvent. All crude products were subsequently analyzed by reversed-phase high pressure liquid chromatography (Waters) using an Acquity UPLCTM BEH130 C18 column. Individual peptide integrity was verified by electrospray ionization-mass spectrometry using a Thermo Scientific TSQ VantageTM (Waltham, MA) liquid chromatography/mass spectrometry system (18). Peptides were supplied as a white powder soluble in water and stored at a concentration of 1 mg/ml.
ELISAs for PA and Peptides
Binding of antibody to PA was measured by ELISA. All incubations were done at 37 °C for 1 h. Briefly, a solution of rPA (1 μg/ml) in phosphate-buffered saline (PBS) was used to coat polystyrene plates (Costar). The polystyrene plates were then blocked with 1% bovine serum albumin/PBS, and either immune sera or hybridoma supernatants were added. Primary antibody binding was detected using alkaline phosphatase labeled goat anti-mouse antibody reagents. After addition of substrate, absorbance was read at 405 nm. Nonlinear regression curve fit (one-site total binding) was used for calculation of dissociation constant values of mAbs 7.5G and 19D9 for PA samples. Plots, curve fits, and statistical analysis were performed using GraphPad Prism version 5.0a, GraphPad Software (San Diego, CA).
For the peptide ELISAs, polystyrene plates were coated with 5 μg/ml streptavidin (100 μl/well) and kept overnight at 37 °C. Then the plates were blocked with 2% bovine serum albumin in PBS (200 μl/well) for 1 h at 37 °C and then washed with 0.1% Tween 20 in PBS (PBST). Subsequently, biotinylated peptides (5 μg/ml) were added and incubated at room temperature for 1 h. After three washes with PBST, mAb or sera were added in dilution 1:100 or 1:500, respectively, in blocking buffer and incubated for 2 h. The plates were again washed with PBST. Alkaline phosphatase-conjugated goat isotype-specific antibody was diluted 1:1000 in blocking buffer, added to the plates, and incubated for 1 h at 37 °C. After another wash, alkaline phosphatase substrate was added to each well and allowed to develop for 20 min, and the absorbance at 405 nm was measured. These experiments were performed at least three times for each mAb. The background of each individual serum or mAb was determined in parallel by using streptavidin-coated, peptide-free wells. The cutoff value for real binding was defined as an absorbance value three times the average background value.
Isotype and IgG subclass analysis was performed as described above, and serial 2-fold dilutions of sera from immunized mice were added. Detection was performed with alkaline phosphatase-labeled goat anti-mouse IgG1, IgG2a, IgG2b, and IgG3 at a 1:1000 dilution (Southern Biotechnology).
Competition ELISAs
mAb-mAb competition ELISAs were used to investigate the specificity of PA mAbs as described (19). Briefly, a variable amount of one mAb was mixed with a constant amount of a second different isotype mAb and allowed to bind to PA immobilized in a polystyrene plate. Binding of the mAbs was detected by isotype-specific alkaline phosphatase-conjugated goat anti-mouse reagent. However, in the case of the same isotypes, one mAb was chosen at random to be labeled with alkaline phosphatase with a commercially available kit, following the manufacturer's instructions. In all instances, incubations were done at 37 °C for 1 h, and absorbances were measured in a microtiter plate reader at 405 nm (Labsystems Multiskan).
Immunization
Six- to 8-week-old female BALB/c mice (The Jackson Laboratory, Bar Harbor, ME) were used for immunizations. The following peptides LKQKSSNSRKKRSTS (MAP-D5) and VKNKRTFLSPWISNI (MAP-E1) were prepared on an eight-branched lysine backbone (peptide on MAP were from W. M. Keck Facility, Yale University, New Haven, CT). Mice were immunized subcutaneously with 100 μg of MAP-peptide in CFA on day 0, followed by subcutaneous booster injections of 100 μg of MAP-peptide in IFA on days 7 and 14. Control mice were immunized with the MAP core (Anaspec, San Jose, CA) in adjuvant and followed the same immunization schedule. Serum was obtained on days 0, 7, 14, 21, 35, and 49. All animal work was done in accordance with regulations of the Institute for Animal Studies at Albert Einstein College of Medicine.
Survival Studies
Six- to 8-week-old female A/JCr mice were injected intraperitoneally with 0.1 mg of mAb 19D9 2 h prior to intravenous infection with 104 bacterial cells of B. anthracis. Mice were monitored daily for mortality and morbidity and deaths recorded. All animal work was done in accordance with regulations of the Institute for Animal Studies at Albert Einstein College of Medicine.
Molecular Modeling
Comparative protein structure models were generated for mAbs 7.5G and 19D9 based on the available similar structures of IgG2b and IgG1 molecules, respectively (Protein Data Bank accession codes 1ibg and 3dvg), using the multiple mapping method (20) to generate a target-template sequence alignment and MODELLER (21) to generate the atomic models. mAbs 7.5G and 19D9 share 42 and 41% identical positions with 1ibg and 3dvg template structures, respectively. Next, 10,000 docking solutions were generated for each of the given antibodies with PA using the program FTDOCK (22). The amino acid compositions of the putative epitopes of PA are known from the peptide library produced in this work, and these sequences can be found linearly displayed on PA. This information was used to screen and locate the most accurate complex structures. In some instances, some of the amino acids of a linear peptide segment are buried within the PA structure, and thus we developed a PERL program that searches the common surface between PA and the antibody for perfect matches with the amino acid composition of the reactive peptides (the PERL program uses VMD version 1.862 for surface recognition). The resulting rigid body docked complexes were used as structural templates to build an optimized atomic structure with MODELLER (21).
Statistical Analysis
All data were analyzed by the Student's t test, and survival analysis was done by log rank censoring long term survivors (SigmaStat, Chicago).
RESULTS
Generation and Identification of PA-binding LeTx-neutralizing mAbs
Two hybridoma clones producing mAbs to PA (7.5G and 10F4) have been described previously (13), and these antibodies will be further characterized in this study. All mice immunized with GalXM-PA conjugate in CFA responded to immunization with a serum antibody response to rPA (Fig. 1). The mouse with the highest antibody titer to PA was selected for spleen harvest and hybridoma generation, and the hybridoma supernatants were screened for reactivity toward PA. The end point ELISA titer with a minimal absorbance reading that was 3-fold higher than background was used as the criterion to select antibodies for the next stage of analysis. Four mAbs were recovered: three IgG1s (29H, 16A12, and 19D9) and one IgM (20G7).
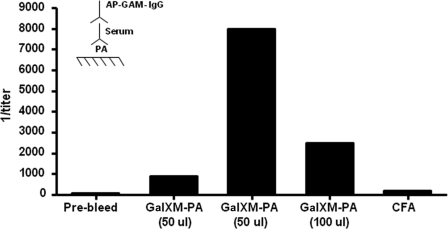
FIGURE 1.
Inverse antibody titers of BALB/c mice immunized with GalXM-PA conjugate as measured by ELISA. Two of the five female BALB/c mice (each bar represents one mouse) were initially immunized with 50 μl and one mouse was immunized with 100 μl of GalXM-PA conjugate in CFA on day 0. All mice were boosted with 50 μl of the conjugate in IFA at day 14. Sera were assayed for IgG anti-PA antibodies by ELISA. Inset, schematic of ELISA configuration used.
Ig Gene Utilization
To determine the variable gene usage of the PA-binding mAbs, the sequences of the heavy and light chain Ig mRNA of each mAb were determined. Total RNA from each hybridoma cell was isolated and reverse-transcribed to generate cDNA. Then the heavy and light chain cDNAs were amplified by PCR using either VH or VK primers, respectively. Analysis of the sequences revealed that all mAbs are similar in molecular construction, using the same germ line VH7183 and VKBD2 gene element (Table 1). For each mAb, the VH and VK domains were deposited in the GenBankTM data base under the respective accession numbers listed.
TABLE 1.
Hybridoma families and mAb VH and VL usage
| Hybridoma | Neutralizing | Accession no.a | Variable gene elementsb |
|||
|---|---|---|---|---|---|---|
| VH | JH | Vk | JK | |||
| 2H9 (IgG1) | No | FJ784743c | 7183 (5) (98.6%) | JH4 (1) (100%) | BD2 (7) (92.7%) | JK1 (93.8%) |
| FJ784747d | ||||||
| 16A12 (IgG1) | No | FJ784744c | 7183 (97%) | JH4 (100%) | BD2 (84.7%) | JK1 (88.1%) |
| FJ784748d | ||||||
| 19D9 (IgG1) | Yes | FJ784745c | 7183 (98%) | JH4 (100%) | BD2 (91.1%) | JK2/JK5 (2) (80%) |
| FJ784749d | ||||||
| 20G7 (IgM) | No | FJ784746c | 7183 (100%) | JH4 (100%) | BD2 (84.0%) | JK1 (90.6%) |
| FJ784750d | ||||||
a Accession numbers are shown for sequences in GenBankTM (Bethesda).
b Variable gene usage was assigned from homology searches. Numbers in parentheses indicate percentage homology to the stated V region elements.
c This indicates heavy chain.
d This indicates light chain.
mAb Specificity for PA Domains
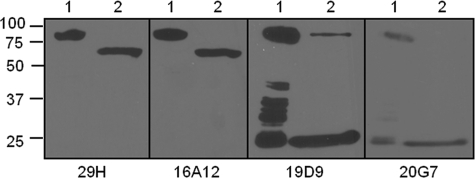
To investigate the domains recognized by the various mAbs to PA, an ELISA detection assay was employed where we studied the binding of the various antibodies to recombinant proteins expressing individual or combinations of different domains of PA as coated protein. We identified two mAbs (19D9 and 20G7) binding to domain 1 of PA and two mAbs (29H and 16A12) binding to expressed protein containing domains 2–4 (data not shown). These results were further confirmed by the binding of mAbs to enzyme-digested PA by Western blot analysis. mAbs 29H and 16A12 bound to PA63, whereas mAbs 19D9 and 20G7 bound to PA20 (Fig. 2). Furthermore, we determined the number of antigenic sites recognized by these antibodies with competition ELISA. The mAb pairs of 19D9 and 20G7, and 29H and 16A12, competed with each other for binding to PA (data not shown). In addition, mAbs 7.5G and 10F4 did not compete with any of the mAbs, implying that each recognized a different epitope on PA. From these experiments we conclude that two mAb sets bind to the same epitopes.
FIGURE 2.
Western immunoblot analysis of mAbs binding to PA. The samples were separated by reduced SDS-10% polyacrylamide gels and then transferred onto nitrocellulose membrane. The membranes were then incubated with mAbs 29H, 16A12, 19D9, or 20G7 as the primary antibody and a horseradish peroxidase-conjugated goat isotype-specific antibody as the secondary antibody. The bands were visualized by ECL chemiluminescence kit. Lane 1, rPA undigested; lane 2, rPA + furin.
Measurement of the affinity of the mAbs for PA was determined by ELISA. The Kd values of the identified antibodies were 5.877 nm for 7.5G and 0.2113 nm for 19D9 (data not shown).
mAbs Effect on LeTx Toxicity on Macrophages
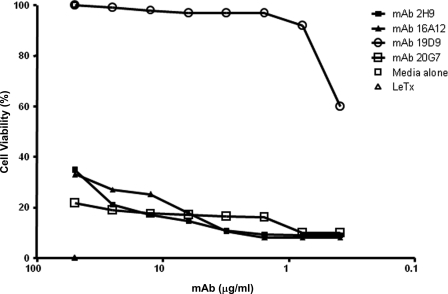
We studied the ability of mAbs to protect macrophages against toxin-mediated cytotoxicity. Each mAb was tested for LeTx neutralization with concentration adjustment (between 75 and 0.5 μg/ml). Therefore, this screening method selected for mAbs that either protected at high levels of antibody or at lower antibody concentrations obtaining high neutralization activity. Only one mAb, 19D9, exhibited significant neutralization activity, and this effect was dose-dependent (Fig. 3).
FIGURE 3.
Analysis of cellular toxicity in the presence of anti-PA mAbs by MTT assay. mAb 19D9 protects J774 macrophage monolayers against LeTx-mediated toxicity. mAbs 2H9, 16A12, and 20G7 had no inhibitory effect against LeTx-mediated toxicity. MTT assays were done three times with similar results.
mAbs Effect on LeTx Toxicity on Mice
To examine further the ability of mAb 19D9 to neutralize LeTx, we tested its protective efficacy in vivo. The mAbs were administered to the mice (5 mice/group) intraperitoneally 2 h prior to intravenous infection with B. anthracis-germinated cells. Control animals received PBS instead of antibody. Administration of mAb 19D9 prolonged the survival of infected mice compared with control mice (data not shown). In addition, mAb 7.5G has also been shown to prolong the survival of BALB/c mice injected with toxin (13).
Mapping of the Epitope Recognized by mAbs
To identify the PA epitopes recognized by the mAbs, 15-mer peptides, overlapping by 10 residues covering the entire sequence of PA, were used for epitope mapping. Three of the six antibodies recognized peptides spanning residues extending from Leu156–Ser170 and Val196–Ile210 in domain 1 of PA (Table 2). These residues surround the furin cleavage site and hence provide insight on the mechanism to which 7.5G and 19D9 mAbs neutralize LeTx. Two other antibodies recognized the same peptide, extending from Ser312–Asn326. However, mAb 10F4 did not react with any peptide. This mAb seems to exhibit binding to a conformational epitope in domain 4 of PA.
TABLE 2.
Peptide specificity of mAbs and mean of ELISA absorbance
| Monoclonal antibody | Neutralizing | Epitope characterization |
|||
|---|---|---|---|---|---|
| PA domain | Position | Sequence | Mean absorbancea | ||
| 7.5G | Yes | 1 | 156–170 | LKQKSSNSRKKRSTS | 0.908 |
| 19D9 | Yes | 1 | 196–210 | VKNKRTFLSPWISNI | 0.569 |
| 20G7 | No | 1 | 196–210 | VKNKRTFLSPWISNI | 0.822 |
| 2H9 | No | 2 | 312–326 | SFFDIGGSVSAGFSN | 2.377 |
| 16A12 | No | 2 | 312–326 | SFFDIGGSVSAGFSN | 2.259 |
| 10F4 | Yes | 4 | NA | Conformational epitope | NA |
a Average of three replicates with a mAb dilution of 1:300 at which the antibody absorbance reading was at least three times the average background value.
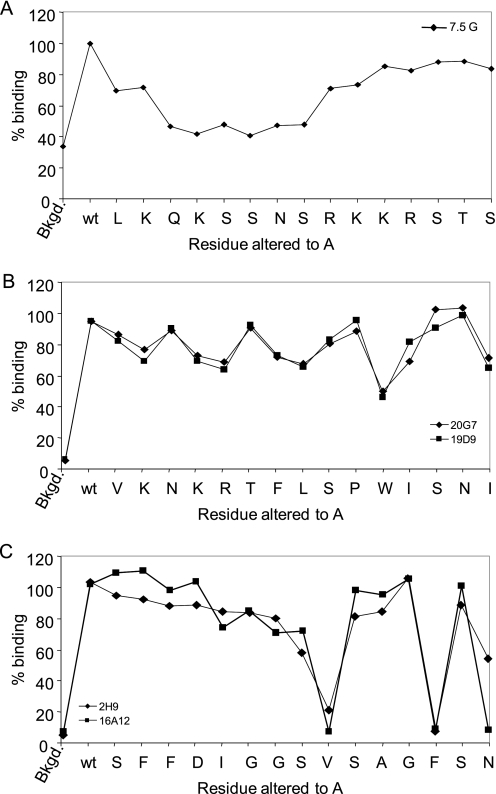
Next, we determined whether there were any specific residues in the identified epitopes critical for the binding of each mAb. Therefore, 15-mer peptides were synthesized such that every residue extending from Leu156–Ser170, Val196–Ile210, or Ser312–Asn326 was changed to alanine with the exception of Ala322, which was changed to Gly. The reactivities with the mAbs, 7.5G, 19D9, 20G7, 16A12, or 29H, were assessed in the same manner described above. The alanine substitution of residues extending from Gln158–Ser163 significantly reduced the binding of mAb 7.5G (Fig. 4A). Interestingly, alanine substitutions along the peptide sequence of Val196–Pro205 had only a modest effect of binding mAbs 19D9 and 20G7, whereas the presence of a bulky Trp residue underscores the importance of this residue in contacting both mAbs (Fig. 4B). In addition, alanine substitution of V320A, F324A, and N326A in peptide Ser312–Asn326 abrogates the binding of mAbs 29H and 16A12 (Fig. 4C). The Val residue in the middle of the epitope and the Phe and Asn residues at the end of the epitope may create a particular structural conformation of the peptide allowing for mAb recognition. However, the G324A replacement did not alter the structural conformation of the peptide, and therefore mAb binding paralleled the binding of wild-type peptide.
FIGURE 4.
An alanine walk to determine the critical residues in three identified epitopes. Each residue extending from Leu156–Ser170 (A), Val196–Ile210 (B), or Ser312–Asn326 (C) was altered in turn to Ala or Gly (in the case if an Ala was present), and their reactivities with mAbs 7.5G, 29H, 19D9, and 20G7 were determined by ELISA. The absorbance readings of relative binding of mAbs to the alanine-substituted peptides is expressed as % binding of each altered peptide with respect to wild-type peptide, where the latter was considered the base-line maximum binding level. The background (Bkgd) of each mAb was determined in parallel, by using streptavidin-coated, peptide-free wells. The average value of three experiments is shown.
Cleavage of PA and Dissociation of PA20
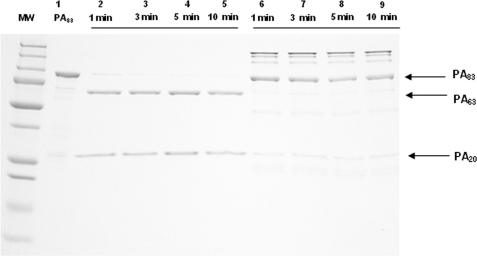
To examine whether mAb 19D9 binding affects dissociation of PA20 from PA63, PA was digested with 2 units of furin for 1-, 3-, 5-, and 10–12-min intervals in the presence and absence of mAb. PA83 and its digested products were separated on a 12% SDS-PAGE (Fig. 5). The amounts of digested products, PA20 and PA63, increased in the absence of mAb 19D9, whereas the amount of undigested PA83 decreased over time. At all time intervals, there were less PA20 and PA63 and more PA83 when mAb 19D9 was present. Hence, mAb 19D9 significantly slows the proteolytic cleavage of PA83 to PA20 and PA63. mAb 7.5G has also been shown to slow furin proteolysis (13).
FIGURE 5.
Cleavage of PA83 with furin in the absence and presence of mAb 19D9. The mAb 19D9 was incubated with PA for 1 h at room temperature prior to the adding of protease. The samples were separated by reduced SDS-10% polyacrylamide gels and stained with Coomassie Blue. MW, molecular weight marker; lane 1, PA undigested; lanes 2–5, PA + furin; lanes 6–9, PA + furin + mAb.
Molecular Modeling
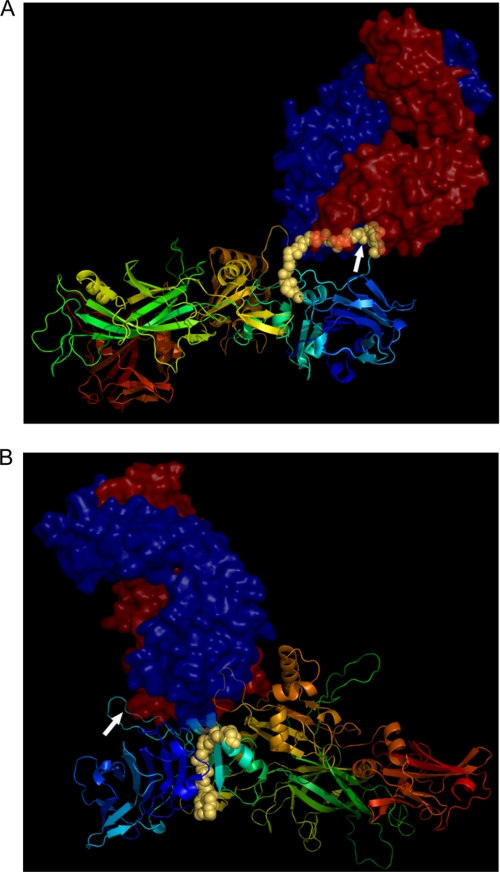
We generated three-dimensional molecular models to study the complex formation between the two regions of PA that are specific to 7.5G- and 19D9-neutralizing mAbs. The resulting docked complexes shown in Fig. 6, A and B, were derived from the crystal structure of PA and the modeled structure of mAbs 7.5G and 19D9. The docked complexes were filtered and refined using the sequence information provided from ELISA experimental results and the alanine-scanning experiments of the epitopes.
FIGURE 6.
Molecular modeling analysis of mAbs 7.5G and 19D9. A, docking of 7.5G mAb on PA protein. B, docking of 19D9 mAb on PA protein. PA protein is depicted as a ribbon representation, whereas the epitopes, which are found in domain 1, are shown as yellow spheres. Arrows represent the furin site. mAbs are shown as space-filling models, where the red represents the heavy chain and the blue represents the light chain of the variable region.
In the case of mAb 7.5G, the reactive epitope 158QKSSNS163 is represented by a long loop on the structure of PA, forming a linear epitope. For that of mAb 19D9, the epitope 206WISNI210 is also exposed but on a smaller loop on the structure of domain 1. Inspection of the molecular surface of the binding sites indicated that the loops could provide an anchor point for binding the epitopes to both mAbs. The docked structures explain why there is no furin cleavage upon mAb binding as the bound complexes in both cases are protected from furin accession. In the case of 7.5G, the mAb directly binds to the loop that contains the furin cleavage site (Fig. 6A, white arrow). For that of 19D9, the mAb binds to a different loop on PA, although able to sterically prohibit furin binding (Fig. 6B).
Reactivity of PA and MAP Peptide-immunized Mouse Sera with Peptides
Using the biotinylated peptides in ELISAs, the prevalence of anti-peptide antibodies in sera of mice immunized with PA was explored. Peptides surrounding the furin cleavage site in domain 1 of PA were recognized by half of the mAbs, indicating that these epitopes are targets for a significant population of anti-PA antibodies. Antibodies to peptides spanning regions Leu156–Ser170 and Val196–Ile210 were found in all sera with anti-PA reactivity (data not shown). None of the control sera, negative for anti-PA antibodies, was found to be reactive with these peptides. Antibodies to the peptide spanning region Ser312–Asn326 in domain 2 of PA were not detected with any sera, thus indicating the rare presence of natural antibodies against this epitope.
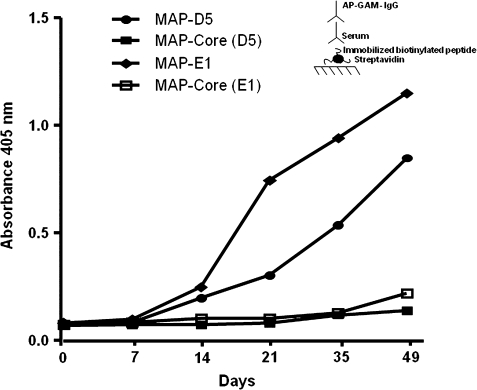
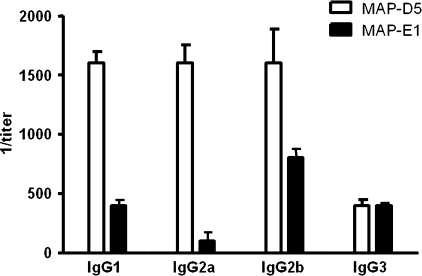
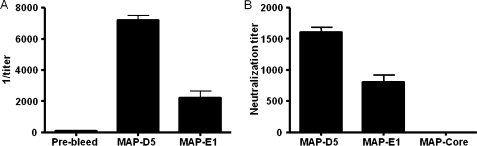
Next, BALB/c mice were immunized with the octameric peptide MAP-LKQKSSNSRKKRSTS (MAP-D5) and MAP-VKNKRTFLSPWISNI (MAP-E1) in CFA, followed by two booster injections of MAP-D5 and MAP-E1 in IFA. This resulted in production of anti-peptide antibodies for both peptides tested. The IgG anti-peptide titer in MAP-D5 and MAP-E1-immunized mice rose by day 21 and continued to increase until day 49 when the mice were killed (Fig. 7). The IgG subclass distribution of anti-peptide antibodies is shown in Fig. 8. Mice immunized with MAP-D5 generated equal amounts of IgG1, IgG2a, and IgG2b antibodies. Immunization with MAP-E1 resulted in the production of predominantly IgG2b anti-peptide antibodies. To further characterize the antigenic specificities present, ELISAs were performed to detect reactivity to PA. Anti-MAP-D5 IgG demonstrated significant titer (1:8,000) within 7 weeks (49 days post-immunization), whereas anti-MAP-E1 IgG reacted with PA at a titer of 1:2,000 (Fig. 9A). No IgG reactivity to PA was found in sera from mice immunized with the MAP core.
FIGURE 7.
Anti-peptide antibodies in MAP-D5- and MAP-E1-immunized mice. Female BALB/c mice (five animals per group) were immunized with 100 μg of MAP-D5 or MAP-E1 in CFA on day 0, and boosted with MAP-D5 or MAP-E1 in IFA on days 7 and 14, whereas control mice were immunized with MAP core in adjuvant. Sera from the different time points were diluted 1:500 and assayed for IgG anti-peptide antibodies by ELISA. Each point represents the average of five mice per treatment group. Inset, schematic of ELISA configuration used.
FIGURE 8.
IgG subclasses of anti-MAP-D5 and anti-MAP-E1 antibodies induced following immunization with MAP-D5 and MAP-E1. The subclasses of the IgG anti-peptide antibodies in day +49 sera from five MAP-D5-immunized mice and five MAP-E1-immunized mice were measured by ELISA.
FIGURE 9.
Anti-PA antibodies in MAP-D5- and MAP-E1-immunized mice and LeTx-neutralizing activity on J774 macrophages. A, sera from day +49 were assayed for IgG anti-PA antibodies by ELISA. The values represent geometric means ± S.D. from five mice per treatment group. B, day +49 serum from mice immunized with MAP-D5 and MAP-E1 confer moderate protection to J774 macrophage monolayers against LeTx-mediated toxicity. Bars represent arithmetic mean of the highest neutralization titers from five mice per treatment group. MTT assays were done three times with similar results. Anti-MAP core sera conferred no protection.
Anti-MAP-D5 and Anti-MAP-E1 Antibody Effect on LeTx Toxicity on Macrophages
The protective efficacy of anti-peptide immune sera from all vaccinated groups against LeTx was evaluated with J774 cells. Cell viability was measured using the MTT method as described under “Experimental Procedures.” The extent of cell death was expressed relative to a control containing LeTx or untreated cells. Day +49 immune sera from mice immunized with MAP-D5 and MAP-E1 conferred protection against the cytotoxic effects of LeTx with moderate protection up to 1:1,600 and 1:800 dilution, respectively (Fig. 9B). Sera from mice immunized with MAP core did not protect macrophages against LeTx cytotoxicity.
DISCUSSION
There is an urgent need for a more effective prophylactic vaccine against anthrax. The treatment of anthrax remains unsatisfactory because of high morbidity and mortality (2), and there are significant drawbacks to the currently licensed vaccine. Consequently, there is considerable interest in the development of passive immune therapies and more effective vaccines. The major drawbacks with the current vaccine have emphasized the importance of improving therapeutic approaches, which include passive immune therapies against B. anthracis. Additionally, anthrax toxin has the ability to impair innate and adaptive immune responses, entering the cytosol of every cell type and altering their signaling pathways, which in turn inhibits the clearance of the bacterium (23).
We have analyzed six hybridoma clones expressing anti-PA mAbs that were generated by cell fusion techniques. We identified two PA-neutralizing antibodies that efficiently protected animals from anthrax toxin challenge in vivo, most likely by preventing furin cleavage. The latter was based on epitope mapping studies, where our approach relied upon the use of overlapping synthetic peptides spanning the entire length of PA. The neutralization epitopes recognized by 7.5G and 19D9 mAbs were mapped to a region of PA comprising Leu156–Ser170 and Val196–Ile210, respectively. In addition, the relevance of these linear epitopes has been confirmed by their reactivity to mouse immune sera with anti-PA antibodies but not with control sera (preimmune sera). To date, three neutralization epitopes in PA have been mapped to regions that contain the sites for cellular receptor binding, LF binding, and heptamer formation (24). In this report, we demonstrate for the first time two neutralization epitopes mapped to regions in PA that are in very close proximity to the furin cleavage site. The three-dimensional complex structures of the mAbs and PA derived from the molecular docking method illustrate a striking complementarity of fit between surfaces of mAb and surface-exposed loops on domain 1 of PA. Peptide mutagenesis studies have highlighted these surface loops as the key regions for mAb interaction with PA and defined critical binding residues on PA. In our docking models, the 7.5G epitope is created by participation of surface loop residues Gln158–Ser163, and the 19D9 epitope is mapped to surface loop residues Trp206–Ile210.
mAbs 7.5G and 19D9 manifest differences in neutralizing activity despite closely related epitopes flanking the furin site. A recent report demonstrates that after mouse injection PA83 is cleaved in vivo, that this cleavage is independent of cell surface binding, and the proteolytic fragments, PA20 and PA63, circulate in the bloodstream (25). In this regard, our finding that mAb 19D9 is more potent in its neutralizing capability than mAb 7.5G suggests that during intoxication free PA20 that is generated and left in the extracellular space may render mAb 7.5G less effective (epitope is located in PA20). In another scenario, binding of mAb 19D9 to the predicted surface-exposed epitope might create steric hindrance that would prevent efficient furin cleavage. Interestingly, we note that the PA-binding affinities of mAbs 7.5G and 19D9 are different from each other, which may also correlate with their neutralization abilities. Peptide mutagenesis studies have delineated some details of mAb interaction with PA, and along with the docked models have confirmed the probable mechanism of protection afforded by mAbs 7.5G and 19D9 against LeTx.
Furthermore, two non-neutralizing mAbs, 2H9 and 16A12, recognized a peptide the spans residues extending from Ser312–Asn326 in domain 2. According to the crystal structure of PA, this region encompasses part of the 2β2–2β3 loop and, as stated above, undergoes structural rearrangements within the PA63 heptamer due to the acidified environment of the endosome, leading to the production of an extended β-barrel that inserts into the endosomal membrane (26–29). In recent studies, the 2β2–2β3 loop was demonstrated to contain a dominant neutralizing epitope with the sequence Ser312–Asp315 (5, 8). However, in our study the epitope recognized by mAbs 2H9 and 16A12 is mapped to 320VSAGFSN326, which is found to be a few amino acids after Ser312–Asp315. This illustrates that such a minor spacing can render an antibody with non-neutralizing capability. Interestingly, mAb 19D9 with strong toxin-neutralizing activity and mAb 20G7 with no neutralizing activity recognized the same epitope within domain 1. The key variation between these mAbs is their isotype; 19D9 is an IgG1, whereas 20G7 is an IgM. To be effective, antibodies must bind the target antigen and also invoke effector functions that will result in the removal of the antigen. The Fc region of the antibody is important in invoking this effector function. Because these two mAbs differ in their Fc region, a possible role could be due to their different interactions with their Fc receptors (FcRs), affording IgG1-FcR interaction more effective in the removal of PA. This further reinforces the concept that antibodies to PA consist of heterogeneous groups.
We note that the four mAbs use the same variable genes in VH and VL construction, yet some differ in epitope specificity and protective efficacy. The finding that these mAbs each use the same gene elements in antibody construction implies that the response to PA is restricted in V region utilization. Given that this mAb set uses the same V regions, yet some differ in fine specificity, implies that the specificity differences arise from somatic mutations.
In this study, we observe that 3 of 6 mAbs investigated bind at or near the furin site. This observation combined with the finding that domain 1 can be highly immunogenic and elicit neutralizing antibodies (3) raises the possibility for a PA immunodominant antigen in the furin region cleavage area. The existence of strong B-cell epitopes near the furin cleavage site that can protect, presumably by interfering with proteolytic cleavage, raises the tantalizing possibility that recognizing epitopes in this area provided an advantage in mammalian evolution from the negative selection of B. anthracis infection. Consequently, we focused on designing a vaccine that directed the immune response on this region. In principle, by selecting only those epitopes that confer protective immunity, it should be plausible to exclude the epitopes responsible for deleterious immune responses. Immunization of BALB/c mice with MAP-D5 (peptide recognized by 7.5G) elicits anti-peptide antibodies. Immunization with peptide MAP-E1 specific for mAb 19D9 led to a peptide-specific response as well. We demonstrated an IgG response to MAP-D5 with the highest anti-MAP-D5 titers to be a mixture of IgG1, IgG2a, and IgG2b, whereas for anti-MAP-E1 titers the response to this peptide was predominantly IgG2b. This suggests a Th2-related response for induction of peptide antibodies. Sera from mice immunized with MAP-D5 and MAP-E1 also bound toxin and neutralized its biological activity. These epitopes thus proved useful to assess the epitope mapping potential of our peptide library and raised the importance of the fact that two selected peptides were indeed able to elicit neutralizing antibodies in the mouse. The ability of antibodies raised against a peptide to bind the cognate protein in its native state further supports our assignments of linear epitopes within PA.
In conclusion, we have successfully identified two independent amino acid sequences within domain 1 of PA that contain neutralizing linear epitopes. These epitopes lie in the N-terminal moiety of the protein, in a region involved in furin cleavage. Identification of these immunodominant B-cell epitopes makes them attractive candidates and represents a first step toward the rational design for epitope-based anthrax vaccines.
This work was supported, in whole or in part, by National Institutes of Health Grant 2U54AI057158-06. This work was also supported by Department of Defense Grant DOD-071640041-002.
- PA
- protective antigen
- EF
- edema factor
- LF
- lethal factor
- LeTx
- lethal toxin
- GalXM-PA
- galactoxylomannan-protective antigen
- MAP-D5
- LKQKSSNSRKKRSTS
- MAP-E1
- VKNKRTFLSPWISNI
- ELISA
- enzyme-linked immunosorbent assay
- mAb
- monoclonal antibody
- PBS
- phosphate-buffered saline
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- CFA
- complete Freund's adjuvant
- IFA
- incomplete Freund's adjuvant
- MAP
- multiple antigenic peptide.
REFERENCES
- 1.Barth H., Aktories K., Popoff M. R., Stiles B. G. (2004) Microbiol. Mol. Biol. Rev. 68, 373–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inglesby T. V., O'Toole T., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Friedlander A. M., Gerberding J., Hauer J., Hughes J., McDade J., Osterholm M. T., Parker G., Perl T. M., Russell P. K., Tonat K. (2002) J. Am. Med. Assoc. 287, 2236–2252 [DOI] [PubMed] [Google Scholar]
- 3.Abboud N., Casadevall A. (2008) Clin. Vaccine Immunol. 15, 1115–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flick-Smith H. C., Walker N. J., Gibson P., Bullifent H., Hayward S., Miller J., Titball R. W., Williamson E. D. (2002) Infect. Immun. 70, 1653–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gubbins M. J., Berry J. D., Corbett C. R., Mogridge J., Yuan X. Y., Schmidt L., Nicolas B., Kabani A., Tsang R. S. (2006) FEMS Immunol. Med. Microbiol. 47, 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reed D. S., Smoll J., Gibbs P., Little S. F. (2002) Cytometry 49, 1–7 [DOI] [PubMed] [Google Scholar]
- 7.Rosovitz M. J., Schuck P., Varughese M., Chopra A. P., Mehra V., Singh Y., McGinnis L. M., Leppla S. H. (2003) J. Biol. Chem. 278, 30936–30944 [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Xu J., Li G., Dong D., Song X., Guo Q., Zhao J., Fu L., Chen W. (2006) Biochem. Biophys. Res. Commun. 341, 1164–1171 [DOI] [PubMed] [Google Scholar]
- 9.Laffly E., Danjou L., Condemine F., Vidal D., Drouet E., Lefranc M. P., Bottex C., Thullier P. (2005) Antimicrob. Agents Chemother. 49, 3414–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson J. W., Comer J. E., Baze W. B., Noffsinger D. M., Wenglikowski A., Walberg K. G., Hardcastle J., Pawlik J., Bush K., Taormina J., Moen S., Thomas J., Chatuev B. M., Sower L., Chopra A. K., Stanberry L. R., Sawada R., Scholz W. W., Sircar J. (2007) Infect. Immun. 75, 3414–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian G. M., Cronin P. W., Poley G., Weinstein A., Stoughton S. M., Zhong J., Ou Y., Zmuda J. F., Osborn B. L., Freimuth W. W. (2005) Clin. Infect. Dis. 41, 12–20 [DOI] [PubMed] [Google Scholar]
- 12.Baillie L., Townend T., Walker N., Eriksson U., Williamson D. (2004) FEMS Immunol. Med. Microbiol. 42, 267–270 [DOI] [PubMed] [Google Scholar]
- 13.Rivera J., Nakouzi A., Abboud N., Revskaya E., Goldman D., Collier R. J., Dadachova E., Casadevall A. (2006) Infect. Immun. 74, 4149–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Jesus M., Nicola A. M., Rodrigues M. L., Janbon G., Casadevall A. (2009) Eukaryot. Cell 8, 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de StGroth S. F., Scheidegger D. (1980) J. Immunol. Methods 35, 1–21 [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 17.Wellings D. A., Atherton E. (1997) Methods Enzymol. 289, 44–67 [DOI] [PubMed] [Google Scholar]
- 18.Beavis R. C., Chait B. T. (1996) Methods Enzymol. 270, 519–551 [DOI] [PubMed] [Google Scholar]
- 19.Casadevall A., Mukherjee J., Scharff M. D. (1992) J. Immunol. Methods 154, 27–35 [DOI] [PubMed] [Google Scholar]
- 20.Rai B. K., Fiser A. (2006) Proteins 63, 644–661 [DOI] [PubMed] [Google Scholar]
- 21.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 22.Gabb H. A., Jackson R. M., Sternberg M. J. (1997) J. Mol. Biol. 272, 106–120 [DOI] [PubMed] [Google Scholar]
- 23.Baldari C. T., Tonello F., Paccani S. R., Montecucco C. (2006) Trends Immunol. 27, 434–440 [DOI] [PubMed] [Google Scholar]
- 24.Brossier F., Lévy M., Landier A., Lafaye P., Mock M. (2004) Infect. Immun. 72, 6313–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moayeri M., Wiggins J. F., Leppla S. H. (2007) Infect. Immun. 75, 5175–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petosa C., Collier R. J., Klimpel K. R., Leppla S. H., Liddington R. C. (1997) Nature 385, 833–838 [DOI] [PubMed] [Google Scholar]
- 27.Benson E. L., Huynh P. D., Finkelstein A., Collier R. J. (1998) Biochemistry 37, 3941–3948 [DOI] [PubMed] [Google Scholar]
- 28.Nassi S., Collier R. J., Finkelstein A. (2002) Biochemistry 41, 1445–1450 [DOI] [PubMed] [Google Scholar]
- 29.Santelli E., Bankston L. A., Leppla S. H., Liddington R. C. (2004) Nature 430, 905–908 [DOI] [PubMed] [Google Scholar]