Abstract
Mycobacterium tuberculosis survival in cells requires mycobactin siderophores. Recently, the search for lipid antigens presented by the CD1a antigen-presenting protein led to the discovery of a mycobactin-like compound, dideoxymycobactin (DDM). Here we synthesize DDMs using solution phase and solid phase peptide synthesis chemistry. Comparison of synthetic standards to natural mycobacterial mycobactins by nuclear magnetic resonance and mass spectrometry allowed identification of an unexpected α-methyl serine unit in natural DDM. This finding further distinguishes these pre-siderophores as foreign compounds distinct from conventional peptides, and we provide evidence that this chemical variation influences the T cell response. One synthetic DDM recapitulated natural structures and potently stimulated T cells, making it suitable for patient studies of CD1a in infectious disease. DDM analogs differing in the stereochemistry of their butyrate or oxazoline moieties were not recognized by human T cells. Therefore, we conclude that T cells show precise specificity for both arms of the peptide, which are predicted to lie at the CD1a-T cell receptor interface.
Pathogens are detected by the host when antigenic molecules directly contact immune receptors during the early stages of infection. The strategy of intracellular infection allows viruses, certain bacteria and protozoa to partially cloak themselves from the immune response by physically encapsulating their antigens within host cells. Intracellular residence also takes advantage of immune tolerance mechanisms that prevent autoimmune destruction of self. T cells play a central role in immunity to intracellular pathogens because they can respond to antigens that are generated inside cells and then transported to the surface of infected cells after binding to antigen-presenting molecules. The antigen-presenting molecules encoded in the major histocompatibility complex are widely known for presenting peptide fragments of proteins (1). More recently, human and mouse members of the CD1 (cluster of differentiation 1) system have been shown to present small amphipathic molecules, including a variety of membrane lipids, glycolipids, and lipopeptides, greatly expanding the molecular structures recognized by the cellular immune system (2, 3).
Among human CD1 proteins (CD1a, CD1b, CD1c, CD1d, and CD1e), each CD1 isoform is expressed on a different spectrum of antigen-presenting cells. Human CD1a proteins are distinguished from other CD1 proteins by high expression levels on the surface of intradermal Langerhans cells, which play a role in barrier immune function (4). Human T cell clones have been shown to directly recognize CD1a proteins in the presence of exogenous foreign antigens (5) or in the presence of sulfatide and other self lipids (6, 7), suggesting a role for CD1a in T cell activation. In addition, mycobacteria and other intracellular pathogens have been shown to increase CD1a expression in lesions found in leprosy and tuberculosis patients, implying a possible role for CD1a in the response to infection, especially at mucosal or skin sites (8–10). Analysis of the molecular target recognized by CD1a-restricted T cell clone (CD8-2) allowed the identification of a foreign antigen presented by CD1a as dideoxymycobactin (DDM) (11).2
Mycobactin binds iron to promote Mycobacterium tuberculosis survival. DDM was initially isolated (11) from antigenic lipid extracts of M. tuberculosis, a pathogen that kills ∼1.7 million humans annually on a worldwide basis (12). The determination of DDM structure was based on mass spectrometric and NMR studies of limiting amounts of natural material derived from the pathogenic organisms, so that not all elements of its chemical structure could be formally determined. Instead, its assigned structure was facilitated by obvious parallels of dideoxymycobactin with mycobactin, a lipopeptide siderophore (13, 14). Iron is required for reduction-oxidation reactions involving respiration and other basic metabolic pathways in bacterial pathogens (13). Environmental mycobacteria have at least two iron uptake pathways, but mycobactin and the related molecule carboxymycobactin represent the only known dedicated iron uptake pathway for pathogenic species like M. tuberculosis (15, 16). Highlighting the physiological importance of the mycobactin pathway, deletion of mycobactin synthase B limits M. tuberculosis survival in cells (13, 14). Also, mammalian innate immune systems produce siderocalin, a 20-kDa lipocalin that binds both ferric and apo siderophores, preventing their uptake and subsequent iron delivery to microbes (17–20). The small available yields of natural material highlighted the need for a straightforward method to synthesize DDM for studies of its role in mycobacterial iron acquisition and testing T cell responses in human populations, as well as to provide authentic standards to investigate unknown aspects of natural DDM stereochemistry. Here we report two syntheses for production of DDM in solution phase and solid phase. Comparison of synthetic and natural DDMs gives unexpected insight into the stereochemical structures of the methylserine, oxazoline, and butyrate moieties of DDM and provides direct evidence that the T cell response is highly specific for a unique aspect of DDM structure that protrudes from the surface of the CD1a-DDM complexes.
EXPERIMENTAL PROCEDURES
Extraction of M. tuberculosis Lipids
We purified DDM using a method described previously (21) that was modified with new reversed phase HPLC systems to obtain peak-to-base line separation of individual molecular species of DDM. Briefly, M. tuberculosis H37 Ra (Difco) or H37Rv was cultivated in biosafety level 3 and serially extracted at 5 mg/ml in chloroform:methanol mixtures at two ratios (2:1 and 1:2). The resulting total lipid extracts were dried, and 1.5 g were resuspended in 3 ml of chloroform/methanol (2:1) and added to 95 ml of cold acetone (0 °C) with 2 ml of 10% MgCl2·6H2O in methanol for 90 min to precipitate phospholipids. The resulting supernatant lipid mixture containing acetone-soluble lipids was separated from the precipitate by centrifugation at 2000 rpm for 15 min to give 60–70% yield from the total lipid extracts. Acetone-soluble lipids (20 mg) were resuspended in 10 ml of 100:25 hexanes:chloroform, loaded into a solid phase extraction silica gel (10 g) column (Altech, Waukegan, IL) and eluted with hexanes:chloroform:2-propanol:acetic acid in the following ratios:100:25:0:0 (40 ml), 100:25:5:0 (40 ml), 100:25:10:0 (40 ml), 100:25:15:0 (40 ml), 50:75:15:0 (40 ml × 2), and 50:75:15:1 (40 ml × 2). DDM was recovered in the final, acetic acid-containing fractions and subjected to reversed phase chromatography on a C4 column (Vydac) with a flow rate of 0.7 ml/min. A linear binary gradient (0 min 20% B, 4 min 20% B, 35 min 60% B, 45 min 60% B, and 50 min 20% B) was used with 50:30:20:0.02 methanol:acetonitrile:water:trifluoroacetic acid (A phase) and 93:7:0.02 2-propanol:hexanes:trifluoroacetic acid (B phase). The liquid chromatography-MS system (Waters Breeze HPLC with Thermo-Finnigan LCQ Advantage) was operated with a 7:1 split interface to allow fraction collection based on UV response in real time. Additional MSnth analysis was completed with nanoelectrospray with an internal stainless steel electrode (600–1000 V) using borosilicate glass pipettes fabricated in-house with a 2-μm orifice (Sutter Instrument Co., Novato, CA).
Derivatization and Gas Chromatography-MS Analysis
A purified DDM-840 sample and amino acid standards were cleaved and esterified by dissolving in 3 n HCl in butanol (prepared by adding 1.0 ml of acetyl chloride to 5.0 ml of n-butanol) and incubating overnight at 100 °C to both cleave all ester and amide linkages and convert all resulting carboxyl groups to butyl esters. The solvent was evaporated (SpeedVac, Savant), and the residue was treated with trifluoroacetic anhydride to derivatize remaining polar functional groups (22). The samples were redissolved (∼100 ng/μl) in dichloromethane for injection (1 μl) into the gas chromatography-MS with a Thermo-Finnigan GCQ mass spectrometer and associated gas chromatograph equipped with a BD-5MS capillary column (30-m × 0.25-mm inner diameter; Alltech Assoc., Waukegan, IL). The column temperature was initially held at 75 °C for 1.0 min, then increased at 5.0 °C/min to 200 °C, held for 1.0 min, and then increased at 10.0 °C/min to a final temperature of 300 °C. The injector was set at 195 °C, and the transfer line to the mass spectrometer was set at 200 °C.
Solution Phase Synthesis of DDM
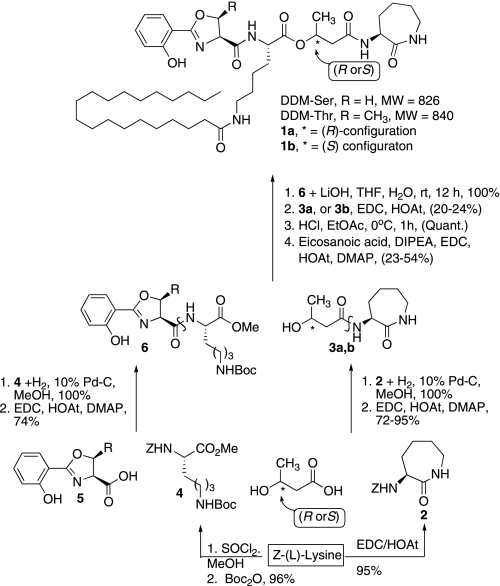
The strategy was based on previous syntheses by which protected forms of the cobactin and mycobactic acid are coupled, deprotected, and acylated (23–27). The synthesis reported here relies on production of deoxymycobactic acid and deoxycobactin (see Fig. 1). The strategy and numerically designated intermediates are shown in Fig. 2, and the detailed methods and chemical description of newly produced intermediates are shown in supplemental Fig. S1. In this way, formation of the diastereomeric cobactin components was initiated by cyclization of Z-(L)-lysine in the presence of EDC/HOAt to provide the Z-protected caprolactam 2 (numbers in bold type refer to Fig. 2) in 95% yield after chromatographic purification. Hydrogenation of the benzyloxycarbamate of 2 was followed by another EDC/HOAt-mediated peptide bond formation with the acid form of the commercially available (R or S)-3-hydroxy butyric acid sodium salt to separately provide the desired pre-cobactin T 3a and pre-cobactin S 3b in high yields. Synthesis of appropriately protected (Boc) pre-mycobactic acid methyl esters 5a,b required the preparation of an orthogonally protected (L)-lysine. Z-(L)-lysine was treated with thionyl chloride in MeOH to form the methyl ester. Without purification, the resulting amine salt was reacted with Boc2O in the presence of excess NaHCO3 to provide orthogonally protected lysine 4 in excellent yield. Hydrogenation of the Z-protecting group revealed the amine, which was then coupled to the known oxazoline acids 5a,b (27) by the action of EDC/HOAt. Completion of the syntheses began with the LiOH-mediated saponification of the methyl esters of 6a,b. Isolation of the acids was followed by an EDC/HOAt-mediated ester bond formation with pre-cobactin T 3a to provide intermediates 7a,b. HCl cleavage of the Boc-protecting group of each diastereomer was followed by another EDC/HOAt-mediated amide bond formation utilizing eicosanoic acid. Purification of each separately synthesized diastereomer provided pre-mycobactins 1a,b.
FIGURE 1.
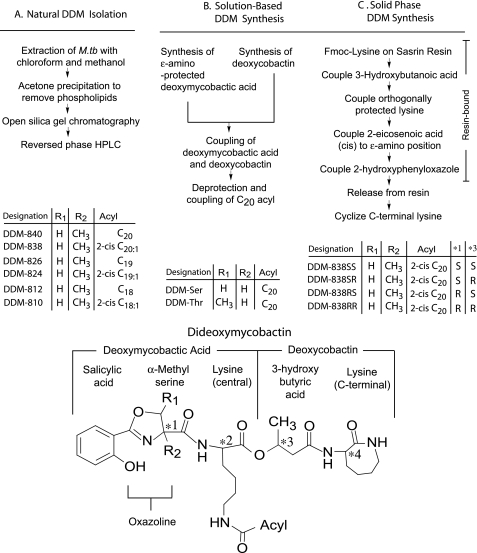
Three approaches used to produce dideoxymycobactin analogs that vary in structure. Dideoxymycobactins are composed of mycobactic acid and cobactin such that they contain four chiral centers in the peptidic backbone, designated *1, *2, *3, and *4. The chiral centers *2 and *4 were associated with the two lysine groups and assumed to have the same stereochemistry as naturally occurring l-lysine. The chiral centers labeled *1 and *3 were previously unknown and required elucidation. DDMs were isolated or produced with varying acyl chains or substitutions at R1 or R2 as indicated.
FIGURE 2.
Solution phase synthesis involves separate preparation of deoxycobactin and deoxymycobactic acid, followed by coupling to produce dideoxymycobactin.
Solid Phase Synthesis of DDM and Addition of 3R-Hydroxybutyric Acid to Fmoc-Lys(Mtt)-Super Acid-sensitive Resin
The synthesis was initiated using commercially available Fmoc-protected lysine bound to super acid-sensitive resin (Bachem Bubendorf, Switzerland) with the ϵ-amino group protected by an Mtt group. Using 11.7 μmol of the resin-bound lysine, the α-amino group was deprotected using 10% piperidine in dimethylformamide for 30 min followed by washing with dimethylformamide followed by dimethylacetamide. 100 μmol of sodium R-3-hydroxybutyrate (Sigma-Aldrich; R isomer with specific optical rotation −14.2° at 24.5 °C, 546 nm) was combined with equimolar amounts of HATU and HOAt in slight excess and dissolved in 3.0 ml of dimethylacetamide (28–30). The reagent solution was then reacted with the resin for 5–10 min followed by washing with dimethylacetamide and dimethylformamide.
Addition of Dde-Lysine(Fmoc)-OH
100 μmol of Dde-lysine(Fmoc)-OH and 120 μmol each of HBTU, HBOt, and collidine were dissolved in 1.5 ml of acetonitrile and reacted with the well washed resin for 2 h. A few microliters of dimethylformamide was added to reduce clumping of the resin in acetonitrile. The best observed yield was 44% relative to starting resin and based on measurement of UV absorbance at 299 nm after Fmoc deprotection.
Synthesis of 2-cis-Eicosenoic Acid (C20)
2-cis-Eicosenoic acid was synthesized using octadecanol as starting material and converting it to the aldehyde with oxalyl chloride, dimethyl sulfoxide, and triethyl amine. Dimethyl sulfoxide (40 mmol in 10 ml of methylene chloride) was added dropwise to a solution of oxalyl chloride (20 mmol) in anhydrous methylene chloride (1.8 ml) under nitrogen at −78 °C. After stirring for 10 min, 18 mmol of octadecanol in 10 ml of methylene chloride was added dropwise, still at −78 °C. The solution was stirred at −78 °C for 30 min, and 12.0 ml of triethylamine was added. The solution was allowed to reach room temperature with continued stirring until reaction completion was reached as determined by thin layer chromatography. The reaction mixture was then washed with saturated ammonium chloride and brine, dried over sodium sulfate, filtered, and evaporated under reduced pressure. Purification was done by chromatography on silica gel, eluting with 2% ethyl acetate in hexanes to give 3.65 g of octadecanal to give 78% yield. The C20 olefin was formed by coupling with the ethyl ester of diethylphosphonoacetic acid (31) with potassium hexamethyldisilazane as the base (32). Potassium hexamethyldisilazane (0.5 m in 2.3 ml of toluene) was added to a solution of the ethyl ester of diethylphosphonoacetic acid (11.62 mmol) and 18-crown-6 (52.9mmol) in 100 ml of anhydrous tetrahydrofuran at −78 °C. Octadecanal (10.57 mmol), dissolved in 5 ml of tetrahydrofuran, was added, and the mixture was stirred at −78 °C until the disappearance of the starting material. Saturated aqueous ammonium chloride (150 ml) was added, and the product was extracted with ethyl acetate (3 × 100 ml). The organic phase was dried over sodium sulfate, filtered, and concentrated. This yielded a mixture of cis- and trans-ethyl 2-eicosenoate in a 5:1 ratio, which could be separated by flash chromatography with 1% ethyl acetate over silica gel with the cis-isomer eluting first. The ethyl ester was hydrolyzed to the free acid using aqueous LiOH in ethanol (1 ml of 1 n LiOH in 10 ml of ethanol) with stirring for 3 days. The reaction mixture was then diluted with 150 ml of ethyl acetate, washed with 10% HCl, saturated NaCl, and then dried over sodium sulfate. The mixture was then filtered and evaporated under reduced pressure to give 1.4 g (80%) of cis-2-eicosenoic acid.
Addition of Acyl Group to the ϵ-Amino Group of Lysine
The ϵ-amino group of the central lysine was first deprotected using 10% piperidine in dimethylformamide for 5 min, avoiding excess reaction time, which begins to slowly remove the α-amino Dde-protecting group. 2-cis-Eicosenoic acid (100 μmol) was combined with 110 μmol of HOBt and dissolved in 200 μl of chloroform and 100 μl of dimethylformamide. The solution was added to the washed resin, and 110 μl of 1 m dicyclohexylcarbodiimide in methylene chloride was added to activate the coupling reaction.
Synthesis of Methylserine- and Salicylate-based Phenyloxazoline
Each enantiomer of methyl 4,5-dihydro-4-methyl-2-(2-benzyloxy)phenyloxazole-4-carboxylate (methylserine-based oxazoline) was separately prepared by converting the benzyl ether of salicylic acid, 2-benzyloxy-benzoic acid to the acid chloride using oxalyl chloride and a drop of dimethylformamide, followed by a reaction with the methyl ester of the separate enantiomers of α-methylserine. The resulting amide was cyclized to the corresponding oxazoline form using bis(2-methoxyethyl)amino-sulfur trifluoride at −78 °C (33). The protecting groups were removed using simultaneous LiOH (10 mg/ml) saponification and catalytic hydrogenation (10 mg of platinum on carbon, 10% by weight) in methanol at room temperature. The product was dissolved in ethyl acetate and extracted with 0.5 n citric acid and then brine. Circular dichroism spectrophotometry (Jasco J-715; Jasco Inc., Easton, MD) was used to confirm the final structure of the phenyloxazoline compounds synthesized by comparison to an R-serine-based oxazoline derivative of known structure (supplemental Fig. S2).
Addition of Methylserine-based Phenyloxazoline
The Dde group was removed using 2% hydrazine in dimethylformamide (stored at 5 °C) for 10 min followed by extensive washing. Deprotected methylserine-based oxazoline (50 μmol) was combined with a 10% excess of HATU and dissolved in 1.5 ml of methylene chloride:acetonitrile:dimethyl formamide (1:1:1) to react with the resin for 4 h with agitation.
Cyclization of Terminal Lysine
The lipopeptide was cleaved from the resin using 2% trifluoroacetic acid in methylene chloride. Cleavage solution was allowed to flow through the resin until the effluent ceased to produce yellow color associated with the free Mtt group. After solvent evaporation, the product was immediately diluted to 1.5 ml in methylene chloride:acetonitrile and treated with a 10% molar excess of HATU with HOAt and collidine added to ensure basic conditions to form the C-terminal cyclic lysine. Reversed phase HPLC on a 1.0 cm × 25 cm C18 column at 2.7 ml/min (Higgins Analytical, Mountain View, CA) was then used to purify the DDM and separate it from reagents. The method used gradient elution starting at 60% B phase and ending at 80% B phase. The A phase was 50:30:20 methanol:acetonitrile:water with 0.1% formic acid, 0.02% trifluoroacetic acid, 10 mm ammonium acetate, 0.02% hexafluoro-2-propanol. The B phase was 10% hexane in 2-propanol with 0.1% formic acid and 0.02% trifluoroacetic acid. Collected DDM was dried under a stream of nitrogen. Final yield was 2.7% relative to the lysine bound to the starting resin based on UV absorbance of the final product. The final yield and complexity of the products was variable and improved if the purification and final removal of acidic mobile phase additives was done quickly. Passage through a short silica gel column and elution with 10% 2-propanol in chloroform was used to help remove polar additives from the final product.
T Cell Assays
CD1a-expressing antigen-presenting cells were isolated from human peripheral blood mononuclear cells by centrifugation over Ficoll-Hypaque, plastic adherence, and treatment with 300 IU/ml granulocyte macrophage colony-stimulating factor and 200 IU/ml interleukin-4 for 72 h, followed by γ-irradiation (5000 Rad) (34). CD1a-restricted, dideoxymycobactin-reactive T cell transfectants (J.RT3.CD8-2) or CD8-2 T cells (5) were tested for interleukin-2 release by using the HT-2 bioassay. A combination of 1 × 105 J.RT3.CD8-2 transfectant cells and 5 × 104 γ-irradiated antigen-presenting cells were incubated in 200 μl of T cell medium containing serial dilutions of lipid antigens; phorbol 12-myristate 13-acetate was added to each well at a final concentration of 10 ng/ml. After 24 h, 50 μl of supernatant was added to wells containing 100 μl of medium and 104 interleukin-2-responsive HT-2 cells. One day later, 1 μCi of [3H]thymidine was added to each well. After 6 h, incorporated [3H]thymidine was measured by counting beta emissions.
For plate-bound CD1 assays, soluble human CD1a fusion proteins covalently linked to human β2m and to the Fc portion of murine IgG2a were constructed, produced, and purified as previously reported (35). Ninety-six-well protein G-coated plates (Pierce) were incubated with 1.25 μg of fusion protein/well in phosphate-buffered saline at pH 7.4 for 24 h at 37 °C. Lipid antigens were then sonicated into phosphate-buffered saline and added to the wells. The plate was incubated for 24 h at 37 °C prior to washing three times with 200 μl/well of sterile phosphate-buffered saline and adding 105 CD8-2 T cells in a total volume of 200 μl of T cell medium/well. The plate was incubated for 24 h at 37 °C, after which culture supernatants were collected for analysis. The assays were performed in triplicate at least three times, and the data are reported as the means ± standard deviation. CD1a plate-bound inhibition assays were performed similarly to standard plate-bound assays, except that nonstimulatory ligands were added to the plate and incubated for 8 h prior to the addition of DDM-838SR. After an additional 12 h of incubation, the plate was washed three times, and T cells were added. Supernatants from blocking assays were tested by interferon γ enzyme-linked immunosorbent assay.
Molecular Modeling
Protein Data Bank coordinates for the DDM-838SR-CD1a complex were prepared using the software suite Insight II (Accelrys). Using the previously determined crystal structure (Protein Data Bank code 1XZ0) (21), DDM-838SR was overlaid on the synthetic molecule to approximate the position of elements bound within the groove. The figures were prepared using the molecular visualization software PyMol (DeLano Scientific LLC), and electrostatic potentials were calculated using an the Poisson-Boltzmann equation as implemented in the software APBS (36).
RESULTS
Isolation of Individual Molecular Species of Dideoxymycobacin
The discovery of DDM was based on mass spectrometric analysis of trace amounts of antigenic material from M. tuberculosis. To generate the larger quantities of DDM needed for complete structural determination of natural molecules and immunological studies with human T cells, we took three approaches (Fig. 1). First, cultivation of liter quantities of M. tuberculosis, along with improvements in extraction and separation techniques, yielded six distinct molecular species of natural DDM. This was accomplished by extraction of intact organisms in chloroform and methanol followed by treatment of lipids with cold acetone to precipitate and remove phospholipids. Passage through a silica gel column provided a simplified subfraction, which was further resolved by molecular size with reversed phase HPLC using a C18 column and 2-propanol with 7% hexanes (21). This method yielded six pure subspecies named according to the nominal masses of their [M + H]+ ion detected in the positive mode: DDM-840, DDM-838, DDM-826, DDM-824, DDM-812, and DDM-810 (11) (Fig. 1A). CID mass spectra of the [M + H]+ ions generated by all six compounds yielded a common product ion at m/z 546 for each compound, which was found to correspond to the mass of the peptidic portion of the DDM (supplemental Fig. S3). Additional product ions enabled determination of the backbone sequence, the location of the acyl group, and its degree of unsaturation (supplemental Fig. S4). The mass differences among individual members of the series were accounted for by their varied substitution with acyl chains of C20, C20:1, C19, C19:1, C18, or C18:1 (Fig. 1 and supplemental Figs. S3 and S4). Although the low microgram yields were insufficient for most biological applications, these natural molecules were important for guiding the synthetic strategy and for use as standards for evaluating the chemical structure and immunological properties of synthetic molecules.
Solution-based DDM Synthesis
The first strategy for DDM synthesis involved separate production of deoxy forms of protected mycobactic acid and cobactin in solution phase (Fig. 1B). These products coupled to form the protected peptide backbone and then deprotected and coupled to the acyl side chain to yield dideoxymycobactin. This strategy was potentially expeditious because problems with certain key intermediates, reaction methods, and extraction procedures had been solved during prior, successful efforts to synthesize mycobactin (23–27). In fact, it might have been possible that mycobactin or other intermediates leading to the synthesis of mycobactin were sufficient to stimulate CD1a-restricted T cells, obviating the need to produce DDM for immunological studies. However, a dideoxymycobactin-reactive T cell line (CD8-2) failed to respond to each of 14 synthetic mycobactin or mycobactin-like compounds (supplemental Fig. S5). These compounds recapitulated many, but not all, aspects of DDM structure, providing evidence that the CD1a-mediated T cell response was specific for the fine structure of the molecule.
Descriptions in the literature suggested that mycobactin synthases incorporate either serine or threonine to form the oxazoline ring (37). To recapitulate these structures in DDM, we used l-serine and l-threonine to make two forms of DDM peptide, designated DDM-Ser and DDM-Thr (Fig. 2 and supplemental Fig. S1). We compared their antigenic activity with that of the mixture of M. tuberculosis dideoxymycobactins. Even though the synthetic molecules mimicked the structure of the two DDMs thought to be present in the mixture of natural compounds, neither synthetic compound stimulated a CD1a-restricted T cell response (supplemental Fig. S6A). Because prior evidence suggested that a single unsaturation at C2–3 in the C20 fatty acid increases antigenic potency, we produced a third molecule, DDM-Ser-2-trans, but it also failed to stimulate a T cell response (supplemental Fig. S6B). Thus, either the initial identification of mycobacterial DDMs as antigens was incorrect, or these synthetic compounds differed from certain naturally produced molecules such that the difference was chemically subtle but immunologically important.
Proton NMR analysis of purified, antigenic DDM-840 suggested a chemical variant of the peptidic chain that was unexpected as compared with prior analysis of mycobactin-like molecules. An isolated set of coupled resonances at 4.70 and 4.27 ppm were observed in both one- and two-dimensional NMR, consistent with two protons located directly adjacent to the oxygen in the oxazoline ring (supplemental Fig. S7). These resonances were observed as simple doublets with no apparent interactions with other neighboring hydrogen atoms, implying that they were derived from geminal hydrogens on an isolated methylene unit. In prior studies of mycobactin incorporating serine or threonine, this simple pattern was not observed. Instead, these amino acids provide one extra proton on the α or β carbon, which causes a more complex coupling pattern (38). The current result implied that the oxazoline ring in DDM-840 from this isolate of M. tuberculosis is formed by incorporation of α-methylserine. This unusual amino acid has been observed in bacterial compounds such as the antibiotic amicetin from Streptomyces fasciculatus (39, 40) and mycobactins from the actinobacteria Nocardia (41) and also in plants (42) but, to our knowledge, not in M. tuberculosis.
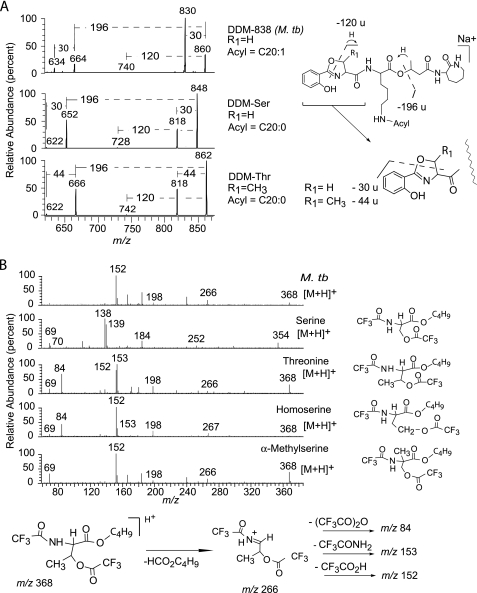
This unexpected molecular variant of natural DDM might explain why synthetic DDMs, which were produced using the more common amino acid structures, were not antigenic. With electrospray ionization MS, DDM-Ser, DDM-Thr, and M. tuberculosis-derived DDM-838 were detected as sodium adducts with nominal [M + Na]+ at m/z 848, 862, and 860, respectively. CID-MS analysis showed prominent product ions corresponding to the loss of either 30 or 44 atomic mass units (Fig. 3A). Because DDM-Thr and DDM-Ser differ only in the presence or absence of a methyl group at position R1 (Fig. 1), these fragment ions could be formally assigned to a product that arises via cleavage through the ring containing this methyl group and therefore correspond to the loss of CH2O or C2H4O from the oxazoline unit, respectively. M. tuberculosis-derived DDM-838 yielded product ions with a loss of 30 atomic mass units, indicating a lack of a methyl group at R1, consistent with the presence of serine or α-methylserine but not threonine (Fig. 3B). In DDM-838 from M. tuberculosis, the fragment corresponding to the loss of the N-terminal butyrate-lysine moiety was observed at m/z 664, indicating that the oxazoline ring was formed either by methylsalicylate and serine or by salicylate and methylserine. These possibilities were resolved by the loss of 120 atomic mass units to generate the product m/z 740, which could be accounted for by a loss of C7H4O2 resulting from cleavage through the oxazoline ring with hydrogen transfer, indicating that salicylate, not methyl salicylate, was present (Fig. 3A). The presence of these fragments could only be explained if the amino acid forming the oxazoline ring were an isomer of threonine, such as homoserine or α-methylserine. In addition, the product resulting from the loss of 30 atomic mass units from the natural DDM (m/z 830) was more abundant than the corresponding ions in the serine- and threonine-based forms, an observation that could be accounted for by relief of the steric crowding at the α carbon, as would be expected for an oxazoline ring incorporating α-methylserine (Fig. 3A).
FIGURE 3.
Synthetic DDM standards allowed identification of α-methylserine as a component of DDM-838 from M. tuberculosis. A, CID products of sodium adducts ([M + Na]+) of synthetic DDM-Ser (m/z 848, middle panel) and synthetic DDM-Thr (m/z 862, bottom panel) were used to deduce the structure of the amino acid forming the oxazoline ring in the natural product DDM-838 (top panel). DDM-Ser and DDM-Thr give product ions at m/z 818, which are, respectively, 30 and 44 atomic mass units less than the parent ions and so correspond to the expected neutral losses of O=CHR1 from the oxazoline rings derived from serine and threonine. The observation that fragmentation of the [M + Na]+ ion of M. tuberculosis DDM-838 yields a predominant product ion via the loss of 30 atomic mass units rules out threonine as a component. The loss of 120 atomic mass units indicates that salicylic acid was incorporated into all three compounds. B, trifluoroacetyl, butyl ester derivatives of chemically digested M. tuberculosis antigenic lipid fraction (M. tb) were compared with amino acid derivatives of known structure using gas chromatography-mass spectrometry. The electron impact spectrum of one of the mycobacterial derivatives matches that of the α-methylserine derivative and is different from the other possible threonine isomers. In particular, the lack of an observable ion at m/z 84 distinguishes the α-methylserine derivative from the other isomers.
After all of the CID-MS experiments suggested the presence of α-methyl serine, we sought to directly identify the amino acids in DDM-840 by acid-catalyzed solvolysis in n-butanol and derivatization with trifluoroacetic anhydride, followed by gas chromatography-MS analysis. The electron impact MS spectra of the derivatives of the DDM-derived amino acid and α-methylserine were nearly identical and were distinct from those of serine, threonine, and homoserine derivatives (Fig. 3B) (52). In particular, the presence and ratio between the abundances of the peaks at m/z 152 and 153 seen in the digest product of M. tuberculosis DDM matched those of α-methylserine but did not match those in the spectra of the derivatives of threonine or serine, which instead exhibited a pair of peaks at m/z 138 and m/z 139. Thus, NMR, CID-MS, and gas chromatography-MS of the derivatized component amino acids all indicated that M. tuberculosis DDMs contain α-methylserine as the building block incorporated into the oxazoline ring. If T cells were capable of discriminating this detailed element of the structure of DDM, these results might explain why synthetic DDMs lacking α-methylserine failed to stimulate a T cell response. Therefore, we conceived a second method for synthesis of α-methylserine containing DDM, which is based on solid phase recovery of intermediates bound to polymeric resins (Fig. 4).
FIGURE 4.
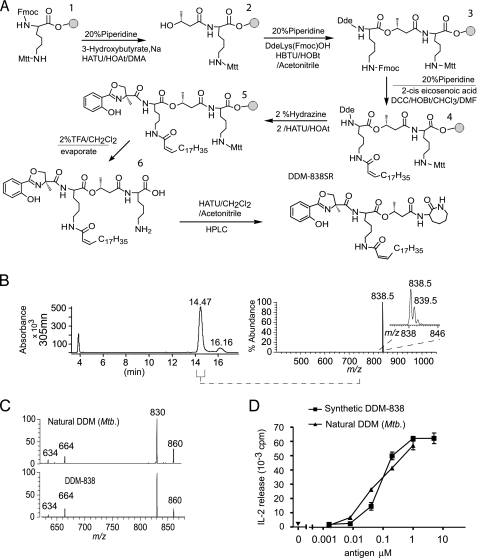
Synthetic DDM-838SR is indistinguishable from M. tuberculosis DDM-838. A, a schematic of solid phase synthesis of DDM-838 is shown with the gray circle representing super acid-sensitive resin. B, HPLC with UV and MS detection was used to collect the fraction from 14.1 to 14.7 min using a split interface for diversion to the mass spectrometer. Positive mode electrospray ionization-MS showed the expected mass and isotope pattern for [M + H]+ of DDM and no evidence of significant contamination by other products. C, MS-MS analysis of M. tuberculosis and synthetic DDM-838SR [M + Na+] ion showed spectra that were nearly identical. The fragment at m/z 830 resulting from cleavage through the oxazoline ring (see supplemental Fig. S1) is clearly observed with similar signal intensity in the spectra of both compounds, indicating that this functional group was unaltered under the conditions used for synthesis and isolation. D, CD1a-restricted T cell activation was measured by interleukin-2 release in response to human monocyte-derived dendritic cells and DDM from synthetic and natural sources.
Solid Phase Synthesis
Resin-based synthesis in the solid phase is a particularly useful technology for stepwise coupling of chemical building blocks in a multi-step synthesis because the insoluble polymer allows each intermediate to be recovered and separated from contaminants by simply washing the resin with solvent. This method also makes it feasible to use a large excess of reagents to drive the reaction to completion and carry out multi-step syntheses with relatively high yields. Solid phase synthesis techniques have been successfully used to make peptides (43–45), including mycobactins (46).
The synthesis started with l-lysine with its α amino group protected by a Fmoc group bound to resin beads with an acid-labile super acid-sensitive resin linker (47, 48). The ϵ-amine was protected with a Mtt group, which was removed under weakly acidic conditions at the final release (Fig. 4A). The synthesis proceeded in a stepwise fashion with coupling of 3-hydroxybutyric acid, orthogonally protected lysine reagent, and 2-cis-eicosenoic acid (C20). The oxazoline formed between salicylic acid and α-methylserine, being accomplished by forming an activated derivative of carboxylic acid and allowing it to react with the NH2 group on the growing chain. Each coupling step was followed by washing by aprotic solvents and deprotection, as needed, prior to the next coupling step as summarized in Fig. 4A. Similar coupling methods were used to cyclize the C-terminal lysine after release.
Nanoelectrospray mass spectrometry confirmed the masses of key synthesis intermediates (supplemental Fig. S8), and HPLC analysis of the final product yielded two peaks (Fig. 3B), one of which contained material with the expected mass (m/z 838.5) and showed a CID-MS spectrum (Fig. 4) and NMR spectra (supplemental Fig. S7) that matched those of natural DDM-838. Importantly, the synthetic DDM-838 stimulated CD1a-restricted T cells with a potency comparable with M. tuberculosis derived DDM-838 (Fig. 4D), consistent with the conclusion that they were identical molecules. Further, recognition of the S-α-methylserine containing molecule provided evidence that this amino acid was present in natural molecules and important for the response. Also, because the yield from a single synthesis exceeded that recoverable from large mycobacterial preparations by ∼100-fold, this biologically active product is of adequate yield for T cell studies in human populations.
Stereocenters Determine the T Cell Response
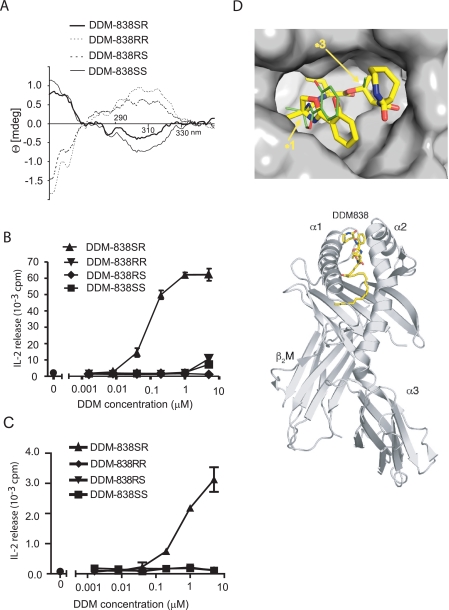
DDM contains four chiral centers: two in each of the lysine moieties, one in the oxazoline ring, and one in the butyrate moiety (designated *1, *2, *3, and [i*4 in Fig. 1). The lysines were assumed to correspond to the naturally occurring L-form. However, the absolute stereochemistry of the α-methylserine and butyrate moieties were not known from prior studies and could not be directly established from trace amounts of DDM available from M. tuberculosis. Analysis of these stereocenters is important for the design of mimics of natural products and provided the opportunity to determine the degree to which T cell recognition of lipopeptides is controlled by the fine structure of the peptide. The solid phase method allowed a modular synthesis of four distinct compounds made using salicyl oxazoline and hydroxybutanoic acid as either R- or S-stereoisomers. Accordingly, we synthesized DDM-838 in four forms that are named according to the R- or S-stereochemistry of the α-methylserine (*1) and butyrate groups (*3), respectively: DDM-838SR, DDM-838RS, DDM-838SS, and DDM-838RR (Fig. 1).
The four DDM-838 compounds were proposed to be pure diastereomers based on the use of optically pure α-methylserine or butyrate precursors and the assumption that these organic acids were not racemized during coupling and isolation of the final DDM product. To test this assumption, the putative DDM-838SR, DDM-838RS, DDM-838SS, and DDM-838RR were subjected to chemical and immunological analysis. MS analysis of [M + Na]+ ions showed that the cationized species had the expected mass (m/z 860.6) and gave nearly indistinguishable CID mass spectra (supplemental Fig. S9). In particular, the signal intensities of the fragment ions at m/z 830, 664, and 634, which correspond to fragments generated by cleavage within or adjacent to the methyl-serine and butyrate moieties, were not affected by the differing stereochemical configuration. However, because the oxazoline ring is part of the DDM chromophore, compounds with differing stereochemistry of methylserine would be expected to exhibit opposite polarity in the CD spectrum. This was indeed the case for the oxazoline precursors (supplemental Fig. S2) and the products (Fig. 5A), confirming the maintenance of chirality during synthesis.
FIGURE 5.
Stereocenters near each end of the peptide moiety control T cell activation. A, circular dichroism spectra acquired for the DDM synthetic analogs show a polar difference for compounds that were predicted to vary in stereochemistry in the oxazoline moiety at the *1 stereocenter, which is part of the oxazoline ring chromophore, confirming that input compounds (supplemental Fig. S9) were not racemized during synthesis. B and C, T cell responses to DDM-838 stereoisomers were compared in assays using intact dendritic cells expressing CD1a (B) or recombinant CD1a proteins bound to plastic plates (C). D, based on a prior crystal structure of CD1a bound to a DDM-like synthetic compound (21), this molecular model was prepared with a molecular surface corresponding to DDM-838SR. Top (upper panel) and side (lower panel) views show the predicted position of DDM-838SR in the CD1a groove with arrows indicating the predicted locations of the stereocenters that affect T cell response. The binding groove is depicted as a molecular surface with negative (red) and positive (blue) electrostatic potentials. (The alternate DDM-838RS configuration was also modeled and is shown as thin green lines.)
Compared with the most active antigenic compound, DDM-838SR, the other compounds with alterations at either stereocenter showed a 1000-fold or greater loss of T cell activation when tested by incubating antigen with T cells and CD1a-expressing antigen-presenting cells (Fig. 5B). The significant effect of slight alterations in peptide structure on T cell activation might be due to alteration of the final structure of CD1a-DDM complexes or due to an effect of altered antigen binding on uptake, transport, or antigen processing by antigen-presenting cells. The results obtained when these T cells were stimulated by DDM incubated with recombinant CD1a-Fc proteins bound to plastic plates (Fig. 5C) indicate that the differing responses were due to CD1a-lipopeptide-T cell interactions, rather than the influence of the lipopeptidic structure on interactions with antigen-presenting cells. Also DDM-838SS inhibits the T cell response to DDM-838SR presented on recombinant CD1a proteins, indicating that this molecule binds to CD1a at or near groove (supplemental Fig. S10A). Inhibition was not seen with a structurally unrelated lipid (glucopsychosine). The titer and extent of inhibition was similar to that of the known CD1a-presented ligand, sulfatide, and is similar to the recently observed effects of CD1c-presented ligands (supplemental Fig. S10, B and C) (49).
Based on a prior co-crystal structure of CD1a bound to a DDM-like analog (21), we prepared a computer model of CD1a bound to DDM-838SR and its stereoisomers for comparison (Fig. 5D). This model shows that the *1 and *3 stereocenters are predicted to lie near the opening of the CD1a groove. Their absolute stereochemical configuration is predicted to affect the orientation of the salicylate and oxazoline and, to a lesser extent, the terminal lysine moieties located near the predicted plane of TCR contact. This computer model provides a specific and plausible mechanism by which such subtle chemical changes in the peptide backbone of DDM could substantially influence the final shape of the CD1a-DDM complex at the site of its predicted interaction with the TCR. Because subtle structural changes at either end of the peptide abrogate T cell recognition, this suggests that the TCR might interact with both arms of the peptide, which are anchored at the center by the lipid-modified lysine. It will be of interest to determine whether these altered lipopeptide ligands have anergy inducing properties, as has been previously shown for peptide mimics of major histocompatibility complex ligands.
DISCUSSION
The synthesis of DDM analogs provides new insight into structures of mycobactin-like compounds and new reagents for immunological studies of CD1a function in humans. After natural products were unavailable in sufficient yield for complete solution of the structure, synthesis of DDM standards allowed a basis for comparison, which verified the presence of the unusual methylserine unit and solved other stereochemical centers in the peptide. Thus, synthesis informed the analytical chemical analysis.
In considering possible differences in the immunological functions of T cells that recognize CD1 or major histocompatibility complex proteins, the biochemical nature of the antigens presented may give some insight. Whereas peptides are highly mutable gene products, lipids are produced by multi-enzyme cascades that cannot be re-engineered in structure and still retain function during the short time course of an epidemic. Also, many of the bacterial antigens recognized by CD1, including DDM, mannosyl phosphomycoketides, and sulfated glycolipids, are lipids that confer increased bacterial virulence by mechanisms unrelated to their capture by CD1. These observations raise the hypothesis that CD1 functions as a failsafe immune recognition mechanism to present antigens that are stable in structure but not dispensable for successful infection.
DDM represents a clear example of this because it participates in the only known iron scavenging mechanism in M. tuberculosis. Prior studies show that alteration of a single stereocenter in mycobactin strongly influences iron acquisition and utilization properties, converting it from a molecule that promotes growth to one that inhibits growth (23). Here we show that CD1a-restricted T cells can precisely scan the part of the peptide structure responsible for iron chelation. Together, these studies show in a detailed way how the immunological specificity for the molecule is related to the same aspects of its chemical substructure that allow this molecule to function as a virulence factor (13, 50). Further, highly precise recognition of only natural DDMs support the hypothesis that this is a natural target of the immune response, and this result indicates that T cell activation requires both arms of the peptide.
Last, production of bioactive DDM overcomes a longstanding hurdle for initiating clinical studies of the function of CD1a in infection. The functions of CD1d and NK T cells have been extensively studied in mouse and human models of infection, autoimmunity, and allergy (51). However, foreign antigens for CD1a have not been recoverable from natural sources in large amounts. CD1a is a prominent, even defining cell surface protein for Langerhans cells. In vitro studies strongly suggest that CD1a activates T cells, but basic information about the function of CD1a-dependent T cells in disease is unknown. Generation of a synthetic ligand for CD1a now allows studies of the abundance and phenotype of such T cells in tuberculosis patients. Studies of such major histocompatibility complex-independent T cell responses in humans might lead to new tests for prior infection or subunits for vaccine formulations.
Supplementary Material
Acknowledgments
We thank Michael Brenner and Manuela Cernadas for providing CD1a-Ig protein and advice. We also appreciate the work of Jaroslav Zajicek with the NMR studies and Shiuyung Chan for assistance with derivatization chemistry.
This work was supported, in whole or in part, by National Institutes of Health Grants AI054193, AR 046832, AI 049313, and AI 074952. This work was also supported by a Cancer Research Institute Investigator Award (LIAI), National Center for Research Resources Grant P41-RR10888, and funds from the Howard Hughes Medical Institute Kwa-Zulu Natal Research Institute for Tuberculosis and HIV (K-RITH).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10.
- DDM
- dideoxymycobactin
- HPLC
- high performance liquid chromatography
- MS
- mass spectrometry
- CID
- collision-induced dissociation
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrocloride
- HOAt
- 1-hydroxy-7-aza-benzotriazole
- Fmoc
- 9-fluorenylmethoxycarbonyl
- Mtt
- 4-methyltrityl
- HATU
- 2-(7-aza-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- HBTU
- 2-(benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- HBOt
- 1-hydroxy-benzotriazole
- Boc
- t-butoxycarbonyl
- Dde
- 1-(4,4-dimethyl-2,6- dioxocyclohexylidene)ethyl.
REFERENCES
- 1.Mellman I., Steinman R. M. (2001) Cell 106, 255–258 [DOI] [PubMed] [Google Scholar]
- 2.De Libero G., Mori L. (2005) Nat. Rev. Immunol. 5, 485–496 [DOI] [PubMed] [Google Scholar]
- 3.Young D. C., Moody D. B. (2006) Glycobiology 16, 103R–112R [DOI] [PubMed] [Google Scholar]
- 4.Hunger R. E., Sieling P. A., Ochoa M. T., Sugaya M., Burdick A. E., Rea T. H., Brennan P. J., Belisle J. T., Blauvelt A., Porcelli S. A., Modlin R. L. (2004) J. Clin. Invest. 113, 701–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosat J. P., Grant E. P., Beckman E. M., Dascher C. C., Sieling P. A., Frederique D., Modlin R. L., Porcelli S. A., Furlong S. T., Brenner M. B. (1999) J. Immunol. 162, 366–371 [PubMed] [Google Scholar]
- 6.Porcelli S., Brenner M. B., Greenstein J. L., Balk S. P., Terhorst C., Bleicher P. A. (1989) Nature 341, 447–450 [DOI] [PubMed] [Google Scholar]
- 7.Shamshiev A., Gober H. J., Donda A., Mazorra Z., Mori L., De Libero G. (2002) J. Exp. Med. 195, 1013–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krutzik S. R., Tan B., Li H., Ochoa M. T., Liu P. T., Sharfstein S. E., Graeber T. G., Sieling P. A., Liu Y. J., Rea T. H., Bloom B. R., Modlin R. L. (2005) Nat. Med. 11, 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roura-Mir C., Catálfamo M., Cheng T. Y., Marqusee E., Besra G. S., Jaraquemada D., Moody D. B. (2005) J. Immunol. 174, 3773–3780 [DOI] [PubMed] [Google Scholar]
- 10.Sieling P. A., Ochoa M. T., Jullien D., Leslie D. S., Sabet S., Rosat J. P., Burdick A. E., Rea T. H., Brenner M. B., Porcelli S. A., Modlin R. L. (2000) J. Immunol. 164, 4790–4796 [DOI] [PubMed] [Google Scholar]
- 11.Moody D. B., Young D. C., Cheng T. Y., Rosat J. P., Roura-Mir C., O'Connor P. B., Zajonc D. M., Walz A., Miller M. J., Levery S. B., Wilson I. A., Costello C. E., Brenner M. B. (2004) Science 303, 527–531 [DOI] [PubMed] [Google Scholar]
- 12.Dye C., Watt C. J., Bleed D. M., Hosseini S. M., Raviglione M. C. (2005) J. Am. Med. Assoc. 293, 2767–2775 [DOI] [PubMed] [Google Scholar]
- 13.De Voss J. J., Rutter K., Schroeder B. G., Su H., Zhu Y., Barry C. E., 3rd (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1252–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snow G. A. (1970) Bacteriol. Rev. 34, 99–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ratledge C. (2004) Tuberculosis 84, 110–130 [DOI] [PubMed] [Google Scholar]
- 16.Ratledge C., Dover L. G. (2000) Annu. Rev. Microbiol. 54, 881–941 [DOI] [PubMed] [Google Scholar]
- 17.Abergel R. J., Wilson M. K., Arceneaux J. E., Hoette T. M., Strong R. K., Byers B. R., Raymond K. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18499–18503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., Akira S., Aderem A. (2004) Nature 432, 917–921 [DOI] [PubMed] [Google Scholar]
- 19.Goetz D. H., Holmes M. A., Borregaard N., Bluhm M. E., Raymond K. N., Strong R. K. (2002) Mol. Cell 10, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 20.Holmes M. A., Paulsene W., Jide X., Ratledge C., Strong R. K. (2005) Structure 13, 29–41 [DOI] [PubMed] [Google Scholar]
- 21.Zajonc D. M., Crispin M. D., Bowden T. A., Young D. C., Cheng T. Y., Hu J., Costello C. E., Rudd P. M., Dwek R. A., Miller M. J., Brenner M. B., Moody D. B., Wilson I. A. (2005) Immunity 22, 209–219 [DOI] [PubMed] [Google Scholar]
- 22.Kaiser F. E., Gehrke C. W., Zumwalt R. W., Kuo K. C. (1974) J. Chromatogr. 94, 113–133 [DOI] [PubMed] [Google Scholar]
- 23.Hu J., Miller M. J. (1997) J. Am. Chem. Soc. 119, 3462–3468 [Google Scholar]
- 24.Maurer P. J., Miller M. J. (1981) J. Org. Chem. 46, 2835–2836 [Google Scholar]
- 25.Maurer P. J., Miller M. J. (1983) J. Am. Chem. Soc. 105, 240–245 [Google Scholar]
- 26.Walz A. J., Möllmann U., Miller M. J. (2007) Org Biomol. Chem. 5, 1621–1628 [DOI] [PubMed] [Google Scholar]
- 27.Xu Y., Miller M. J. (1998) J. Org. Chem. 63, 4314–4322 [Google Scholar]
- 28.Bijvoet J. M., Peerdeman A. F., Bommel A. J. (1951) Nature 168, 271–272 [Google Scholar]
- 29.Serck-Hanssen K., Stallberg-Stenhagen S., Stenhagen E. (1953) Arkiv. Kemi. 5, 203–221 [Google Scholar]
- 30.Peerdeman A. F., van Bommel A. J., Bijvoet J. M. (1951) Proc. R. Acad. Amsterdam 54, 16–19 [Google Scholar]
- 31.Wadsworth W. S., Jr., Emmons W. D. (1961) J. Am. Chem. Soc. 83, 1733–1738 [Google Scholar]
- 32.Still W. C., Gennari C. (1983) Tetrahedron Lett. 24, 4405–4408 [Google Scholar]
- 33.Phillips A. J., Uto Y., Wipf P., Reno M. J., Williams D. R. (2000) Org. Lett. 2, 1165–1168 [DOI] [PubMed] [Google Scholar]
- 34.Moody D. B., Guy M. R., Grant E., Cheng T. Y., Brenner M. B., Besra G. S., Porcelli S. A. (2000) J. Exp. Med. 192, 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gumperz J. E., Roy C., Makowska A., Lum D., Sugita M., Podrebarac T., Koezuka Y., Porcelli S. A., Cardell S., Brenner M. B., Behar S. M. (2000) Immunity 12, 211–221 [DOI] [PubMed] [Google Scholar]
- 36.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snow G. A., White A. J. (1969) Biochem. J. 115, 1031–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greatbanks D., Bedford G. R. (1969) Biochem. J. 115, 1047–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flynn E. H., Hinman J. W., Caron E. L., Woolf D. O. (1953) J. Am. Chem. Soc. 75, 5867–5871 [Google Scholar]
- 40.Takamura N., Terashima S., Achiwa K., Yamada S. (1967) Chem. Pharm. Bull. 15, 1776–1784 [DOI] [PubMed] [Google Scholar]
- 41.Tsukamoto M., Murooka K., Nakajima S., Abe S., Suzuki H., Hirano K., Kondo H., Kojiri K., Suda H. (1997) J. Antibiot. 50, 815–821 [DOI] [PubMed] [Google Scholar]
- 42.Matsutani H., Setogawa K., Wakamiya T., Kobayashi Y., Oda Y., Shiba T. (1988) Phytochemistry 27, 931–932 [Google Scholar]
- 43.Chang C. D., Meienhofer J. (1978) Int. J. Pept. Protein Res. 11, 246–249 [DOI] [PubMed] [Google Scholar]
- 44.Fields G. B., Noble R. L. (1990) Int. J. Pept. Protein Res. 35, 161–214 [DOI] [PubMed] [Google Scholar]
- 45.Merrifield R. B. (1964) Biochemistry 3, 1385–1390 [DOI] [PubMed] [Google Scholar]
- 46.Poreddy A. R., Schall O. F., Marshall G. R., Ratledge C., Slomczynska U. (2003) Bioorg. Med. Chem. Lett. 13, 2553–2556 [DOI] [PubMed] [Google Scholar]
- 47.Mergler M., Tanner R., Gosteli J., Grogg P. (1988) Tetrahedron Lett. 29, 4005–4008 [Google Scholar]
- 48.Mergler M., Tanner R., Gosteli J., Grogg P. (1988) Tetrahedron Lett. 29, 4009–4012 [Google Scholar]
- 49.Van Rhijn I., Young D. C., De Jong A., Vazquez J., Cheng T. Y., Talekar R., Barral D. C., León L., Brenner M. B., Katz J. T., Riese R., Ruprecht R. M., O'Connor P. B., Costello C. E., Porcelli S. A., Briken V., Moody D. B. (2009) J. Exp. Med. 206, 1409–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quadri L. E., Sello J., Keating T. A., Weinreb P. H., Walsh C. T. (1998) Chem. Biol. 5, 631–645 [DOI] [PubMed] [Google Scholar]
- 51.Tupin E., Kinjo Y., Kronenberg M. (2007) Nat. Rev. Microbiol. 5, 405–417 [DOI] [PubMed] [Google Scholar]
- 52.Leimer K. R., Rice R. H., Gehrke C. W. (1977) J. Chromatogr. 141, 121–144 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.