Abstract
We have developed a system to reconstitute all of the proposed steps of Okazaki fragment processing using purified yeast proteins and model substrates. DNA polymerase δ was shown to extend an upstream fragment to displace a downstream fragment into a flap. In most cases, the flap was removed by flap endonuclease 1 (FEN1), in a reaction required to remove initiator RNA in vivo. The nick left after flap removal could be sealed by DNA ligase I to complete fragment joining. An alternative pathway involving FEN1 and the nuclease/helicase Dna2 has been proposed for flaps that become long enough to bind replication protein A (RPA). RPA binding can inhibit FEN1, but Dna2 can shorten RPA-bound flaps so that RPA dissociates. Recent reconstitution results indicated that Pif1 helicase, a known component of fragment processing, accelerated flap displacement, allowing the inhibitory action of RPA. In results presented here, Pif1 promoted DNA polymerase δ to displace strands that achieve a length to bind RPA, but also to be Dna2 substrates. Significantly, RPA binding to long flaps inhibited the formation of the final ligation products in the reconstituted system without Dna2. However, Dna2 reversed that inhibition to restore efficient ligation. These results suggest that the two-nuclease pathway is employed in cells to process long flap intermediates promoted by Pif1.
Eukaryotic cellular DNA is replicated semi-conservatively in the 5′ to 3′ direction. A leading strand is synthesized by DNA polymerase ϵ in a continuous manner in the direction of opening of the replication fork (1, 2). A lagging strand is synthesized by DNA polymerase δ (pol δ)3 in the opposite direction in a discontinuous manner, producing segments called Okazaki fragments (3). These stretches of ∼150 nucleotides (nt) must be joined together to create the continuous daughter strand. DNA polymerase α/primase (pol α) initiates each fragment by synthesizing an RNA/DNA primer consisting of ∼1-nt of RNA and ∼10–20 nt of DNA (4). The sliding clamp proliferating cell nuclear antigen (PCNA) is loaded on the DNA by replication factor C (RFC). pol δ then complexes with PCNA and extends the primer. When pol δ reaches the 5′-end of the downstream Okazaki fragment, it displaces the end into a flap while continuing synthesis, a process known as strand displacement (5, 6). These flap intermediates are cleaved by nucleases to produce a nick for DNA ligase I (LigI) to seal, completing the DNA strand.
In one proposed mechanism for flap processing, the only required nuclease is flap endonuclease 1 (FEN1). pol δ displaces relatively short flaps, which are cleaved by FEN1 as they are created, leaving a nick for LigI (7–9). FEN1 binds at the 5′-end of the flap and tracks down the flap cleaving only at the base (5, 10, 11). Because pol δ favors the displacement of RNA-DNA hybrids over DNA-DNA hybrids, strand displacement generally is limited to that of the initiator RNA of an Okazaki fragment (12). In addition, the tightly coordinated action of pol δ and FEN1 also tends to keep flaps short. However, biochemical reconstitution studies demonstrate that some flaps can become long (13, 14). Once these flaps reach ∼30 nt, they can be bound by the eukaryotic single strand binding protein replication protein A (RPA) (15). Binding by RPA to a flap substrate inhibits cleavage by FEN1 (16). The RPA-bound flap would then require another mechanism for proper processing.
This second mechanism is proposed to utilize Dna2 (16) in addition to FEN1. Dna2 is both a 5′-3′ helicase and an endonuclease (17, 18). Like FEN1, Dna2 recognizes 5′-flap structures, binding at the 5′-end of the flap and tracking downward toward the base (19, 20). Unlike FEN1, Dna2 cleaves the flap multiple times but not all the way to the base, such that a short flap remains (20). RPA binding to a flap has been shown to stimulate Dna2 cleavage (16). Therefore, if a flap becomes long enough to bind RPA, Dna2 binds and cleaves it to a length of 5–10 nucleotides from which RPA dissociates (21). FEN1 can then enter the flap, displace the Dna2, and then cleave at the base to make the nick for ligation (16, 18, 22). The need for this mechanism may be one reason why DNA2 is an essential gene in Saccharomyces cerevisiae (23, 24). It has been proposed that, in the absence of Dna2, flaps that become long enough to bind RPA cannot be properly processed, leading to genomic instability and cell death (23).
In reconstitution of Okazaki fragment processing with purified proteins, even though some flaps became long enough to bind RPA, FEN1 was very effective at cleaving essentially all of the generated flaps (13, 14). Evidently, FEN1 could engage the flaps before binding of RPA. However, these reconstitution assays did not include the 5′-3′ helicase Pif1 (25, 26). Pif1 is involved in telomeric and mitochondrial DNA maintenance (26) and was first implicated in Okazaki fragment processing from genetic studies in S. cerevisiae. Deletion of PIF1 rescued the lethality of dna2Δ, although the double mutant was still temperature-sensitive (27). The authors of this report proposed that Pif1 creates a need for Dna2 by promoting longer flaps. Further supporting this conclusion, deletion of POL32, which encodes the subunit of pol δ that interacts with PCNA, rescued the temperature sensitivity of the dna2Δpif1Δ double mutant (12, 27). Importantly, pol δ exhibited reduced strand displacement activity when POL32 was deleted (12, 28, 29). The combination of pif1Δ and pol32Δ is believed to create a situation in which virtually no long flaps are formed, eliminating the requirement for Dna2 flap cleavage (27).
We recently performed reconstitution assays showing that Pif1 can assist in the creation of long flaps. Inclusion of Pif1, in the absence of RPA, increased the proportion of flaps that lengthened to ∼28–32 nt before FEN1 cleavage (14). With the addition of RPA, the appearance of these long flap cleavage products was suppressed. Evidently, Pif1 promoted such rapid flap lengthening that RPA bound some flaps before FEN1 and inhibited cleavage. The RPA-bound flaps would presumably require cleavage by Dna2 for proper processing.
Only a small fraction of flaps became long with Pif1. However, there are hundreds of thousands of Okazaki fragments processed per replication cycle (30). Therefore, thousands of flaps are expected to be lengthened by Pif1 in vivo, a number significant enough that improper processing of such flaps could lead to cell death.
Our goal here was to determine whether Pif1 can influence the flow of Okazaki fragments through the two proposed pathways. We first questioned whether Pif1 stimulates strand displacement synthesis by pol δ. Next, we asked whether Pif1 lengthens short flaps so that Dna2 can bind and cleave. Finally, we used a complete reconstitution system to determine whether Pif1 promotes creation of RPA-bound flaps that require cleavage by both Dna2 and FEN1 before they can be ligated. Our results suggest that Pif1 promotes the two-nuclease pathway, and reveal the mechanisms involved.
EXPERIMENTAL PROCEDURES
Materials
Radioactive nucleotides [γ-32P]ATP, [α-32P]dCTP, and [α-32P]dGTP were obtained from PerkinElmer Life Sciences. Oligonucleotide primers were synthesized by Midland Certified Reagents Co. (Midland, TX) or Integrated DNA Technologies (Coralville, IA). Primer sequences are listed in Table 1. Polynucleotide kinase, Klenow fragment, and streptavidin were obtained from Roche Applied Science. Other reagents were the best grade commercially available.
TABLE 1.
Oligonucleotide sequences
| Primer | Length | Sequence |
|---|---|---|
| nt | ||
| Upstream (5′–3′) | ||
| U1 | 20 | GTCCACCCGACGCCACCTCC |
| U2 | 44 | GTCCACCCGACGCCACCTCCTGCCTTCAATGTGCTGGGATCCTA |
| Downstream (5′–3′) | ||
| D1 | 29 | ACCGTGCCAGCCTAAATTTCAATCCACCC |
| D2 | 60 | AGACGAATTCCGGATACGACGGCCAGTGCCGACCGTGCCAGCCTAAATTTCAATCCACCC |
| D3 | 90 | TTCTACTTCCAATTGATACGCGCTATAACCAGACGAATTCCGGATACGACGGCCAGTGCCGACCGTGCCAGCCTAAATTTCAATCCACCC |
| D4 | 95 | TTCTACTTCCAATTGATACGCGCTATAACCAGACGAATTCCGGATACGACGGCCAGTGCCGACCGTGCCAGCCTAAATTTCAATCCACCCTGACT |
| Template (3′–5′) | ||
| T1 | 42 | CAGGTGGGCTGCGGTGGAGGGTCGGATTTAAAGTTAGGTGGG |
| T2a | 110 | CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACGACCCTAGGATGTTGGTTCTGCTTAAGGCCTATGCTGCCGGTCACGGCTGGCACGGTCGGATTTAAAGTTAGGTGGG |
a Template T2 is tinylated at both the 5′ and 3′ ends.
Enzyme Expression and Purification
S. cerevisiae wild-type pol δ was overexpressed and purified from S. cerevisiae as previously described (28). S. cerevisiae RFC was overexpressed and purified from Escherichia coli as previously described (31). S. cerevisiae Rad27 (32) (FEN1) and PCNA (13) were cloned into the T7 expression vector pET-24b (Novagen/EMD Biosciences, Madison, WI), expressed in E. coli BL21(DE3) codon plus strain (Stratagene and Novagen/EMD Biosciences, respectively) and purified as previously described. S. cerevisiae Pif1 was cloned into the pET-28b bacterial expression vector (Novagen/EMD Biosciences), expressed in the E. coli Rosetta strain (Novagen/EMD Biosciences), and purified as previously described (33). S. cerevisiae RPA was overexpressed and purified from E. coli as previously described (34). S. cerevisiae Dna2 was overexpressed and purified from baculovirus High Five cells as previously described (23). S. cerevisiae LigI was overexpressed and purified from S. cerevisiae as previously described (7).
Oligonucleotide Substrates
The oligonucleotide primers were used to design substrates that simulate Okazaki fragment processing intermediates. Primers U2, D1, and D2 were radiolabeled at the 5′-end with [γ-32P]ATP using polynucleotide kinase. Primers D2 and D3 were annealed at the 3′-end to a 20-nt labeling template with a 5′-GCTA overhang and radiolabeled with [α-32P]dCTP using Klenow polymerase. Radiolabeled primers were separated by electrophoresis on a 15%/7 m urea polyacrylamide gel and then gel-purified. Substrates were then created by annealing primer components in annealing buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm dithiothreitol), heating at 95 °C for 5 min, transferring to 70 °C, and slowly cooling to room temperature. When the upstream primer was labeled, the annealing ratio was 1:2:4 of upstream primer to template to downstream primer. When the downstream primer was labeled, the annealing ratio was 1:2:4 of downstream primer to template to upstream primer.
The first substrate was designed to examine cleavage by FEN1 and Dna2 and ligation by LigI following strand displacement by pol δ. This substrate consists of a 44-nt upstream primer (U2) (see Table 1) and a 60-nt downstream primer (D2) annealed to the 3′- and 5′-ends, respectively, of a 110-nt template (T2) and is referred to as the standard substrate in the text. The second substrate was designed to examine cleavage by Dna2 on a short flap in the absence and presence of Pif1. This substrate consists of a 2-nt upstream primer (U1) and a 29-nt downstream primer (D1) annealed to the 3′- and 5′-ends, respectively, of a 42-nt template (T1). This creates a fixed, double-flap substrate with a 6-nt 5′-flap and a 1-nt 3′-flap, which is referred to as the 6-nt flap substrate in the text. The third substrate was designed to examine cleavage by FEN1 and Dna2 and ligation by LigI during strand displacement in the presence of a 3-nt pre-created flap. This consists of a 44-nt upstream primer (U2) and a 9-nt downstream primer (D3) annealed to the 3′- and 5′-ends, respectively, of a 11-nt template (T2). The 3′-end of the downstream primer anneals to only 6-nt on the template and contains a 3-nt unannealed region, creating the flap. This substrate simulates an intermediate created during strand displacement of the standard substrate and is referred to as the pre-created flap substrate in the text. The fourth substrate was designed to examine synthesis by pol δ and ligation by LigI. This substrate consists of a 44-nt upstream primer (U2) and a 95-nt downstream primer (D4) annealed to the 3′- and 5′-ends, respectively, of a 11-nt template (T2), creating a substrate identical to the pre-created flap substrate, except the downstream primer is extended by 5 nt at the 3′-end. This substrate is referred to as the extended pre-created flap substrate in the text. Specific substrates used in each figure are indicated in the legends and pictured above the figures. The location of the radiolabel (either 5′ or 3′) on either the upstream or downstream primer is indicated by an asterisk in the respective figures.
Enzyme Assays
Prior to starting strand-displacement reactions, 5 fmol of radiolabeled biotinylated substrate was incubated on ice with 500 fmol of streptavidin for 20 min. Streptavidin complexes with biotin on the substrate template, effectively blocking the ends of the substrate and requiring that RFC load PCNA onto the substrate. The blocked ends are not depicted in the figures. Substrate was then incubated with various amounts of pol δ, PCNA, RFC, FEN1, Dna2, LigI, RPA, and Pif1 for 10 min at 30 °C in a total volume of 20 μl. The reaction buffer contained 50 mm Tris-HCl, pH 7.5, 2 mm dithiothreitol, 25 μg/ml bovine serum albumin, 0.5 mm MgCl2, 1 mm ATP, 50 μm of each dNTP, and 75 mm NaCl. Reactions were stopped by adding 20 μl of 2× termination dye (90% formamide (v/v), 10 mm EDTA, and 0.01% bromphenol blue, and xylene cyanole), followed by heating at 95 °C for 5 min. Products were separated by electrophoresis on a 22.5%/7 m urea polyacrylamide gel for 1 h and 20 min at 85 watts. The gel was placed on filter paper and dried on a gel drier (Bio-Rad). Dried gels were exposed to a phosphor screen, which was scanned using an Amersham Biosciences PhosphorImager and analyzed with ImageQuant version 1.2 software.
For the fixed, double-flap cleavage reactions, 5 fmol of radiolabeled 6-nt flap substrate was incubated with 500 fmol of streptavidin for 20 min on ice prior to starting each reaction. Since free streptavidin may have had an effect on the reaction, streptavidin was included even though the substrate was not biotinylated. Substrate was incubated with Dna2 and various amounts of Pif1 for 10 min at 30 °C in a total volume of 20 μl. The reaction buffer was the same as above. Reactions were stopped, separated by electrophoresis, and analyzed as described above.
For reactions shown in Fig. 6B, 5 fmol of unlabeled extended pre-created flap substrate was incubated with 500 fmol of streptavidin for 20 min on ice prior to starting each reaction. Substrate was incubated with pol δ, PCNA, RFC, FEN1, Dna2, LigI, and RPA for 10 min at 30 °C in a total volume of 20 μl. For this experiment, the reaction buffer contained 50 mm Tris-HCl, pH 7.5, 2 mm dithiothreitol, 25 μg/ml bovine serum albumin, 0.5 mm MgCl2, 1 mm ATP, 20 μm dTTP, dCTP, and dATP, 0.5 μm [α-32P]dGTP, and 75 mm NaCl. Reactions were then stopped as described above and passed through Micro Bio-Spin 30 chromatography columns (Bio-Rad) to remove unincorporated [α-32P]dGTP. 20 μl of 2× termination dye was added, and reactions were incubated for 3 min at 95 °C, separated by electrophoresis, and then analyzed as described above.
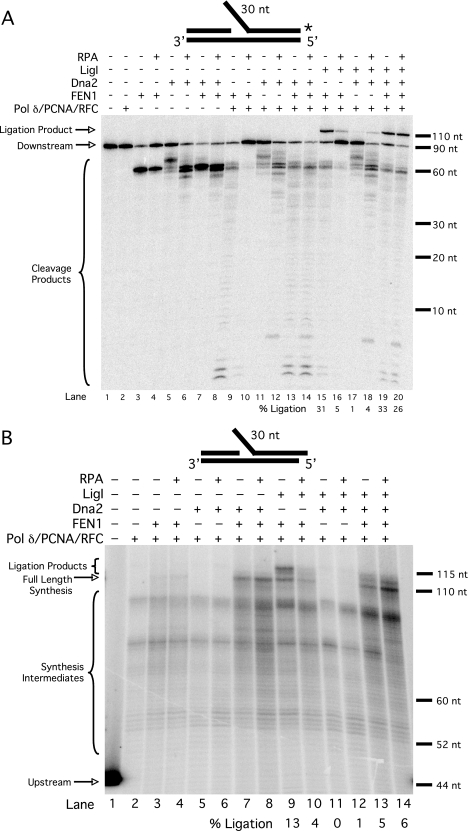
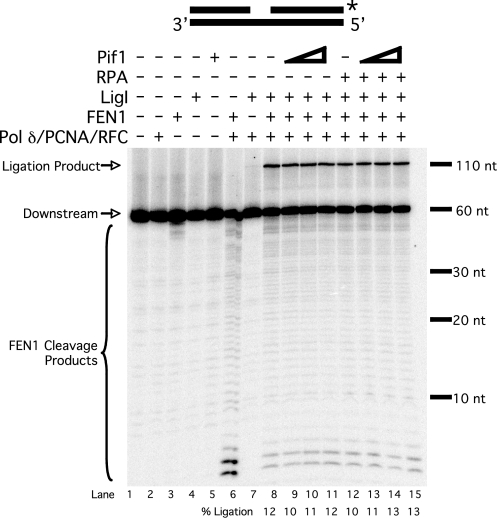
FIGURE 6.
Dna2 rescues RPA-inhibited ligation of long flaps. A, cleavage by FEN1 (5 fmol) and Dna2 (50 fmol) and ligation by LigI (25 fmol) were assayed on the pre-created flap substrate (U2:T2:D3) in the presence of various combinations of pol δ (23 fmol), PCNA (25 fmol), RFC (25 fmol), and RPA (200 fmol) as indicated in the figure and as described under “Experimental Procedures.” B, synthesis by pol δ (23 fmol) and ligation by LigI (25 fmol) were assayed on unlabeled extended pre-created flap substrate (U2:T2:D4) in the presence of 0.5 μm [α-32P]dGTP and various combinations of PCNA (25 fmol), RFC (25 fmol), FEN1 (5 fmol), Dna2 (50 fmol), and RPA (200 fmol) as indicated in the figure and as described under “Experimental Procedures.” Lane 1 contains upstream primer (U2) radiolabeled at the 5′-end. Substrate depictions and designations are as in Fig. 1.
The amount of each protein used in each experiment is given in the appropriate figure legend. All enzyme assays were repeated at least in triplicate with a representative gel shown in each figure.
RESULTS
Reconstitution of Okazaki fragment processing showed that a distinct portion of flaps displaced in the presence of Pif1 bound sufficient RPA to inhibit FEN1 (14). Experiments presented here are designed to test the hypothesis that inclusion of Dna2 compensates for Pif1, improving both flap cleavage and the ultimate ligation to form a continuous strand.
Pif1 Stimulates Synthesis by pol δ
We showed previously that Pif1 lengthens the distribution of flaps displaced by pol δ during strand displacement synthesis, and these flaps can be cleaved by FEN1. It has also been shown that FEN1 activity permits pol δ to displace the downstream strand more effectively (7, 13). Here we asked whether inclusion of Pif1, in the absence of FEN1, stimulates upstream primer elongation further into the annealed region of the downstream primer (Fig. 1). Such a result would be consistent with the interpretation that Pif1 promotes strand displacement synthesis by facilitating unwinding of the downstream primer.
FIGURE 1.
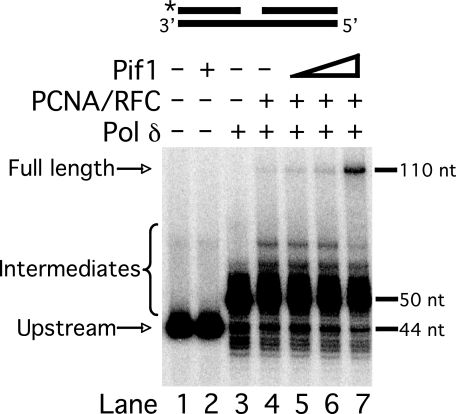
Pif1 stimulates strand displacement synthesis by pol δ. Strand displacement synthesis by pol δ (23 fmol) was assayed on the standard substrate (U2:T2:D2) in the presence of various combinations of PCNA (25 fmol), RFC (25 fmol), and varying amounts of Pif1 (25, 50, or 100 fmol) as indicated in the figure and as described under “Experimental Procedures.” The substrate is depicted above the figure, with the location of the radiolabel indicated by the asterisk. Plus (+) denotes presence and minus (−) denotes absence of the given enzyme.
The upstream primer of the standard substrate was radiolabeled at the 5′-end for visualization of synthesis products. pol δ alone added ∼6 nt efficiently, which represents synthesis through the gap, but was ineffective at subsequent strand displacement (lane 3). When PCNA and RFC were included in the reaction (lane 4), pol δ partially displaced the downstream primer, extending the upstream primer to more than 5-nt but less than the full-length 110 nt. Synthesis to the maximum length is indicative of complete displacement of the downstream primer. When Pif1 was titrated into the reaction (lanes 5–7), pol δ was stimulated to synthesize substantial full-length product. Notably, the bands representing the intermediate length primer extension faded while the band of full-length extension intensified. This indicates that most full-length product was produced by further strand displacement synthesis past the 5-nt pause point. Evidently, Pif1 bound and lengthened short flaps already displaced by pol δ. A small fraction of flaps were lengthened by Pif1, consistent with our previous results (14). Overall, these results suggest that pol δ displaced short flaps, allowing binding of Pif1. Then the polymerase and helicase worked together for further displacement.
Pif1 Permits Dna2 Cleavage by Lengthening Short Flaps
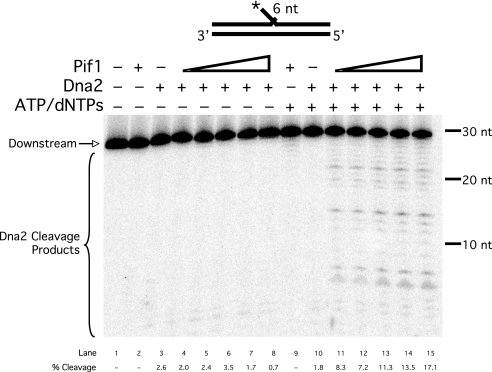
A key aspect of our proposed mechanism is that Pif1 lengthens short flaps so that they become substrates for Dna2. Dna2 requires at least a 1-nt long flap for cleavage, and has been shown to reduce flaps to lengths of 3–6 nt (16). We designed a 6-nt fixed flap substrate and asked whether Pif1 can stimulate Dna2 cleavage on this flap (Fig. 2). Dna2 alone displayed minimal cleavage on this substrate (lanes 3 and 10). When Pif1 was titrated into the reaction in the presence of ATP (lanes 11–15), Dna2 cleavage products were both more abundant and longer, suggesting that Pif1 unwound the flap substrates to produce longer flaps that could be cleaved by Dna2. Stimulation was ∼10-fold at the highest level of Pif1. Stimulation by Pif1 was greater here than in the more complex reconstitution, when the Pif1 was augmenting flap creation by the polymerase. When we prevented Pif1 helicase activity by excluding ATP and dNTPs (lanes 4–8), Pif1 failed to stimulate cleavage by Dna2. This result shows that active helicase function is required to provide a suitable substrate for Dna2. Apparently, the combination of Pif1 and Dna2 creates a steady-state situation in which flaps are lengthened by Pif1 and shortened by Dna2 so that they maintain a length of ∼6 nt. This length is too short to support RPA binding so that the flaps remain clear for entry by FEN1.
FIGURE 2.
Pif1 stimulates Dna2 cleavage on a short flap. Cleavage by Dna2 (50 fmol) was assayed on the 6-nt flap substrate (U1:T1:D1) in the presence or absence of ATP and dNTPs and varying amounts of Pif1 (5, 10, 25, 50, or 100 fmol) as indicated in the figure and as described under “Experimental Procedures.” dNTPs were included to reflect the exact conditions of the full reconstitution assays. Substrate depiction and designations are as in Fig. 1.
Pif1 Stimulates Dna2 Cleavage during Strand Displacement Synthesis
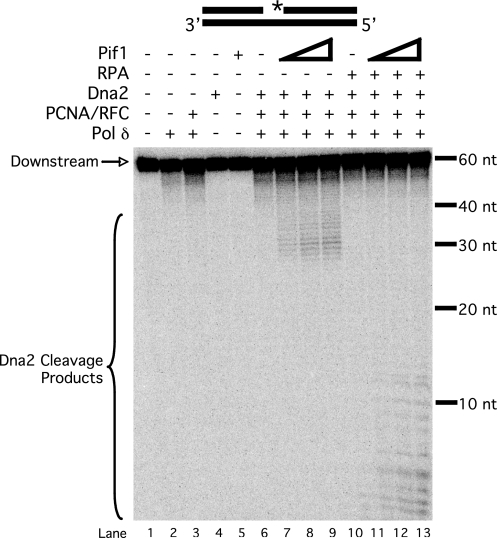
The previous experiment was done with a static flap substrate. We asked whether Pif1 could similarly accelerate the lengthening of flaps during displacement by pol δ to promote Dna2 cleavage. We used the standard substrate labeled at the 5′-end of the downstream primer to observe cleavage products. Dna2 was not able to cleave the substrate in the absence of synthesis (Fig. 3, lane 4), because there was no flap for Dna2 to bind. Even in the presence of pol δ, PCNA, and RFC, no cleavage occurred (lane 6). Previous studies have shown that pol δ does not readily displace DNA to produce flaps longer than a few nucleotides, too short for Dna2 to bind and cleave (8). However, when Pif1 was titrated into the reaction, a distribution of long cleavage products appeared (lanes 7–9). Again, Pif1 acted on a small fraction of substrate, consistent with previous reconstitution experiments (Fig. 1 and Ref. 14). When RPA was also included in the reactions (lanes 10–13), the distribution shifted to shorter products. This is consistent with results showing that RPA stimulates Dna2 cleavage efficiency, such that the Dna2 cleaved sooner as it tracked from the 5′-end of the flap (21). As a control, the reactions with Pif1 were repeated in the absence of pol δ. In this case, Pif1 did not stimulate Dna2 cleavage (data not shown), suggesting that initial strand displacement by pol δ is required before Pif1 can create a Dna2 substrate.
FIGURE 3.
Pif1 stimulates Dna2 cleavage on a strand-displaced flap. Cleavage by Dna2 (25 fmol) was assayed on the standard substrate (U2:T2:D2) in the presence of various combinations of pol δ (23 fmol), PCNA (25 fmol), RFC (25 fmol), RPA (100 fmol), and varying amounts of Pif1 (25, 50, or 100 fmol) as indicated in the figure and as described under “Experimental Procedures.” Substrate depiction and designations are as in Fig. 1.
Pif1 and RPA Together Inhibit Ligation When FEN1 Is the Sole Nuclease
Previous reconstitution of flap creation and cleavage indicated that Pif1 promotes a population of long flaps that bind RPA, which blocks tracking and cleavage by FEN1 (14). We wanted to know whether this inhibition would impact ultimate joining of the fragments. To accomplish this, reconstitution reactions were performed with DNA LigI and a standard substrate labeled at the 3′-end of the downstream primer. This ensured that the label would not be lost during flap cleavage, such that the ligated product would be visible. In a system including pol δ, PCNA, RFC, FEN1, and LigI, we observed a 11-nt full-length product (Fig. 4, lane 8) dependent on the presence of LigI (lane 6). When Pif1 was titrated into the reaction, there was virtually no change in the amount of ligation product (lanes 9–11). Surprisingly, when RPA was also included in the reaction, there was again no change in the amount of ligation product (lanes 12–15). We interpret this to mean that Pif1 promoted RPA inhibition of ligation on only a small fraction of the substrate. From our previous results (14), only ∼1–2% of substrate flaps are both lengthened by Pif1 and bound by RPA. We simply could not detect a reduction of that amount from the density of product bands in the ligation assay.
FIGURE 4.
Assessing completion of Okazaki fragment processing via ligation. Ligation by LigI (25 fmol) was assayed on the standard substrate (U2:T2:D2) in the presence of various combinations of pol δ (23 fmol), PCNA (25 fmol), RFC (25 fmol), FEN1 (20 fmol), RPA (200 fmol), and varying amounts of Pif1 (25, 50, or 100 fmol) as indicated in the figure and as described under “Experimental Procedures.” Substrate depiction and designations are as in Fig. 1.
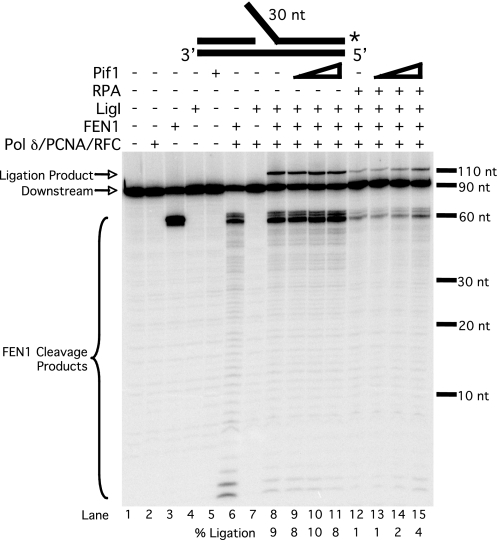
To assess the fate of that fraction of flaps that are lengthened in the presence of Pif1 such that they bind RPA before FEN1 cleavage, we altered the assay to make 100% of the flap intermediates into such long flaps. A new substrate was designed that simulates a flap already lengthened by Pif1, the pre-created flap substrate, identical to the standard substrate but with a 3-nt unstructured flap on the 5′-end of the downstream primer. We then performed the same reactions as in Fig. 4 using the pre-created flap substrate labeled at the 3′-end of the downstream primer (Fig. 5). The cleavage product observed when FEN1 was incubated alone with the substrate (lane 3) results from FEN1 cleavage at the base of the pre-created flap. When pol δ, PCNA, and RFC were included (lane 6), the primary cleavage product was 1 nt shorter and is due to cleavage following pol δ strand displacement, making the flap longer by 1 nt. This creates a double flap, which is the preferred substrate for FEN1 (32). A ligation product was observed when the system was reconstituted with pol δ, PCNA, RFC, FEN1, and LigI (lane 8). Ligation occurred mostly on substrates in which synthesis passed far into the downstream annealed region, as is evident from the decrease in short cleavage products. We again observed no change in the amount of ligation product when Pif1 was titrated into the reaction (lanes 9–11). This is expected, because the substrate has a built-in long flap, so further flap lengthening by Pif1 should have no effect. However, when RPA was included in the reaction (lanes 12–15), both cleavage and ligation decreased significantly, with approximately equivalent extents. This is clear evidence that a substantial portion of the flaps bound RPA such that subsequent cleavage by FEN1 was inhibited, ultimately inhibiting flap joining. Titration of Pif1 into the reaction in the presence of RPA resulted in a slight increase in cleavage and ligation, suggesting that Pif1 can stimulate FEN1 or LigI activity. These observations indicate the pre-created flap substrate is effectively simulating the small fraction of long flap intermediates promoted by Pif1.
FIGURE 5.
RPA inhibits processing and ligation of long flaps in the absence of Dna2. Ligation by LigI (25 fmol) was assayed on the pre-created flap substrate (U2:T2:D3) in the presence of various combinations of pol δ (23 fmol), PCNA (25 fmol), RFC (25 fmol), FEN1 (20 fmol), RPA (200 fmol), and varying amounts of Pif1 (25, 50, or 100 fmol) as indicated in the figure and as described under “Experimental Procedures.” Substrate depiction and designations are as in Fig. 1.
Dna2 Rescues RPA Inhibition of the Final Ligation Product
The two-nuclease model of fragment processing proposes that once a flap has been bound by RPA, cleavage by Dna2 is required for proper processing and ligation (16). We asked whether Dna2 could rescue the observed RPA-inhibited ligation using the pre-created flap substrate. As with other ligation assays, the substrate was labeled at the 3′-end of the downstream primer. Previous results have shown that FEN1 can cleave a flap structure with a gap between the upstream and downstream primers (10). FEN1 was indeed able to cleave the pre-created flap substrate (Fig. 6A, lane 3). As expected, RPA inhibited FEN1 cleavage (lane 4) ∼50%. Dna2 was also able to cleave the substrate (lane 5), and RPA stimulated Dna2 to cleave past the base of the flap (lane 6), as previously reported (21). The combination of FEN1 and Dna2 resulted in more cleavage than either nuclease alone (lane 7), also expected because Dna2 stimulates FEN1 (16). Because the radiolabel is at the 3′-end of the downstream primer, only the final cleavage product will be visible, which in this case is produced by FEN1. RPA did not inhibit cleavage by both nucleases together (lane 8). This is consistent with a recent report describing the coordinated action of FEN1, Dna2, and RPA (35). During strand displacement synthesis, we observed cleavage by FEN1 (lane 9), and that cleavage was inhibited by RPA (lane 10). When Dna2 was the sole nuclease present during displacement synthesis (lane 11), we observed a cleavage pattern similar to that observed when Dna2 was incubated alone with the substrate (lane 5). This is expected because pol δ should not be able to displace flaps long enough to be cleaved by Dna2. After Dna2 cleaves the pre-created flap, no long flaps are displaced, as shown in Fig. 3. When RPA was included, we again saw stimulation of Dna2 cleavage further into the downstream primer (lane 12). This likely resulted from RPA strand melting activity, which has been shown to cause flap elongation (21). When FEN1 and Dna2 were both present during displacement synthesis, we again observed a high level of cleavage (lane 13), which was not inhibited by RPA (lane 14). When FEN1 and LigI were both included during displacement synthesis (lane 15), we observed efficient formation of the ligation product. Both ligation and FEN1 cleavage were inhibited by RPA (lane 16), as shown in Fig. 5. With Dna2 and LigI present during displacement synthesis, there was very little ligation product observed (lane 17), as expected because Dna2 does not cleave at the base of the flap (16). With RPA present, a small amount of ligation product was observed (lane 18). The ligation observed in lanes 17 and 18 likely results from a combination of pol δ flap displacement and Dna2 cleavage, followed by pol δ 3′ exonuclease activity to form a ligatable nick (9). RPA may stimulate this process, although this has not been directly examined. With FEN1, Dna2, and LigI present during displacement synthesis, ligation was efficient (lane 19) and not inhibited by RPA (lane 20). Significantly, the combination of FEN1 and Dna2 provided the highest level of ligation, highlighting the importance of both nucleases in processing a long flap.
The important comparison is among lanes 15, 16, and 20. In lane 15, a significant amount of ligation occurred during displacement synthesis when only FEN1 was present. When RPA was then included, cleavage and ligation were inhibited, as expected (lane 16). When Dna2 was then added (lane 20), cleavage and ligation were rescued, to an amount even greater than when Dna2 and RPA were absent. This indicates that, when a long flap is bound by RPA, Dna2 rescues both cleavage and ligation. Because we had shown that Pif1 has only a minor effect with this substrate (Fig. 5), as it already simulates the Pif1 helicase product, we did not include Pif1 in this experiment. When these reactions were repeated in the presence of Pif1, similar results were obtained (data not shown). These findings are consistent with our hypothesis that Pif1 shuttles fragment processing intermediates into the two-nuclease pathway for flap removal.
Evidence That Ligation Occurred during Strand Displacement Synthesis
In genuine Okazaki fragment processing, ligation must occur during the strand displacement synthesis process. Because our substrate used a pre-created flap, it is possible that our reconstitution system did not fulfill this criterion. On any substrate, the coordinated actions of FEN1, Dna2, and RPA may have removed the flap before the upstream primer was extended by pol δ to form a nick with the downstream primer. If this were the case, the system would not represent dynamic flap creation and removal expected in vivo. To address whether the system is dynamic, we used an unlabeled extended pre-created flap substrate similar to that used in Fig. 6A with radiolabeled dGTP included in the assay. The template oligonucleotide sequence in the gap of the pre-created flap substrate did not contain any cytosine residues, but there were cytosines in the template complementary to the downstream primer, the first being 2 nt past the base of the flap. Consequently, we would not observe a product on our gel unless pol δ synthesized through the gap and added nucleotides during strand displacement. If we observed a ligation product, it must have resulted from dynamic strand displacement synthesis followed by flap processing by FEN1, Dna2, and RPA. Additionally, to distinguish between a ligated product and full-length synthesis through the entire downstream primer, we extended the downstream primer in our pre-created flap substrate by 5 nt at the 3′-end. The ligation product would then migrate slower on electrophoresis than a full-length synthesis product. The results of this experiment are shown in Fig. 6B. We observed synthesis intermediates in all lanes, but substantial full-length synthesis products appeared only when the system was reconstituted with pol δ, PCNA, RFC, FEN1, and Dna2 (lane 7). This indicates that, when both nucleases are present, pol δ could fully extend the upstream primer, such that the entire downstream primer was displaced. When LigI is included in the reaction, we observe a product band that ran higher than the full-length synthesis band (lane 9). This band represents the ligation product. In addition, there were minor bands migrating between the full-length synthesis and ligation products. These bands likely result from pol δ 3′-5′ exonuclease cleavage of the ligation product. RPA inhibits ligation in the absence of Dna2, as expected (lane 10). When the entire system is reconstituted with all proteins (lane 14), we also observe ligation products. This strongly suggests that with the pre-created flap substrate, pol δ strand displaces into the downstream annealed region while flaps are being processed by FEN1, RPA, and Dna2.
Overall, our results indicate that, even in the presence of Pif1, the number of flaps requiring the two-nuclease pathway represents a small percentage of the total. Nevertheless, flaps that grow long enough by strand displacement to bind RPA can be effectively processed by this alternative pathway.
DISCUSSION
We previously reconstituted Okazaki fragment processing to understand the roles of individual proteins in carrying out this essential pathway while maintaining genome integrity. The reconstituted system uses purified proteins and DNA substrates that simulate reaction intermediates. Results showed that RNA primers were removed through flap intermediates, some of which became long before necessary cleavage by FEN1 (13). Pif1, a helicase known to influence the processing reaction, was shown to promote displacement such that RPA bound the flaps and inhibited FEN1 (14). In this report, we show that a two-nuclease pathway employing Dna2 and FEN1 is necessary to resolve these RPA-bound flaps, such that the fragments can be efficiently joined.
Flap creation is a consequence of strand displacement synthesis catalyzed by pol δ. The efficiency of this reaction depends on the ability of the polymerase to disrupt hydrogen bonding of the downstream primer with the template. In our system, PCNA-clamped pol δ alone could not fully extend the upstream primer to the end of the template. Previous biochemical studies suggest that FEN1 cleavage stimulates extension of the upstream primer by pol δ (7, 13). A reasonable explanation is that removal of short flaps eliminated the possibility of reformation of hydrogen bonds that would provide a physical barrier to forward motion by the polymerase. However, in the context of reconstituted Okazaki fragment processing, strand displacement synthesis promoted by FEN1 probably does not accelerate long flap formation. This is because it is the very cleavage of a flap that is expected to promote the displacement. Here we demonstrate a similar stimulation of full-length extension when PCNA-clamped pol δ was engaged in strand displacement synthesis in the presence of Pif1 (Fig. 1). Significantly, there was a proportionately greater reduction in the intensity of bands representing extension products of lengths intermediate between the pause site at the start of the downstream primer and the full-length product, compared with the intensity of the pause site band. This suggests that pol δ displaced an initial short flap, which Pif1 bound. Pif1 then unwound the downstream annealed region, clearing the way for full extension by pol δ.
Promotion of strand displacement by Pif1 is different from the effects of FEN1 in three key ways. Firstly, it occurs through a helicase mechanism rather than a nuclease mechanism, and so should be fundamentally independent of the actions of FEN1. Secondly, the helicase mechanism produces long flaps. Thirdly, displacement is not inhibited by RPA interaction with the flap. This latter conclusion is evident from results showing that Pif1 stimulates Dna2 activity in the presence of RPA (Fig. 3). Presumably, FEN1 and Pif1 do not necessarily collaborate in promoting strand displacement synthesis in vivo. Instead, with some Okazaki fragments, we envision that Pif1 elongates flaps quickly enough that they can bind RPA and exclude the entry of FEN1 at the 5′-end and tracking to the flap base for cleavage.
Fortunately, lengthening of short displaced flaps by Pif1 also promoted cleavage by Dna2. Dna2 cannot efficiently cleave flaps shorter than 6 nt (16). However, in the presence of Pif1, Dna2 cleavage was stimulated (Fig. 2). This stimulation was dependent on Pif1 helicase activity. During dynamic strand displacement by PCNA-clamped pol δ, Pif1 similarly stimulated Dna2 cleavage (Fig. 3). In the absence of Pif1, pol δ did not displace very many flaps long enough for Dna2 to cleave. This is consistent with our previous results indicating that pol δ alone does not readily carry out long distance strand displacement synthesis (14). The stimulation of Dna2 cleavage by RPA binding to the Pif1-lengthened flaps (Fig. 3) was expected from similar observations with fixed length flaps (21). Evidently, a steady state exists between flap lengthening by Pif1 and flap shortening by Dna2 cleavage, maintaining a short flap. Viewed in this manner, the steady state would be facilitated by the substrate specificity of Dna2, which disallows cleavage of short flaps, and the ability of RPA to stimulate Dna2 cleavage of long flaps. In addition, it is likely that binding of RPA prevents formation of secondary structure in the flap that would inhibit both FEN1 and Dna2 (20, 21).
Although a 5′ helicase, Pif1 was not anticipated to directly inhibit ligation, because it cannot displace a fully annealed segment and so would not be expected to disrupt a nicked site. However, we expected the combination of Pif1 and RPA to inhibit ligation in the presence of PCNA-clamped pol δ and FEN1. This is because Pif1 lengthening of flaps and RPA binding to the long flaps would inhibit FEN1 and thus inhibit the production of a nicked intermediate. However, this was not observed, presumably because our electrophoresis assays were not sensitive enough to detect the small change in ligation (Fig. 4). To assess the influence of long flap creation on ligation, we constructed a substrate with a pre-created 3-nt flap. A 3-nt flap length is within the distribution of cleavage products observed during Okazaki fragment processing reconstitutions with the standard substrate (14) and therefore appropriately simulates a flap already lengthened by Pif1. Using this substrate in the fully reconstituted system, we found that RPA significantly inhibited both cleavage and ligation (Fig. 5). This shows that flap lengthening by Pif1 in our system does indeed lead to inhibition of the ultimate ligated product via RPA flap binding. With Pif1 present, FEN1 is not efficient enough to successfully process all flaps.
We observed a slight stimulation of cleavage and ligation in the reconstituted system (Fig. 5), including RPA but not Dna2, as more Pif1 was added. Possibly Pif1 displaced RPA from a fraction of flaps. Pif1 has been shown to displace telomerase from telomeric substrates (33), so Pif1 may be able to displace other proteins from DNA as well. It is also possible that Pif1 directly stimulated the cleavage or ligation reactions. Either mechanism would increase the amount of product observed, because the level of RPA used was not fully inhibitory of FEN1. A thorough investigation of Pif1 functional interactions with combinations of the processing proteins may reveal additional ways in which it influences processing.
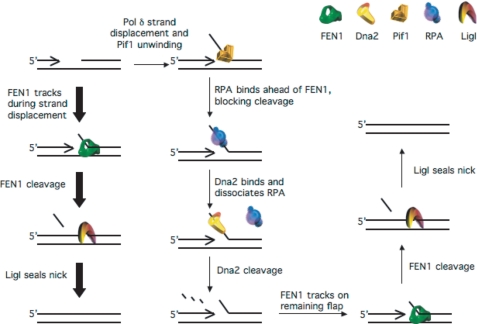
Addition of Dna2 to the system recovered ligation in the presence of RPA (Fig. 6A). The amount of cleavage and ligation in the presence of Dna2 and RPA was slightly greater than in their absence, indicating that the system may be optimized when FEN1, Dna2, and RPA coordinate to process the flap. Based on these findings, we propose a modified model for Okazaki fragment processing in vivo (Fig. 7). On a large majority of Okazaki fragments, FEN1 cleaves short flaps as pol δ displaces them. Continuous displacement and cleavage of short flaps removes the entire primer, eventually leaving a nick for LigI to seal. On a small fraction of fragments, Pif1 binds short flaps displaced by pol δ before FEN1 can act. Pif1 actively lengthens those flaps allowing stable binding of RPA. This inhibits FEN1 cleavage but permits Dna2 cleavage. Dna2 displaces RPA (21) and cleaves, leaving a short flap. FEN1 then displaces Dna2 (35) and cleaves the short flap. LigI then seals the nick to create the double-stranded product. Pif1 activity effectively shuttles flaps into the two-nuclease pathway. Although Pif1 is active on only a small portion of flaps, there are hundreds of thousands of Okazaki flaps processed per yeast replication cycle (35). Therefore, thousands of flaps are most likely processed by the two-nuclease pathway.
FIGURE 7.
Pif1 directs a small subset of flaps into the two-nuclease pathway for flap processing. A majority of flaps (denoted by the thick arrows) displaced by pol δ are bound and cleaved by FEN1. Continuous displacement of short flaps by pol δ and cleavage by FEN1 eventually produces a nick that LigI seals. On a minority of flaps (denoted by the thin arrows), Pif1 binds the short flap displaced by pol δ prior to FEN1. Pif1 activity lengthens the flap, allowing binding of RPA. RPA inhibits FEN1 but stimulates Dna2. Dna2 then displaces RPA and cleaves the flap, remaining bound to a short RPA-free flap. FEN1 displaces Dna2 and cleaves the short flap, producing the nicked intermediate that LigI seals.
We emphasize two additional ways in which our reconstituted system appropriately emulates the native Okazaki fragment processing reaction. The first is that it represents the dynamic interaction of synthesis, cleavage, and ligation reactions. However, use of one of our reconstitution substrates, having the pre-created flap, brought up the concern that synthesis was uncoupled from cleavage in time and distance. To address this issue, we devised an assay in which a radiolabel is incorporated into the ligation product only if strand displacement occurs. Under these conditions we still observed a ligation product (Fig. 6B). This is strong evidence that our system indeed represents dynamic flap creation and cleavage. Another notable observation was that the intensity of the full-length synthesis bands suggested that pol δ synthesized through the entire downstream primer at a frequency similar to that of ligation. However, ligation products shorter than full length were also observed that resulted from pol δ 3′-5′ exonuclease cleavage of the full-length ligation product. The intensity of the full-length synthesis band when the entire system was reconstituted (Fig. 6B, lane 14) was likely a combination of full-length synthesis and exonuclease cleavage of the full-length ligation products. Therefore, the density of the band over-represented the actual level of full-length synthesis. Instead, based on the abundance of ligation products obtained with the pre-created flap substrate, we believe that flap processing and ligation via the two-nuclease pathway are occurring at a similar rate as the FEN1-only pathway. This would be anticipated if the two-nuclease pathway were employed in cells in preference to elimination of long flap intermediates by simply displacing the entire Okazaki fragment.
The second respect is that our system considers the short 5′ RNA segment on native Okazaki fragments (4). We have shown in two previous reports that a full DNA substrate acts equivalently to a substrate with an RNA-initiated downstream primer in the reconstituted system (13, 14). Additionally, Pif1 does not bind RNA (33). Therefore, it is likely the RNA component of the primer is removed by other mechanisms such as the FEN1-only pathway, because FEN1 can cleave RNA flaps (36), or a combination of RNase HI and FEN1 cleavage (37), without the influence of Pif1. Pif1 can bind as the DNA portion of the flaps is displaced by pol δ. Therefore, our experiments should be accurately simulating the influence of Pif1.
We have provided biochemical evidence that Pif1 helicase activity promotes the two-nuclease pathway for flap removal. It would appear that Pif1 involvement imposes an unnecessary energy requirement on the cell, because virtually all Okazaki flaps can be processed by the FEN1-only pathway in the absence of Pif1. However, several lines of evidence indicate that Pif1 is specifically involved in DNA replication. Genetic studies in S. cerevisiae show that dna2Δpif1Δ mutants are viable, suggesting that Pif1 creates a need for Dna2 (12, 27). Presumably, in the absence of Pif1, Dna2 and the two-nuclease pathway are no longer required, although the dna2Δpif1Δ mutants still displayed temperature-sensitive growth. Upon deletion of POL32, the gene encoding the subunit of pol δ that interacts with PCNA, growth at the restrictive temperature was restored. Similarly, deletion of POL32 or PIF1 suppressed the lethality or growth defects of certain rad27Δ mutants that tend to produce long flaps due to lack of efficient short flap processing by FEN1 (12). Thus Pif1 genetically interacts with several proteins known to be involved in Okazaki fragment processing. Genetic studies in Schizosaccharomyces pombe showed that pfh1, the homolog of Pif1, is required in strains that lack telomeres and mitochondrial DNA (38), suggesting that Pif1-family helicases have an essential role in genome maintenance beyond the roles in telomeric and mitochondrial DNA stability. Therefore Pif1 has likely evolved to work with the replication machinery, but what is its positive role? It is possible that Pif1 is required for proper primer removal and flap processing at specific sequences such as GC-rich regions. The strong hydrogen bonding between the strands of GC-rich regions may present a barrier to sufficient pol δ strand displacement synthesis unless Pif1 is present. Pif1 may also be important in sequence regions that could potentially block flap processing by either pathway, such as self-complementary sequences that can form fold-back flaps. Further biochemical studies with GC-rich or fold-back substrates will be helpful in determining whether this interpretation has validity. Moreover, studies in vivo to examine the genomic location of Pif1 may help reveal whether Pif1 localizes to specific sequences other than telomeres. Finally, reconstitution experiments using mutant versions of the proteins, particularly Pif1 and Dna2, may further explain the genetic results and provide additional insight into the role of Pif1.
Acknowledgments
We thank the members of the Bambara laboratory for helpful discussions and suggestions. We thank Dr. Marc Wold for providing us with purified RPA.
This work was supported by National Institutes of Health Grant GM024441 (to R. A. B.), with additional support from Grants GM087666 (to J. L. C.), and GM32431 (to P. M. B.).
- pol δ
- DNA polymerase δ
- nt
- nucleotide(s)
- pol α
- DNA polymerase α/primase
- PCNA
- proliferating cell nuclear antigen
- RFC
- replication factor C
- LigI
- DNA ligase I
- FEN1
- flap endonuclease 1
- RPA
- replication protein A.
REFERENCES
- 1.Kunkel T. A., Burgers P. M. (2008) Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pursell Z. F., Isoz I., Lundström E. B., Johansson E., Kunkel T. A. (2007) Science 317, 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornberg A., Baker T. A. (1992) in DNA Replication, 2nd Ed., pp. 113–195, W. H. Freeman, New York [Google Scholar]
- 4.Bambara R. A., Murante R. S., Henricksen L. A. (1997) J. Biol. Chem. 272, 4647–4650 [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Kao H. I., Bambara R. A. (2004) Annu. Rev. Biochem. 73, 589–615 [DOI] [PubMed] [Google Scholar]
- 6.Rossi M. L., Purohit V., Brandt P. D., Bambara R. A. (2006) Chem. Rev. 106, 453–473 [DOI] [PubMed] [Google Scholar]
- 7.Ayyagari R., Gomes X. V., Gordenin D. A., Burgers P. M. (2003) J. Biol. Chem. 278, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 8.Garg P., Stith C. M., Sabouri N., Johansson E., Burgers P. M. (2004) Genes Dev. 18, 2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Y. H., Ayyagari R., Resnick M. A., Gordenin D. A., Burgers P. M. (2003) J. Biol. Chem. 278, 1626–1633 [DOI] [PubMed] [Google Scholar]
- 10.Harrington J. J., Lieber M. R. (1994) EMBO J. 13, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murante R. S., Rust L., Bambara R. A. (1995) J. Biol. Chem. 270, 30377–30383 [DOI] [PubMed] [Google Scholar]
- 12.Stith C. M., Sterling J., Resnick M. A., Gordenin D. A., Burgers P. M. (2008) J. Biol. Chem. 283, 34129–34140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi M. L., Bambara R. A. (2006) J. Biol. Chem. 281, 26051–26061 [DOI] [PubMed] [Google Scholar]
- 14.Rossi M. L., Pike J. E., Wang W., Burgers P. M., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fanning E., Klimovich V., Nager A. R. (2006) Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae S. H., Bae K. H., Kim J. A., Seo Y. S. (2001) Nature 412, 456–461 [DOI] [PubMed] [Google Scholar]
- 17.Bae S. H., Choi E., Lee K. H., Park J. S., Lee S. H., Seo Y. S. (1998) J. Biol. Chem. 273, 26880–26890 [DOI] [PubMed] [Google Scholar]
- 18.Bae S. H., Seo Y. S. (2000) J. Biol. Chem. 275, 38022–38031 [DOI] [PubMed] [Google Scholar]
- 19.Kao H. I., Campbell J. L., Bambara R. A. (2004) J. Biol. Chem. 279, 50840–50849 [DOI] [PubMed] [Google Scholar]
- 20.Kao H. I., Veeraraghavan J., Polaczek P., Campbell J. L., Bambara R. A. (2004) J. Biol. Chem. 279, 15014–15024 [DOI] [PubMed] [Google Scholar]
- 21.Stewart J. A., Miller A. S., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 31356–31365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart J. A., Campbell J. L., Bambara R. A. (2006) J. Biol. Chem. 281, 38565–38572 [DOI] [PubMed] [Google Scholar]
- 23.Budd M. E., Choe W., Campbell J. L. (2000) J. Biol. Chem. 275, 16518–16529 [DOI] [PubMed] [Google Scholar]
- 24.Lee K. H., Kim D. W., Bae S. H., Kim J. A., Ryu G. H., Kwon Y. N., Kim K. A., Koo H. S., Seo Y. S. (2000) Nucleic Acids Res. 28, 2873–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bessler J. B., Torredagger J. Z., Zakian V. A. (2001) Trends Cell Biol. 11, 60–65 [DOI] [PubMed] [Google Scholar]
- 26.Boulé J. B., Zakian V. A. (2006) Nucleic Acids Res. 34, 4147–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budd M. E., Reis C. C., Smith S., Myung K., Campbell J. L. (2006) Mol. Cell. Biol. 26, 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgers P. M., Gerik K. J. (1998) J. Biol. Chem. 273, 19756–19762 [DOI] [PubMed] [Google Scholar]
- 29.Johansson E., Garg P., Burgers P. M. (2004) J. Biol. Chem. 279, 1907–1915 [DOI] [PubMed] [Google Scholar]
- 30.Burgers P. M. (2009) J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerik K. J., Gary S. L., Burgers P. M. (1997) J. Biol. Chem. 272, 1256–1262 [DOI] [PubMed] [Google Scholar]
- 32.Kao H. I., Henricksen L. A., Liu Y., Bambara R. A. (2002) J. Biol. Chem. 277, 14379–14389 [DOI] [PubMed] [Google Scholar]
- 33.Boulé J. B., Vega L. R., Zakian V. A. (2005) Nature 438, 57–61 [DOI] [PubMed] [Google Scholar]
- 34.Sibenaller Z. A., Sorensen B. R., Wold M. S. (1998) Biochemistry 37, 12496–12506 [DOI] [PubMed] [Google Scholar]
- 35.Stewart J. A., Campbell J. L., Bambara R. A. (2009) J. Biol. Chem. 284, 8283–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murante R. S., Rumbaugh J. A., Barnes C. J., Norton J. R., Bambara R. A. (1996) J. Biol. Chem. 271, 25888–25897 [DOI] [PubMed] [Google Scholar]
- 37.Turchi J. J., Huang L., Murante R. S., Kim Y., Bambara R. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9803–9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J. Q., Qi H., Schulz V. P., Mateyak M. K., Monson E. K., Zakian V. A. (2002) Mol. Biol. Cell 13, 2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]