Abstract
The actin nucleation factors Spire and Cappuccino interact with each other and regulate essential cellular events during Drosophila oogenesis in a cooperative fashion. The interaction blocks formin actin nucleation activity and enhances the Spire activity. Analogous to Spire and Cappuccino, the mammalian homologs Spir-1 and formin-2 show a regulatory interaction. To get an understanding of the nature of the Spir-formin cooperation, we have analyzed the interaction biochemically and biophysically. Our data shows that the association of Spir-1 and formin-2 is not significantly mediated by binding of the Spir-1-KIND domain to the formin FH2 core domain. Instead, a short sequence motif C-terminal adjacent to the formin-2-FH2 domain could be characterized that mediates the interaction and is conserved among the members of the Fmn subgroup of formins. In line with this, we found that both mammalian Spir proteins, Spir-1 and Spir-2, interact with mammalian Fmn subgroup proteins formin-1 and formin-2.
Basic cell biological functions such as proliferation, migration, division, and vesicle transport rely on the organization of the actin cytoskeleton. The initiation of actin polymerization from free actin monomers is regulated by actin nucleation factors (NF),2 which help to overcome the kinetic barrier of spontaneous G-actin nucleation and, thus, catalyze the formation of filamentous actin structures and networks (1). To date, three different classes of NFs are described, the ARP2/3 complex, FH2 domain containing NFs of the formin superfamily, and NFs containing one or multiple WH2 domains (Spire/Cordon-bleu/Leiomodin) (2). The formin superfamily is subdivided into seven subfamilies (Dia, FRL, DAAM, Delphilin, INF, FHOD, Fmn) (3). The mechanisms of actin nucleation as well as the regulation of the NFs vary significantly between the three classes (and also show variances in between the distinct superfamilies). Spire and Cappuccino are NFs that belong to the Spire subfamily of WH2 containing nucleators and to the Fmn subfamily of the FH2 domain containing formins, respectively. In contrast to the Arp2/3 complex that nucleates branched filaments, Spire and the formin Cappuccino nucleate unbranched actin filaments (4).
Almost two decades ago it was found that mutants of the two Drosophila NFs (Spire/Cappuccino) have an identical phenotype in early Drosophila oogenesis, i.e. both induce premature ooplasmic streaming (5, 6). Later it was shown that both proteins cooperate in the generation of a dynamic actin mesh in the oocyte that prevents premature ooplasmic streaming (7). Spire and Cappuccino do not solely have the same mutant phenotype; the proteins also physically interact and cross-regulate each other. The Cappuccino C-terminal half, encoding the FH2 domain and flanking sequences, enhances the nucleation activity of Spire, whereas the nucleation activity of Cappuccino is decreased in the presence of the Spire-KIND domain (8).
Cappuccino belongs to the Fmn subgroup of formins (3, 9). In mammals, two Fmn subgroup members (formin-1, formin-2) and two Spir proteins (Spir-1, Spir-2) exist (3, 10). The formin-2 and spir-1 genes are coexpressed in the developing and adult nervous system, and the proteins interact analogous to their Drosophila counterparts Spire and Cappuccino (8, 10). Several reports showed the importance of formin-2 in mouse oogenesis and here especially in the positioning of the meiotic spindle (11–14). Recently it was found that a dynamic actin mesh, as during Drosophila oogenesis, is also required for mouse oogenesis (11, 14). The correct localization of the meiotic spindle during mouse oogenesis and the resulting asymmetric division depends on an actin mesh that is built up by formin-2. Myosin-2 generates the pulling forces required for spindle movement (14). Beside the evolutionary conserved roles for the formins Cappuccino and formin-2, Spire family proteins also seem to be evolutionary conserved regulators of oocyte development. Spire genes of the African clawed frog Xenopus (pEg6) and the sea squirt Ciona savignyi (Pem-5) have been identified as maternal genes in the oocyte in analogy to its Drosophila homolog and are proposed to function in polarity during early embryogenesis (15, 16).
In an initial characterization it was found that the KIND domains of Spir-1/dSpire interact with the C-terminal sequences of formin-2/Cappuccino, which encode the FH2 domains and flanking sequences (8). To gain a further understanding of the interaction and cross-regulation of the two proteins, we investigated this interaction in detail. The objective of the study was the dissection of the formin-2/Spir-1 interaction and the determination of the structural elements that are responsible for the binding. The dissection revealed a high affinity Spir-1 interaction site of formin-2, which could be mapped to the very C terminus of formin-2 adjacent to its core FH2 domain (formin Spir interaction (FSI) sequence). The FSI sequence is conserved among the members of the Fmn subgroup. Consistently we found that all mammalian members of the two distinct nucleator families, Spir-1/2 and Fmn-1/2, interact with each other.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
All Spir and formin constructs were generated by standard cloning techniques using Pfu DNA polymerase from Promega® and restriction enzymes and T4-DNA ligase from New England Biolabs® according to the manufacturers' recommendations. Expression vectors were from GE Healthcare® (pGEX), Invitrogen (pProExHTb, pcDNA3), QIAgen® (pQE80L), and Takara/Clontech® (pEGFP-C1, pAcGFP-C1). The Spir and formin proteins correspond to Swiss-Prot entry numbers Q08AE8–2 (hs Spir-1 (II)), Q8WWL2 (hs Spir-2), Q05860 (mm formin-1 (IV)), and Q9JL04 (mm formin-2). Supplemental Table S1 summarizes constructs, a brief description, fragment boundaries, purification scheme, and their purpose in this study (supplemental Table S1). For prokaryotic expression of proteins, Escherchia coli Rosetta pLysS bacteria were transformed with plasmids encoding for glutathione S-transferase (GST) and His6-tagged Spir and formin proteins, respectively. Bacteria were grown in LB (+100 mg/liter ampicillin and 30 mg/liter chloramphenicol) at 37 °C to an A600 of 0.4–0.6. Protein expression was initiated by induction with 100–300 μm isopropyl 1-thio-β-d-galactopyranoside and allowed to proceed at 20 °C for 12–18 h. Bacteria were lysed by ultrasonication, and soluble proteins were purified with an ÄKTA purifier system (GE Healthcare) using nickel-nitrilotriacetic acid affinity (Ni-NTA HP, GE Healthcare), GSH affinity (GSH-Sepharose FF, GE Healthcare), cation exchange (SP-Sepharose XL, GE Healthcare), and size exclusion (Sephadex G200, GE Healthcare) chromatography according to the manufacturers' recommendations. His and GST tags were cleaved where indicated by tobacco etch virus protease. Proteins were concentrated by ultrafiltration using Amicon Ultra-4 ultracentrifugation devices with molecular mass cut offs of 3,000, 10,000, and 30,000 Da (Millipore®). The final purity of the constructs was estimated by SDS-Tris-PAGE and SDS-Tricine-PAGE using Image J gel analysis software (rsb.info.nih.gov) (supplemental Fig. S4).
Fluorophore Labeling
For anisotropy measurements, proteins and peptides were labeled on cysteines with the thiol-reactive maleimidocaproyl-linked BodipyFLTM-fluorophore (Probes®). Proteins were buffer-exchanged into labeling buffer (10 mm Hepes, pH 7.0, 100 mm NaCl, 0.25 mm Tris(2-carboxyethyl)phosphine) using Nap10 columns (GE Healthcare). A 2-fold molar excess of dye was coupled with the protein (1–5 g/liter) for 16–18 h at 4 °C. Excess non-reacted dye was quenched with 20 mm dithioerythritol and subsequently removed using Nap10 columns. The degree of labeling was ∼ 0.2–0.25 for the Fmn peptides and ∼0.75 for the KIND domain.
Cell Culture and Transfection
HeLa and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% fetal calf serum (FCS, HiClone®), 2 mm l-glutamate (Glu), and 100 units/ml penicillin and 100 μg/ml streptomycin at 37 °C and 10% CO2. Cells were transfected with LipofectamineTM/Lipofectamine 2000TM according to the manufacturer's recommendations. Briefly cells were seeded in DMEM supplemented with FCS but without Glu and penicillin/streptomycin 24 h before transfection. At the day of transfection, cells reached a confluency of 80–90%. For HeLa cells (∼8 × 105 cells) 6 μl of Lipofectamine were incubated with 1.4 μg of DNA in DMEM for 20 min; HEK293 and HeLa cells (∼8 × 105 cells) were transfected alternatively with 7.5 μl of Lipofectamine 2000/4 μg of DNA in DMEM). After 5 h the medium was changed to DMEM containing FCS, penicillin/streptomycin, and Glu. Cells were allowed to express proteins for 24–36 h.
GST Pulldown Assay
For GST Pulldown experiments with the GST-KIND domains and enhanced green fluorescent protein (eGFP)-tagged formin proteins, ∼50 μg of purified GST/GST fusion proteins bound to 20 μl of GSH-Sepharose 4B resin (GE Healthcare) were used. Cell lysates were prepared by lysing ∼3 × 106 HEK293 cells in 900 μl of pulldown buffer 1 (25 mm Tris, pH 7.4, 150 mm NaCl, 0.1% Nonidet P-40, 10% (v/v) glycerol, 1 mm EDTA, inhibitor mixture (Roche Applied Science)) for 20 min at 4 °C. The lysate was centrifuged at 20,000 × g for 20 min. 50 μg of GSH-Sepharose 4B-bound GST fusion proteins were incubated with the high speed supernatant of the cell lysate for 2 h at 4 °C. Beads were washed 5 times with pulldown buffer 1 and analyzed by PAGE and subsequent Western blot analysis. GFP-tagged proteins were detected with a rabbit A.V-living colors antibody (1 μg/ml; Takara/Clontech), a horseradish peroxidase-linked anti-rabbit secondary antibody (1:5000; GE Healthcare), and the enhanced chemiluminescence kit from GE Healthcare.
Analytical Gel Filtration
Analytical Gel filtration experiments were done in analytical gel filtration buffer (20 mm Tris, pH 8.0, 50 mm KCl, 2 mm dithioerythritol) using a computer-controlled ÄKTATM purifier high performance liquid chromatography equipped with a Sephadex G200 15/30 column (∼23-ml volume) at 6–8 °C. The injection volume of the proteins was 100 μl with concentrations ranging from ∼2 g/liter for the Spir-KIND domains, ∼3.5 g/liter for the formin FH2 domains, and ∼0.6 or 2 g/liter for the formin peptides. The flow rate was kept constant at 0.4 ml/min. Eluted proteins were collected in 1-ml fractions and analyze by PAGE to verify the identity of the proteins in the distinct peak fractions. Data analysis and processing were done with the Unicorn evaluation software (GE Healthcare) and Sigma Plot 9.0 (Systat Software®). Molecular weight calibration of the analytical gel filtration column was done using the Sigma gel filtration molecular mass standard kit 12,000–200,000 Da (cytochrome c, 12.4 kDa; carbonic anhydrase, 29 kDa; bovine serum albumin, 66 kDa; alcohol dehydrogenase, 150 kDa; b-amylase, 200 kDa). The observed molecular masses for the individual proteins and protein complexes can be found in Table 1.
TABLE 1.
Observed masses of Spir/formin constructs and complexes from the analytical gel filtration
Theoretical masses of the complexes were calculated according to a 1:2 stoichiometry of Spir:Fmn, as densitometric analysis of Coomassie Brilliant Blue G250/R250-stained SDS-PAGE gels of complex fractions (the insets to Fig. 2 and supplemental Fig. S1A) and molecular masses deduced from the gel filtration suggested a 1:2 stoichiometry of the complexes. Numbers in parentheses are the cumulative weights of the observed masses according to a 1:2 stoichiometry.
| Protein | Theoretical mass | Observed mass | Oligomer state/stoichiometry |
|---|---|---|---|
| Da | Da | ||
| Spir-1-KIND | 25.8 | 40.4 | Monomer |
| Spir-2-KIND | 21.2 | 43.3 | Monomer |
| Fmn-2-FH2-FSI | 52.7 | 123.8/239.5 | Dimer/tetramer |
| Fmn-2-FH2 | 51.2 | 113.4/262.3 | Dimer/tetramer |
| Fmn-2-eFSI | 7.1 | 16.6 | Dimer |
| Fmn-2-FSI | 3.8 | 8.0 | Dimer |
| Fmn-1-FH2 | 50.8 | 100.4 | Dimer |
| Spir-1-KIND and Fmn-2-FH2-FSI | 157.0 (164.2) | 164.5 | 1:2 |
| Spir-1-KIND and Fmn-2-eFSI | 32.9 (57.0) | 47.4 | 1:2 |
| Spir-2-KIND and Fmn-2-eFSI | 28.3 (59.9) | 51.9 | 1:2 |
| Spir-1-KIND and Fmn-2-FSI | 29.6 (48.4) | 39.0 | 1:2 |
Fluorescence Anisotropy
Fluorescence anisotropy measurements were performed in a Horriba Jobin Yvon® Fluoromax-4 spectrophotometer in anisotropy buffer (10 mm Hepes, pH 7.0, 100 mm NaCl) at 20 °C. The BodipyFLTM fluorophore was excited at 495 nm, and emission was collected at 510 nm, with an integration time of 2 s for the BodipyFLTM-labeled formin peptides (∼100 nm) and 4 s for the BodipyFLTM-labeled Spir-1-KIND domain (∼100 nm). The slit width of the emission and excitation monochromators was set to 2 nm for the BodipyFLTM-labeled formin peptides and 3 nm for the labeled Spir-1-KIND domain, respectively. Each data point of the binding curve is the mean of at least eight collected polarization signals. Data analysis and processing was done with Sigma Plot 9.0 (Systat Software).
Equilibrium binding data were fitted according to the equation
 |
with a representing the maximum amplitude (Bmax), and b representing the equilibrium constant (Kd). Competition binding experiments were performed as described by Vinson et al. (17).
Competition binding curve was fitted according to the equation
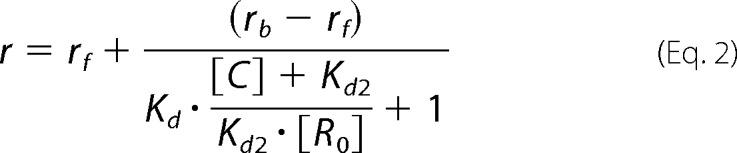 |
where is the anisotropy, rb if the anisotropy of the Spir-1-KIND·Bodipy-Fmn-2-eFSI complex, rf is the anisotropy of the free Bodipy-labeled eFSI, [C] is the concentration of the unlabeled eFSI, [R0] is the concentration of the free Spir-1-KIND when [C] = 0, Kd is the dissociation constant for the binding of the Bodipy-labeled eFSI to Spir-1-KIND, and Kd2 is the dissociation constant for the binding of the unlabeled eFSI to Spir-1-KIND.
Immunostaining and Fluorescence Microscopy
LipofectamineTM- transfected HeLa cells seeded on 13-mm round glass slides were fixed with 3.7% (w/v) paraformaldehyde for 20 min at 4 °C and permeabilized with 0.2% (v/v) Triton X-100 in phosphate-buffered saline for 3.5 min at 20–22 °C. Immunostaining was done by incubating the cells with the mouse anti-Myc 9E10 antibody (Santa Cruz®) at a concentration of 4 μg/ml in phosphate-buffered saline supplemented with 1% (v/v) FCS at room temperature for 60 min. TRITC-conjugated donkey anti-mouse secondary antibody (Dianova®, 1:200 in phosphate-buffered saline + 1% (v/v) FCS) was allowed to react with the primary antibody for 60 min at room temperature. Stained cells were mounted in moviol solution (15% (w/v) moviol, 30% (v/v) glycerol, and 2.25% (w/v) N-propyl gallate in phosphate-buffered saline) and analyzed with a Leica® AF6000LX imaging system equipped with a Leica® HCX PL APO 63×/1.30 glycerol objective and a Leica® DFC350FX camera. Images were deconvoluted (blind) with the deconvolution software from Leica® (Leica Application Suite Advanced Fluorescence) and further processed with Adobe® Photoshop CS.
Native Polyacrylamide Gel Electrophoresis
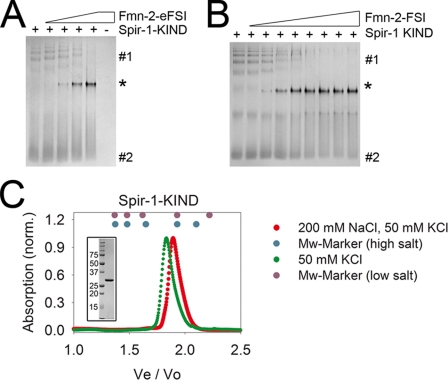
Native polyacrylamide gel electrophoresis analysis was performed with a Laemmli native PAGE system. 20 μm Spir-KIND was incubated for 20 min in the presence or absence of increasing amounts of formin-2 peptides (0–45 μm Fmn-2-eFSI; 0–80 μm Fmn-2-FSI) in anisotropy buffer at 20–22 °C. Subsequent gel electrophoresis was conducted at 150 V for 20 min followed by a 300-V step for 50 min for Spir-1-KIND. Gel electrophoresis for Spir-2-KIND was performed as for Spir-1 except that the second voltage step lasts only 30 min due to the higher negative charge of the Spir-2-KIND domain.
RESULTS
The Spir-1-KIND Domain Binds to the Extreme C Terminus of Formin-2
The Spir-1 and formin-2 proteins interact directly with each other (8). The Spir-1-KIND domain and the C-terminal part of the formin-2 protein encoding the FH2 domain and flanking the C-terminal sequences mediate the interaction. Formin-2 actin nucleation activity is blocked by the interaction of the formin with the Spir-KIND domain (8). To gain more insights into the interaction between the Spir-1-KIND domain and the formin-2 C-terminal half encoding the actin nucleating FH2 domain and flanking sequences, we have investigated this interaction structurally and biochemically.
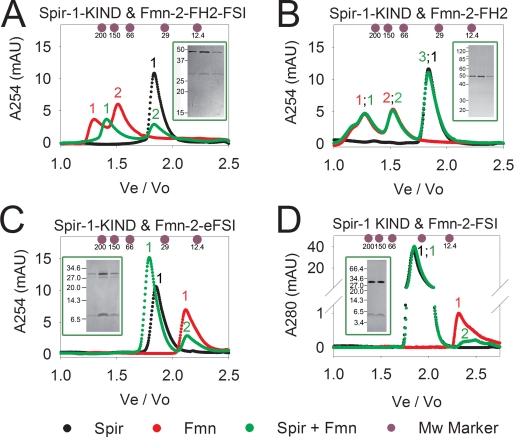
Purified bacterially expressed recombinant proteins have been employed in analytical gel filtration experiments to analyze the complex formation and dissect the sequences necessary for the interaction. In agreement with previous findings (8), we found that the KIND domain of Spir-1 (Fig. 1B, KIND short) does not adopt a globular shape, as it migrates on the analytical gel filtration faster than expected from its molecular mass (Fig. 2A, black curve, peak 1, Table 1). The purified formin-2 C-terminal part encoding the FH2 domain and flanking C-terminal sequences (Fig. 1A, Fmn-2-FH2-FSI) eluted in fractions corresponding to dimeric and tetrameric proteins as judged by the elution volumes of marker proteins (Fig. 2A, red curve, 1 and 2, Table 1). Mixing the two proteins resulted in a complex formation, which is nicely documented by a shift of the lower molecular mass form in peak 2 in the elution profile toward higher molecular masses (Fig. 2A, green curve, peak 1) and a co-elution of the Spir-1-KIND domain and the Fmn-2-FH2-FSI protein (Fig. 2A, inset, corresponding to green peak 1). Mixing of the two proteins was also accompanied by partial precipitation of the complex, explaining the decreased signal intensity of the peak fraction containing the complex.
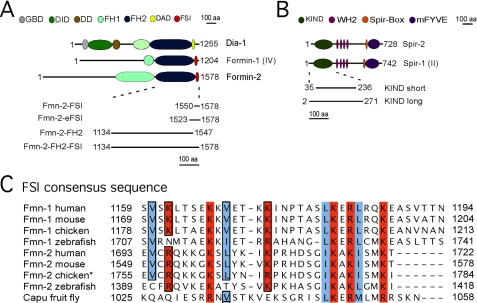
FIGURE 1.
Domain organization of Spir and formin proteins and sequence of the conserved newly identified FSI sequence. A, domain assembly of the formins mDIA1, formin-1, and formin-2. GBD, GTPase binding domain; DD, dimerization domain; FH1, formin homology 1 domain; FH2, formin homology 2 domain. B, domain assembly of the Spir-1 and Spir-2 proteins. KIND, kinase non-catalytic C-lobe domain; WH2, WASP homology-2 domain; mFYVE, modified-FYVE zinc finger. aa, amino acids. C, sequence alignment of the extreme C termini of Fmn subfamily members of various species; for the alignment, the following gi-accession numbers were used: Fmn-1 human (gi:157168329), Fmn-1 mouse (gi:122889638), Fmn-1 zebrafish (gi:68395057), Fmn-1 chicken (gi:45383319), Fmn-2 human (gi:160707881), Fmn-2 mouse (gi:160707879), Fmn-2 chicken (gi 25933298), Fmn-2 zebrafish (gi:189526476), and Capu Drosophila (gi:1061334). The asterisk indicates a discrepancy between genomic sequence and an EST clone. For the alignment the chicken EST-clone sequence was used.
FIGURE 2.
Analytical gel filtration analysis of Spir-1-KIND and formin-2 C-terminal constructs. The Spir-1-KIND domain interacts with formin-2-FH2-FSI (A) but not with a formin-2-FH2 construct lacking the FSI (B). The last 55 amino acids of the C terminus of formin-2 (eFSI) comprising the last predicted α-helix of the FH2 domain and the FSI bind to the KIND domain of Spir-1 (C). Even the last 29 amino acids of the C terminus outside the core FH2 domain of formin-2 (FSI) still bind to the KIND domain of Spir-1 (D). Insets show the presence of Spir- and formin proteins in the peak fractions (depending on the width of the peak 2 or 3 peak fractions, respectively) of the complexes (insets A, C, and D) and no Spir-1-KIND when mixing Spir-1-KIND and Fmn-2-FH2 lacking the conserved FSI (inset B, green curve, peak 2). Black elution curve, Spir; red elution curve, formin; green elution curve, Spir and formin. Colors of the peak numbering and elution profiles correspond. Masses of molecular weight standards: 12.4 kDa. cytochrome c; 29 kDa, carbonic anhydrase; 66 kDa, bovine serum albumin; 150 kDa, alcohol dehydrogenase; 200 kDa, b-amylase. Observed molecular masses for the Spir/formin proteins and complexes can be found in the Table 1; mAU, milliabsorption units; Ve/Vo, elution volume/void volume.
Formins nucleate actin polymerization by the stabilization of an actin oligomer with a dimeric FH2 domain ring structure. Because the interaction of Spir-1-KIND and the formin-2 protein blocks the actin nucleation activity of the formin, we next tested if the Spir-1-KIND domain binds the formin-2-FH2 core domain (Fig. 1A, Fmn-2-FH2). In contrast to the Fmn-2-FH2-FSI protein that formed a complex with the Spir-1-KIND domain, we could not detect a stable complex formation of the formin-2-FH2 core domain with the Spir-1-KIND domain in our gel filtration experiments (Fig. 2B), which is most likely because of a significant decrease in affinity. The elution profile of the mixed proteins was identical with that of the two individual proteins (Fig. 2B, red, black, and green curves). No Spir-1-KIND protein was shifted into the fraction of Fmn-2-FH2 (Fig. 2B, inset, corresponding to green peak 2).
This suggests that flanking sequences of the FH2 core domain mediate the interaction. As no stable complex formation of the Spir-1-KIND domain and the core FH2 domain of formin-2 could be detected on the analytical gel filtration, an interaction sequence of the C terminus of formin-2 outside the core FH2 domain was assumed. To test this, we expressed and purified a peptide comprising the last 56 amino acids of formin-2 (Fmn-2-eFSI). This peptide coeluted with the Spir-1-KIND domain on the analytical gel filtration column, indicating the formation of a complex of the Spir-1-KIND domain and the formin-2 C terminus (Fig. 2C, inset corresponding to green peak 1). An even shorter peptide comprising the last 29 amino acids of the C terminus of formin-2 (Fmn-2-FSI) also coeluted with the core KIND domain of Spir-1 in the analytical gel filtration, indicating that the last 29 amino acids provide the main binding site for the Spir-1-KIND domain (Fig. 2D, inset corresponding to green peak 1). One should note that in the case of the small FSI peptide, no shift of the KIND domain to a higher molecular mass could be observed. This indicates that the core binding motif of Fmn-2 either fills a cavity in the KIND domain or tightly wraps around the KIND domain. This assumption is supported by the fact that the complex of the KIND domain and the Fmn-2-eFSI also elutes at lower molecular weights than expected from the cumulative observed masses for the two proteins (see Table 1). In contrast to the results of Quinlan et al. (8), who deduced from their analytical ultracentrifugation studies a 2:2 stoichiometry of the Spir-formin complex, our analytical gel filtration analysis along with the densitometric analysis of the peak complex fractions on the SDS-PAGE (Fig. 2; insets) suggest a 1:2 stoichiometry of the KIND-formin complex (see Table 1). The discrepancy between the work done by Quinlan et al. (8) and our work concerning the stoichiometry of the complex may be because of the different methods used and subsequent data interpretation.
It is interesting to note that alignments of the very C-terminal sequences of Fmn subgroup proteins from flies, fish, birds, and mammals show a conserved sequence motif within the C-terminal region which we have mapped as an FSI sequence (Fig. 1C). The conserved sequence is unique for Fmn subgroup proteins and could not be detected in other formins. This suggests that next to Drosophila Cappuccino and vertebrate formin-2, the third member of this protein subgroup, the formin-1 protein, also may interact with Spir proteins (see below).
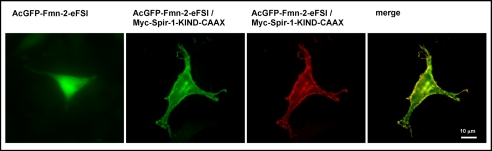
To further analyze the interaction of the C terminus of formin-2 with the KIND domain of Spir-1, colocalization studies in HeLa cells were performed in which Spir-1-KIND and formin-2-eFSI were transiently overexpressed (Fig. 3). Analogous to the assays by Quinlan et al. (8), a membrane-targeted Myc-tagged Spir-1-KIND domain (Myc-Spir-1-KIND-CAAX) was used to relocate a green fluorescence protein-tagged formin-2 C-terminal peptide (pAcGFP-Fmn2-eFSI) to the membrane in HeLa cells. When expressed alone, the GFP-fused Fmn-2-eFSI construct was evenly distributed in the cytoplasm (Fig. 3). Coexpression of AcGFP-Fmn-2-eFSI and the membrane-targeted KIND domain results in a relocalization of the GFP-tagged formin-2 peptide to the plasma membrane, indicating an interaction of the proteins.
FIGURE 3.
Colocalization of membrane-targeted Myc-Spir-1-KIND and AcGFP-Fmn-2-eFSI in HeLa cells. Proteins were visualized by monitoring the intrinsic fluorescence of the AcGFP-tag (green) or indirect immunostaining of the Myc-tagged KIND domain with anti-Myc antibody and a TRITC-conjugated secondary antibody (red). When expressed alone, AcGFP-Fmn-2-eFSI (green) is distributed evenly in the cytoplasm. Coexpression of AcGFP-Fmn-2-eFSI (green) and a membrane-targeted version of the KIND domain of Spir-1 (Myc-Spir-1-KIND-CAAX (red)) translocates AcGFP-Fmn-2-eFSI to the membrane. Images are deconvoluted (blind) and were further processed by Adobe Photoshop CS.
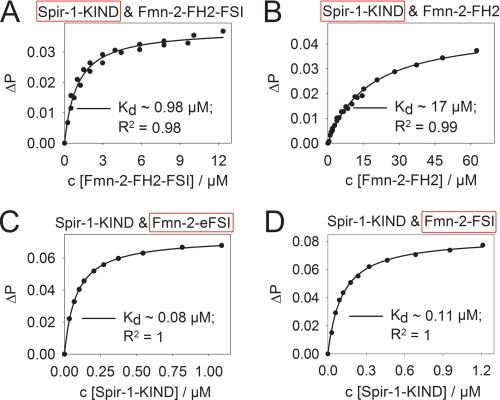
Fluorescence anisotropy measurements provide a very sensitive tool to detect and quantify protein interactions. As we cannot exclude from our gel filtration experiments that the FH2 core domain does not contribute to the overall binding of the KIND domain, we have analyzed the interaction of the Spir-1-KIND domain with the various C-terminal formin constructs. Using a fluorescent BodipyFl-labeled version of the core Spir-1-KIND domain, the interaction of the KIND domain with the core FH2 domain of formin-2 and the FH2 domain plus its C-terminal extension (Fmn-2-FH2, Fmn-2-FH2-FSI) was probed (Fig. 4, A and B). To monitor the interaction of the C terminus of formin-2 (Fmn-2-eFSI, Fmn-2-FSI) and the Spir-1 KIND domain, BodipyFl-labeled C-terminal peptides of formin-2 were used (Fig. 4, C and D). In these fluorescence anisotropy measurements the affinities of the Spir-1/formin-2 interaction for the core and the extended FH2 domain as well as for the C-terminal peptides of formin-2 could be determined (Table 2). The affinity of the Spir-1/formin-2 interaction sharply decreases when the C terminus of formin-2 is removed (∼1 μm → ∼17 μm). In accordance with this, we measured a high affinity interaction of the Spir-1-KIND domain and the 55-amino acid-long eFSI peptide of formin-2 (∼80 nm). The even shorter FSI peptide which comprises only the last 29 amino acids of the C terminus of formin-2 binds with an ∼1.5–2-fold lower affinity to the KIND domain of Spir-1 (∼110 nm) than the eFSI peptide. The anisotropy data along with the analytical gel filtration data strongly suggest that the last 29 amino acids of formin-2 represent the key binding motif of the Spir-1/formin interaction. Additional contacts in the proximity of this binding motif may further increase binding strength, as can be seen by the 1.5–2-fold higher affinity of the 26-amino acid-long eFSI peptide and the residual binding affinity of the core FH2 domain for the KIND domain. The values of the affinities measured for the interaction of the KIND domain and the core and extended FH2 domains of formin-2 should be regarded as estimates, as the FH2 domains showed a decreased stability and tendency to form higher oligomeric complexes under several conditions. We observed that the tendency of the FH2 domain of formin-2 to self-associate increases with increasing ionic strength, suggesting that the oligomerization of the FH2 domain is driven by hydrophobic interactions. Reduction of the ionic strength reduces oligomerization at low temperatures (4–8 °C) but also negatively influences the stability at elevated temperatures (20–25 °C). Thus, the “true” affinity of the Spir-1-KIND formin-2-FH2-FSI interaction is probably higher than observed (∼1 μm) and would be better reflected by the affinity of the Spir-1-KIND domain and the extended C-terminal formin-2 peptide (eFSI; Kd ∼ 80 nm), which shows no tendency to oligomerize or precipitate. To exclude that the isolated KIND domain binds to the BodipyFL fluorophore instead of the formin peptide, we performed competition binding experiments with non-labeled formin-2-eFSI (supplemental Fig. S5). The non-labeled eFSI peptide could efficiently compete the binding of the BodipyFL-labeled eFSI peptide to the Spir-1-KIND domain showing the validity of the newly discovered interaction.
FIGURE 4.
Fluorescence anisotropy/polarization measurements probing the interactions of the Spir-1-KIND domain and formin-2 C-terminal constructs. Binding of formin-2-FH2-FSI (A) and formin-2-FH2 (B) to BodipyFl labeled Spir-1-KIND (100 nm). The binding of the Spir-1 KIND domain to formin proteins was quantified by fitting the anisotropy data according to a one-site ligand binding model. The affinity of the KIND domain to formin-2-FH2 constructs significantly drops when the core FSI sequence is removed. Spir-1-KIND binds to the BodipyFl-labeled eFSI sequence (100 nm) (C) and BodipyFL-labeled core FSI-sequence (100 nm) (D) with high affinity. Residual contacts outside the core FSI sequence enhance the binding, as can be seen by the 1.5–2-fold higher affinity of the eFSI peptide compared with the FSI peptide and the residual binding affinity of the FH2 construct lacking the core FSI. ΔP, change in polarization; red boxes mark the BodipyFl-labeled proteins.
TABLE 2.
Affinities of the Spir-KIND/formin-FSI interaction determined by fluorescence-anisotropy/polarization measurements
| Protein interaction | Kd |
|---|---|
| μm | |
| Spir-1 KIND and Fmn-2-FH2-FSI | 0.98 ± 0.09 |
| Spir-1 KIND and Fmn-2-FH2 | 17.20 ± 0.98 |
| Spir-1 KIND and Fmn-2-eFSI | 0.082 ± 0.001 |
| Spir-1 KIND and Fmn-2-FSI | 0.113 ± 0.004 |
| Spir-2 KIND and Fmn-2-eFSI | 0.060 ± 0.003 |
| Spir-2 KIND and Fmn-2-FSI | 0.125 ± 0.009 |
In the analysis of the Spir-1-KIND/Fmn-2-eFSI interaction by basic native PAGE, we made an interesting observation which led us to the assumption that the conformation of the Spir-1-KIND domain may get stabilized by the interaction with the formin-2 protein. The separation by the basic native PAGE relies on a mixture of charge and size exclusion of the proteins that were analyzed. In the basic PAGE only anionic species are visualized on the polyacrylamide gel. As no SDS is present in the system, the molecules move either to the anode or cathode, depending on their charge at an ambient pH. At nearly neutral pH (pH 6.8–7.0) the KIND domains possess a high negative charge because of their low isoelectric points (Spir-1-KIND, pI 5.1; Spir-2-KIND, pI 4.4), whereas both Fmn-2 peptides are charged positive at neutral pH (Fmn-2-eFSI, pI 9.8; Fmn-2-FSI, pI 10.5). Thus, only the KIND domains and the complex comprising the KIND domain and Fmn-2 peptide (complex, pI 5.8 and 6.0, respectively) are visible on the native gel. With increasing amounts of either the eFSI peptide (Fig. 5A) or FSI peptide (Fig. 5B) of Fmn-2, one can clearly observe the appearance of a novel band in the middle of the gel which corresponds to the complex of the Spir-1-KIND domain and the Fmn-2 peptide. We judge the distinct band pattern of the Spir-1-KIND domain in the absence of the Fmn-2 peptides as different conformations, as no evidence for oligomerization or impurities was observed (Fig. 5C, inset). Interestingly, in the presence of the formin-2 peptides, the band pattern of the Spir-1-KIND domain sharpens and focuses to the one distinct protein band representing the complex, which might be indicative of a structural stabilization of a certain Spir-1-KIND conformation by the Fmn-2 peptides.
FIGURE 5.
Laemmli basic native PAGE analysis of the Spir-1-KIND/Fmn-2-eFSI (A) and Spir-1-KIND/Fmn-2-FSI interaction (B). 20 μm Spir-1-KIND was mixed with increasing amounts of eFSI peptide (0, 4, 8, 15, 47 μm) (A) or FSI peptide (0, 10, 20, …, 80 μm) (B) and incubated for 20 min before native PAGE. 2 μg of Spir-1-KIND/lane (A) and 2.8 μg of KIND/lane (B) were applied to the gel. In the absence of the formin-2 peptides, Spir-1-KIND exhibits a complex band pattern (#1, #2) that sharpens (*) with increasing amounts of peptide. The multiple bands at higher molecular masses (#) do not correspond to oligomers or impurities as judged by analytical gel filtration and SDS-PAGE (C, C inset).
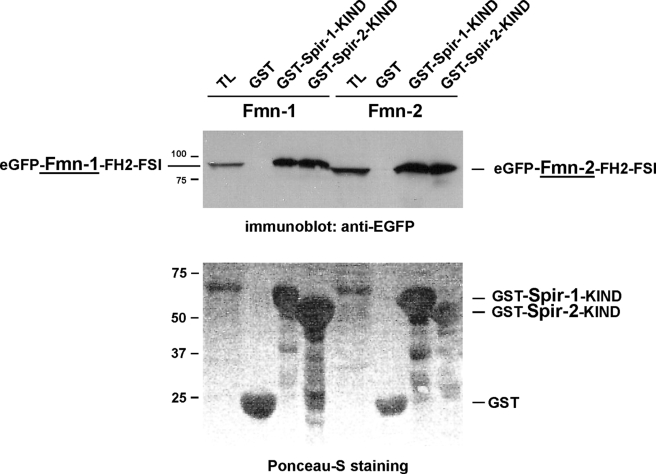
As already pointed out above, the C terminus of formin-1 shows significant similarity to the C termini of formin-2 and Cappuccino (Fig. 1C). The similarity suggests that also formin-1 could directly interact with the KIND domain of Spir-1, and because of the high conservation of the Spir KIND domains, Spir-2 is also a potential interaction partner for Fmn subgroup proteins. We, therefore, have analyzed if in analogy to the Spir-1 and formin-2 interaction, the Spir-2 protein interacts with formin-2 and if both mammalian Spir proteins interact with formin-1. To address this question, a GST pulldown approach was performed. Purified GST-tagged KIND domains of Spir-1 and Spir-2 were assayed for their interaction with the extended FH2 domains of formin-1 and formin-2 (eGFP-Fmn-1-FH2-FSI; eGFP-Fmn-2-FH2-FSI). As depicted in Fig. 6, both GST-tagged KIND domains bind to eGFP-tagged formin1-FH2-FSI and formin-2-FH2-FSI that were transiently overexpressed in HEK293 cells.
FIGURE 6.
Interaction of Spir and formin proteins. GST pulldown experiments have been performed to analyze the interaction of Spir and formin proteins. GST protein alone or GST protein fused to the Spir-1-KIND domain (GST-Spir-1-KIND) or the Spir-2-KIND domain (GST-Spir-2-KIND) have been coupled to glutathione-Sepharose beads and incubated with HEK293 cell lysates either expressing the formin-1-FH2-FSI domain fused to the eGFP (eGFP-Fmn-1-FH2-FSI) or a eGFP formin-2-FH2-FSI domain fusion protein (eGFP-Fmn-2-FH2-FSI). The HEK293 lysates (TL) and the GST fusion proteins were separated by SDS polyacrylamide gel electrophoresis and blotted onto a nitrocellulose membrane. A Ponceau S staining of the separated proteins is shown. eGFP fusion proteins expressed in the lysates and pulled down in the pulldown experiments were detected by immunoblotting with an eGFP-specific antibody. The migration of the proteins of different molecular sizes in kilodaltons is indicated.
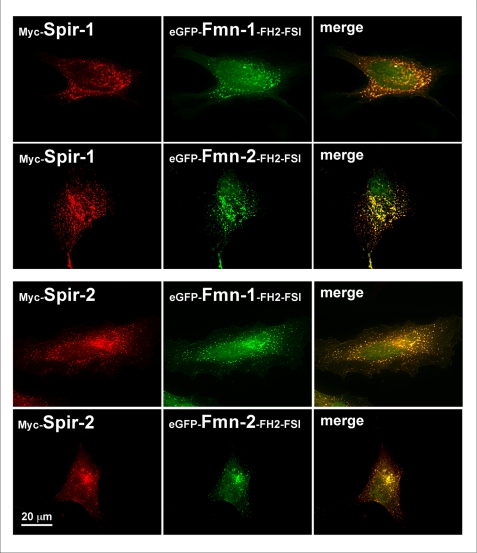
The pulldown interaction data were validated by colocalization studies in HeLa cells. In these studies the colocalization of Myc-tagged Spir proteins with the eGFP-tagged formin-FH2-FSI domains was analyzed (Fig. 7). Previously it was shown that exogenously expressed Spir proteins localize to the trans-Golgi network and post-Golgi vesicles in NIH3T3 fibroblasts and that coexpression of Spir-1 with the extended FH2 domain of formin-2 leads to a recruitment of the cytosolic FH2 domain into the defined Spir-1 spots. In agreement with the pulldown experiments, one observes a colocalization of Spir-1 and the FH2-FSI domain of formin-2 and formin-1 as well as a colocalization of Spir-2 with the FH2-FSI domains of formin-1 and formin-2, respectively. As controls, we coexpressed eGFP with Spir-1 and Spir-2, respectively, and did not find the Spir-proteins to colocalize with eGFP (supplemental Fig. S6).
FIGURE 7.
Colocalization of Spir and formin proteins. HeLa cells which transiently coexpress Myc-tagged Spir-1 (Myc-Spir-1) or Spir-2 (Myc-Spir-2) with enhanced green fluorescent protein-tagged formin-1-FH2-FSI (eGFP-Fmn-1-FH2-FSI) or formin-2-FH2-FSI (eGFP-Fmn-2-FH2-FSI) domains have been immunostained with a Myc-epitope specific antibody and a TRITC-conjugated secondary antibody (red). The localization of proteins has been analyzed by fluorescence microscopy. The red immunofluorescence of the Myc-tagged proteins and the green autofluorescence of the eGFP fusion proteins are shown. In addition an overlay (merge) of the two fluorescence channels is shown. Images are deconvoluted (blind) and further processed by Adobe Photoshop CS.
To further validate the finding that the FSI sequence mediates binding to the KIND domains of both mammalian Spir proteins, we performed key experiments for the dissection of the KIND/FSI interaction and also for the KIND domain of Spir-2 and formin. Analytical gel filtration experiments were done with the extended FSI peptide of formin-2 (eFSI) and the KIND domain of Spir-2. As was found for the KIND domain of Spir-1, Spir-2-KIND migrates faster in the analytical gel filtration than expected from its molecular mass (supplemental Fig. S1A, black curve, 1 and Table 1). Spir-2-KIND coelutes in analytical gel filtration experiments together with the eFSI peptide (supplemental Fig. S1A, green curve, inset corresponding to green peak 1), giving further evidence for the validity of the conserved FSI/KIND interaction. Fluorescence anisotropy measurements as were performed for the eFSI/FSI/Spir-1-KIND interaction revealed that Spir-2-KIND binds with a similar affinity to the eFSI peptide (Kd ∼ 60 nm) or to the FSI peptide (Kd ∼ 130 nm) compared with Spir-1-KIND (supplemental Fig. S2, A and B). Native PAGE-analysis of the Spir-2-KIND/Fmn-2-eFSI/FSI interaction showed that Spir-2-KIND interacts both with the eFSI-peptide and the core FSI (supplemental Fig. S3, A and B), in agreement with the previous findings. Compared with the KIND domain of Spir-1, Spir-2-KIND did not exhibit such a complex band pattern in the absence of Fmn-2 peptide, as was observed for Spir-1-KIND in the absence of the Fmn peptides. However, on the native PAGE one can clearly see a novel band appearing (the asterisk in supplemental Fig. S3, A and B) representing the complex.
In line with the analytical gel filtration data for the interaction with the formin-2 core FH2 domain (Fig. 2B), which did not interact with the Spir-1 KIND domain, we could show that neither KIND domain of Spir-1 and Spir-2 interacts with a formin-1-FH2 construct lacking the obligatory FSI sequence (supplemental Fig. S1, B and C).
Summarizing, we showed that the Spir-1/formin-2 interaction is mediated by a binding of the KIND domain to the extreme C terminus of formin-2 rather than to its core FH2 domain. A newly identified conserved sequence in the C terminus of Fmn subfamily members could be determined that is responsible for the high affinity binding to the core KIND domain of Spir-family proteins. In accordance with this we also found that formin-1 interacts with Spir-1 and Spir-2 and that the interaction also depends on an intact C terminus of the formin.
DISCUSSION
The dissection of the Spir-1/formin-2 interaction revealed a high affinity Spir binding site at the very C terminus of formin-2 (FSI sequence) adjacent to its core FH2 domain. We found the FSI sequence to be highly conserved within the Fmn subfamily of formin proteins and absent in other formin subfamilies. The conserved amino acids within the FSI are basic and aliphatic (Fig. 1C), which indicates combined electrostatic and hydrophobic interaction forces. Our data show that the FSI interacts with the KIND domains of both Spir-1 and Spir-2 with a similar affinity but not with KIND domains in general, as formin-2 does not interact with the KIND domain of the KIND family protein very-KIND (8). Additionally we could show the interaction of the second mammalian Fmn subgroup protein, formin-1, with both Spir proteins.
In Drosophila, Spire and Cappuccino mutant phenotypes are indistinguishable (5, 9, 18). Both actin nucleators cooperate in the regulation of an actin mesh in the oocyte (7). The data presented here strongly indicate that this functional cooperation is conserved among flies and mammals and that both mammalian formin proteins can cooperate with both mammalian Spir proteins. Not very much is known about the cell biological function of the mammalian Fmn subgroup and Spir proteins. Best studied here is the formin-2 protein, which was recently shown to function similarly to its Drosophila homolog Cappuccino in the regulation of an actin meshwork in the mouse oocyte, which is required for the positioning of the meiotic spindle (11, 14). Obvious studies to analyze if as in Drosophila formin-2 cooperates with one of the mammalian Spir proteins in this process still have to be done.
Several members of the formin superfamily have been shown to contain an autoregulatory peptide in their C termini (DAD, diaphanous autoregulatory domain) that is responsible for the intramolecular binding of an autoinhibitory domain located in the N terminus (diaphanous inhibitory domain (DID)) (19–22) (Fig. 1A). The intramolecular interaction between the C termini and the N termini is mediated by the DAD/DID interaction and results in an autoinhibited conformation of the formin proteins. The release of this inhibited conformation is regarded as an essential step in formin activation. Homologous DAD/DID sequences could not be found in the C terminus of the Fmn subfamily members. Therefore, it seems possible that Fmn subfamily members are regulated by a distinct mechanism. Nevertheless it cannot be excluded that the FSI of Fmn subfamily formins bind to a cryptic DID-like domain in the N terminus and contribute to an autoinhibited conformation of the Fmn subgroup proteins. The validity of intramolecular interactions within the Fmn subgroup of formins is, therefore, still an open issue and should be addressed in future experiments.
However, the data shown by Quinlan et al. (8) that the addition of excess Spir-1-KIND to the extended Fmn-2-FH2 domain could significantly decrease the nucleation activity of Fmn-2 may account for a different mode of regulating formin nucleation activity apart from any intramolecular interaction. The results presented here raise the possibility that the newly identified conserved FSI at the C terminus of Fmn subfamily members may function analogous to the DAD/DID interaction but mechanistically not in cis but in trans by employing the Spir-KIND domains. Further structural, biochemical, and cell biological studies will be needed for a detailed understanding of the interesting interaction and cross-regulation of the two distinct actin nucleators.
Supplementary Material
Acknowledgments
C. Makbul and Ch. Hermann are acknowledged for the pGEX4T1-NTEV expression vector.
This work was supported by the Bavarian Genome Research Network (BayGene) and the Deutsche Forschungsgemeinschaft (SPP 1150, KE 447/4-3).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
- NF
- nucleation factor
- FSI
- formin Spir interaction
- eFSI
- extended FSI
- FCS
- fetal calf serum
- DMEM
- Dulbecco's modified Eagle's medium
- TRITC
- tetramethylrhodamine isothiocyanate
- DAD
- diaphanous autoregulatory domain
- DID
- diaphanous inhibitory domain
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- eGFP
- enhanced GFP
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Pollard T. D. (2007) Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 [DOI] [PubMed] [Google Scholar]
- 2.Chesarone M. A., Goode B. L. (2009) Curr. Opin. Cell Biol. 21, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgs H. N., Peterson K. J. (2005) Mol. Biol. Cell 16, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinlan M. E., Heuser J. E., Kerkhoff E., Mullins R. D. (2005) Nature 433, 382–388 [DOI] [PubMed] [Google Scholar]
- 5.Manseau L. J., Schüpbach T. (1989) Genes Dev. 3, 1437–1452 [DOI] [PubMed] [Google Scholar]
- 6.Theurkauf W. E. (1994) Science 265, 2093–2096 [DOI] [PubMed] [Google Scholar]
- 7.Dahlgaard K., Raposo A. A., Niccoli T., St Johnston D. (2007) Dev. Cell 13, 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinlan M. E., Hilgert S., Bedrossian A., Mullins R. D., Kerkhoff E. (2007) J. Cell Biol. 179, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emmons S., Phan H., Calley J., Chen W., James B., Manseau L. (1995) Genes Dev. 9, 2482–2494 [DOI] [PubMed] [Google Scholar]
- 10.Schumacher N., Borawski J. M., Leberfinger C. B., Gessler M., Kerkhoff E. (2004) Gene Expr. Patterns 4, 249–255 [DOI] [PubMed] [Google Scholar]
- 11.Azoury J., Lee K. W., Georget V., Rassinier P., Leader B., Verlhac M. H. (2008) Curr. Biol. 18, 1514–1519 [DOI] [PubMed] [Google Scholar]
- 12.Dumont J., Million K., Sunderland K., Rassinier P., Lim H., Leader B., Verlhac M. H. (2007) Dev. Biol. 301, 254–265 [DOI] [PubMed] [Google Scholar]
- 13.Leader B., Lim H., Carabatsos M. J., Harrington A., Ecsedy J., Pellman D., Maas R., Leder P. (2002) Nat. Cell Biol. 4, 921–928 [DOI] [PubMed] [Google Scholar]
- 14.Schuh M., Ellenberg J. (2008) Curr. Biol. 18, 1986–1992 [DOI] [PubMed] [Google Scholar]
- 15.Le Goff C., Laurent V., Le Bon K., Tanguy G., Couturier A., Le Goff X., Le Guellec R. (2006) Biol. Cell 98, 697–708 [DOI] [PubMed] [Google Scholar]
- 16.Satou Y., Satoh N. (1997) Dev. Biol. 192, 467–481 [DOI] [PubMed] [Google Scholar]
- 17.Vinson V. K., De La Cruz E. M., Higgs H. N., Pollard T. D. (1998) Biochemistry 37, 10871–10880 [DOI] [PubMed] [Google Scholar]
- 18.Wellington A., Emmons S., James B., Calley J., Grover M., Tolias P., Manseau L. (1999) Development 126, 5267–5274 [DOI] [PubMed] [Google Scholar]
- 19.Higgs H. N. (2005) Trends Biochem. Sci. 30, 342–353 [DOI] [PubMed] [Google Scholar]
- 20.Li F., Higgs H. N. (2003) Curr. Biol. 13, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 21.Liu W., Sato A., Khadka D., Bharti R., Diaz H., Runnels L. W., Habas R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 210–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaillant D. C., Copeland S. J., Davis C., Thurston S. F., Abdennur N., Copeland J. W. (2008) J. Biol. Chem. 283, 33750–33762 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.