Abstract
The phylum Apicomplexa are a group of obligate intracellular parasites responsible for a wide range of important diseases. Central to the lifecycle of these unicellular parasites is their ability to migrate through animal tissue and invade target host cells. Apicomplexan movement is generated by a unique system of gliding motility in which substrate adhesins and invasion-related proteins are pulled across the plasma membrane by an underlying actin-myosin motor. The myosins of this motor are inserted into a dual membrane layer called the inner membrane complex (IMC) that is sandwiched between the plasma membrane and an underlying cytoskeletal basket. Central to our understanding of gliding motility is the characterization of proteins residing within the IMC, but to date only a few proteins are known. We report here a novel family of six-pass transmembrane proteins, termed the GAPM family, which are highly conserved and specific to Apicomplexa. In Plasmodium falciparum and Toxoplasma gondii the GAPMs localize to the IMC where they form highly SDS-resistant oligomeric complexes. The GAPMs co-purify with the cytoskeletal alveolin proteins and also to some degree with the actin-myosin motor itself. Hence, these proteins are strong candidates for an IMC-anchoring role, either directly or indirectly tethering the motor to the cytoskeleton.
Apicomplexan parasites cause a multitude of illnesses through infection of both human and livestock hosts. Members of this phylum include the opportunistic human parasites Toxoplasma gondii and Cryptosporidium parvum, pathogens of livestock, including Theileria annulata and Eimeria tenalla, and most notably the Plasmodium species, the causative agents of malaria in humans. Infection with P. falciparum results in ∼1–3 million deaths and a further 500 million infections annually (1).
During various stages of the Apicomplexan lifecycle the parasites require motility to migrate through their insect and vertebrate hosts and to invade and internalize themselves within targeted host cells (2–4). The parasite's unique mechanism of gliding motility is powered by an Apicomplexan-specific motor complex termed the actin-myosin motor (5), which resides between the outer plasma membrane and inner membrane complex (IMC)4 (6). The IMC is a continuous patchwork of flattened vesicular cisternae located directly beneath the plasma membrane and overlying the cytoskeletal network (7, 8). The IMC appears to arise from Golgi-associated vesicles flattened during parasite maturation to form large membranous sheets, which envelope the parasite and leave only a small gap at the extreme parasite apex (9).
The myosin component of the actin-myosin motor has previously been defined as a tetrameric complex consisting of a class XIV myosin termed Myo-A (10), a myosin tail interacting protein (also called myosin light chain) (7) and the two glideosome-associated proteins GAP45 and GAP50 (11). These motor components are linked to the outer IMC membrane via the membrane proteins GAP45/50 (11). Between the plasma membrane and the IMC are actin filaments held in place through aldolase-mediated contact with the C-terminal tails of plasma membrane-spanning adhesive proteins whose ectodomains bind substrate and host cells (2). To power the forward movement of apicomplexan zoite stages, myosin pulls the actin filaments and their attached adhesins rearward. For this to succeed the GAP-myosin complex must presumably be fixed to the IMC, possibly via interactions with unidentified proteins linking the motor to the underlying cytoskeleton. Studies of fluorescently tagged GAP50 confirm it is relatively immobile within the IMC, however attempts to identify potential anchoring proteins have not been successful and have instead indicated that GAP50 may be immobilized by the lipid-raft like properties of the IMC membranes (12).
The actin-myosin complex is confined to the outer IMC membrane while the opposing innermost IMC membrane is studded with 9 nm intramembranous particles, revealed by electron microscopy of freeze fractured Toxoplasma tachyzoites and Plasmodium ookinetes (13, 14). The size of these particles suggests that the proteins involved are likely to form high molecular weight complexes that overlay the parasite's cytoskeletal network and possibly anchor the IMC to the cytoskeleton (12–15). Due to the close apposition of the inner and outer IMC membranes (14, 16), it is possible that the intramembranous particles could bridge the IMC lumen and interact with the GAP-myosin complex contributing to its stabilization within the IMC.
To identify putative proteins that might be components of the intramembranous particles, we examined data from the detergent-resistant membrane (DRM) proteome of schizont-stage P. falciparum parasites containing developing merozoites (17, 18). DRMs, or lipid-rafts, were of considerable interest, because they appeared to harbor proteins involved in host cell invasion such as glycosylphosphatidylinositol (GPI)-anchored merozoite surface proteins. Our data also indicated that P. falciparum schizont-stage DRMs contained the IMC proteins PfGAP45/50 (17), and recent studies in T. gondii have also suggested that the IMC is enriched in DRMs (12). Another study indicated that when P. falciparum DRM protein complexes were separated by blue native gel electrophoresis, a band was produced containing PfGAP45/50 and PfMyo-A as well as a novel six-pass transmembrane protein (PlasmoDB: PFD1110w, GenBankTM: CAD49269) (18). This protein was related to another six-pass transmembrane DRM protein (PlasmoDB: MAL13P1.130, GenBankTM: CAD52385) we had previously identified in P. falciparum schizont-stage DRMs (17).
We show here that MAL13P1.130 and PFD1110w, termed PfGAPM1 and PfGAPM2 (glideosome-associated protein with multiple-membrane spans), respectively, belong to a family of proteins specific to the Apicomplexa and demonstrate that P. falciparum GAPM proteins, and their orthologues in T. gondii, localize to the parasite IMC. The GAPMs form high molecular weight complexes that are resistant to dissociation and solubilization by a variety of common detergents and could therefore be components of the intramembranous particles seen in electron microscopy. When isolated by immunoprecipitation, the GAPM complexes co-purify with components of the actin-myosin motor and particularly the parasite cytoskeletal network suggesting GAPMs could anchor the IMC to the cytoskeleton and perhaps even play a role in tethering the motor to cytoskeleton.
EXPERIMENTAL PROCEDURES
Sequence and Phylogenetic Analysis
Orthologues of PfGAPM1, -2, and -3 were identified by BLASTP searches (E-value <0.01) of the following databases; NCBI Non-Redundant data base, PlasmoDB, GeneDB, and OrthoMclDB and are listed in supplemental Table S1. The GAPM proteins were aligned with MUSCLE (19), evolutionary distances were estimated under the JTT model, and neighbor joining trees were constructed with Phylip (J. Felsenstein, Phylogeny Inference Package version 3.6, Dept. of Genome Sciences, University of Washington, Seattle). Bootstrap support was also calculated.
GAPM Plasmid Construction
gapm genes were PCR-amplified using cDNA extracted from either strain 3D7 P. falciparum parasites or strain RH T. gondii parasites (primers listed in supplemental Table S2). Products were ligated into the pGEM T-easy vector (Promega) and were verified by sequencing. The P. falciparum gapm genes were then excised with PstI and ligated into the PstI site of a modified version of pTGFP-GPI (20, 21), called pTM2HA and pTM2GFP to create gene fusions with a double HA epitope and GFP, respectively, called pPfGAPM1/2/3-HA and pPfGAPM1/2/3-GFP (supplemental Fig. S2). Tggapm1a, Tggapm2b, and Tggapm3 were excised from pGEM T-easy with BglII and AvrII and ligated into pCTCh3H to create a gene fusion with mCherry and the triple HA epitope called pTggapm1/2/3-ChFP-HA (supplemental Fig. S2). pCTCh3H is a derivative of pCTG in which GFP has been replaced with the mCherry/HA fusion (22).
Parasite Culture
P. falciparum strain 3D7 parasites were transfected with either 100 μg of pPfGAPM1/2/3-HA or pPfGAPM1/2/3-GFP and cultured continuously (23). Transfectants were selected with 10 nm WR99210 (24). T. gondii strain-RH parasites were transfected with pTgGAPM1/2/3-ChFP-HA plasmids using standard conditions (25) and cultured in human foreskin fibroblasts and vero cells. Addition of 20 μm chloramphenicol to cultures permitted selection of stable transgenic parasites.
Western Blot Analyses
Schizont stage P. falciparum parasites were treated with 0.15% saponin in RPMI media to release hemoglobin from the red blood cells. The hemoglobin was removed by washing the parasite pellet three times in cold phosphate-buffered saline. Parasites were solubilized at room temperature for 10 min in either 1% Triton X-100, RIPA (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 25 mm Tris-HCl, pH 8, in phosphate-buffered saline), 2% SDS, or two-dimensional sample buffer (7 m urea, 2 m thiourea, 2% ASB-14). Soluble proteins were separated from the insoluble by centrifugation in a microcentrifuge at 13,000 rpm for 10 min. A portion of the soluble fractions as well as the insoluble pellet fractions (washed in phosphate-buffered saline) were incubated in two-dimensional sample buffer overnight at room temperature to further extract the GAPMs. Prior to electrophoresis protein samples were mixed with reducing SDS-PAGE sample buffer at 1× concentration (0.05 m Tris-HCl, pH 6.8, 10% glycerol, 2 mm EDTA, 2% SDS, 0.05% bromphenol blue, 100 mm dithiothreitol). Samples not containing urea were heated at 80 °C for 10 min prior to SDS-PAGE. T. gondii tachyzoites were solubilized by sonication in RIPA buffer and soluble fractions were subsequently treated with either reducing SDS-PAGE sample buffer or reducing two-dimensional sample buffer. Samples were electrophoresed in pre-cast 4–12% acrylamide gradient bis-tris gels (Invitrogen) and blotted onto nitrocellulose or polyvinylidene difluoride prior to probing with specific primary antibodies; mouse anti-HA (mAb 12CA5) (1:500), rabbit anti-ALV repeat (ARILKPLIQEKIVEIMKPEIEEKIIEVPQVQYIEKLVEVPHVILQEKLIHIPKPVIHERIKKCSKTIFQEKIVEVPQIKVVDKIVEVPQYVYQEKIIEVPKIMVQERIIPVPKKIVKEKIVEIPQIELKNIDIEKVQEIPEYIPE, 1:100), rabbit anti-GFP (a gift from Emanuella Handman, 1:500), rabbit anti-PfGAP45 (1:200) (26), rabbit anti-PfGAP50 (1:100) (26), rabbit anti-aldolase (1:500) (26), rabbit anti-SAG1 (1:1000) (a gift from David Sibley), or rabbit anti-TgGAP45 (1:2000) (a gift from Con Beckers). 700 and 800 nm secondary antibodies were all from Rockland Immunochemicals. Bound antibody probes were detected with LiCor Odyssey infrared imager.
Microscopy
Live cell microscopy was performed on P. falciparum transfectants expressing PfGAPM1/2-GFP and T. gondii RH-strain transfectants expressing TgGAPM1/2/3-ChFP-HA stained with 1 mm 4,6-diamidino-2-phenylindole. For immunofluorescence microscopy, PfGAPM1/3-HA parasites were fixed in either ice-cold acetone/methanol or 4% paraformaldehyde and probed with the following primary antibody combinations; mouse anti-HA (mAb 12CA5, 1:50) and either rabbit anti-GAP45 (1:50) (26) or rabbit anti-MSP119 (1:50). Secondary antibodies were Alexa Fluor 568 nm goat anti-mouse IgG (1:2000, Molecular Probes) and Alexa Fluor 488 nm goat anti-rabbit IgG (1:2000, Molecular Probes). TgGAPM-ChFP-HA transfectants were treated with Clostridium septicum α-toxin as previously described (27). Treated parasites were fixed in 4% paraformaldehyde and probed with mouse anti-HA mAb 12CA5 (1:200) and rabbit anti-SAG1 (1:200) (a gift from David Sibley).
Affinity Purification Assays
For co-immunoprecipitation assays, saponin-lysed PfGAPM1-HA, PfGAPM2-GFP, PfGAPM3-HA, and control 3D7 parasites as well as TgGAPM1/2/3-HA-ChFP and control RH strain parasites (100 μl pellets), were solubilized by sonication in RIPA buffer (2 ml) with Complete protease inhibitor mixture (Roche Applied Science). Insoluble material was pelleted, and either goat-polyclonal anti-HA agarose beads (AbCam, 50–100 μl) or rat anti-GFP-agarose beads (Medical and Biological Laboratories, 50–100 μl) were added to the supernatant, and the mixture was incubated at 4 °C for 3–4 h. For immunoprecipitation assays with parasite-specific antibodies, soluble 1% Triton X-100 lysates of the PfGAPM1-HA, PfGAPM2-GFP, and PfGAPM3-HA parasites were prepared as described above. Rabbit anti-PfGAP45 (10 μl) or rabbit anti-ALV repeat (10 μl) was added to the lysate for 16 h at 4 °C. Sheep anti-rabbit Dynal beads (Invitrogen) were added to the lysates, and the mixture was incubated at 4 °C for 3 h. Bound proteins were eluted, reduced in two-dimensional sample buffer or 2% SDS, and analyzed by Western blotting as described as above.
Cross-linking Analyses
Saponin-lysed P. falciparum PfGAPM1-HA, PfGAPM2-GFP, or PfGAPM3-HA schizont stage parasites were prepared as described above (∼20-μl pellets) and resuspended in 0.4 ml of phosphate-buffered saline containing 0.5 mm dithiobis succinimidyl propionate for 30 min. Cross-linkers were later quenched in 100 mm NaCl, 25 mm Tris, pH 7.5. Cross-linked pellets were resuspended in two-dimensional sample buffer, and soluble fractions were subsequently either reduced (addition of 100 mm dithiothreitol) or left non-reduced.
RESULTS
The GAPM Family Is Restricted to the Apicomplexa and Comprises Three Phylogenetically Distinct Groups
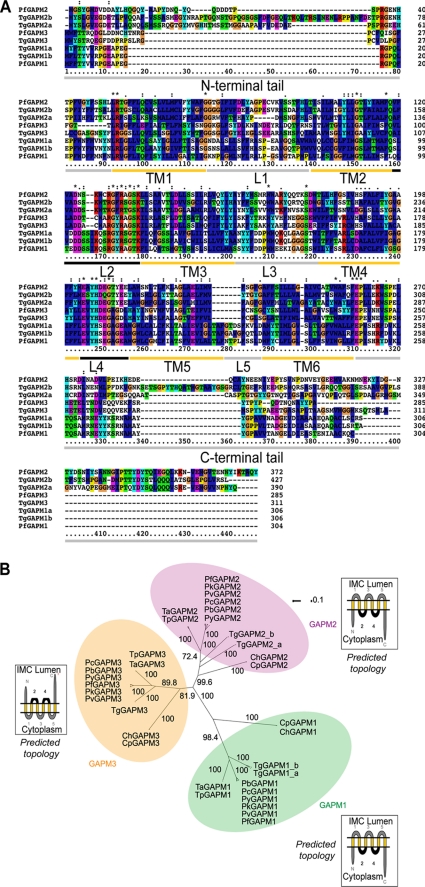
Two hypothetical proteins PfGAPM1 (MAL13P1.130) and PfGAPM2 (PFD1110w), with six transmembrane domains were previously identified in DRMs extracted from developing P. falciparum schizont/merozoite lysates (17, 18). Alignment of the amino acid sequences of these proteins revealed a low to moderate degree of similarity (29.9% similar and 18.1% identical) (Fig. 1A). Sequence similarity searches of the Plasmodium genome data base (28) indicated that P. falciparum encoded an additional member of the family termed PfGAPM3 (PlasmoDB: PF14_0065, GenBankTM: AAN36677). All three proteins were predicted by TMHMM (29) to have six transmembrane domains with the N- and C-terminal tails and loops 2 and 4 facing the cytoplasmic side of the membrane for the GAPM1 and -2 and the opposite orientation for GAPM3 (Fig. 1B).
FIGURE 1.
GAPMs form an Apicomplexan-specific protein family characterized by six transmembrane domains and short cytoplasmic loops/tails. A, a multiple alignment of the protein sequences of the P. falciparum and T. gondii GAPM sequences with their amino acids shaded according to their side chain characteristics. Transmembrane domains were identified using TMHMM (version 2.0) (29). TM, transmembrane domain; L, loop. B, neighbor-joining tree constructed using all available amino acids sequences of GAPM family members. Evolutionary distances are estimated under the JTT model. Calculated bootstrap values are indicated on significant branches. Each GAPM protein falls into one of three distinct orthologous groups; GAPM1, -2 or -3. Diagrams indicating predicted GAPM membrane orientation predicted by TMHMM are shown beside each GAPM group.
Subsequent searches of the Apicomplexan genome data base (30) revealed that most other Apicomplexan genomes also encode three of the GAPM proteins. Phylogenetic analysis of their sequences indicated that each of these proteins falls into one of three distinct orthologous groups, which we have called GAPM1, GAPM2, and GAPM3 (Fig. 1B). Interestingly, T. gondii encodes five GAPMs, two each in groups GAPM1 (TgGAPM1a and TgGAPM1b; GenBankTM: EEB00395 and EEB00396) and GAPM2 (TgGAPM2a and TgGAPM2b; GenBankTM: EEB03551 and EEB00710) and one GAPM3 protein (TgGAPM3; GenBankTM: EEA98700) (Fig. 1). Protein similarity searches of GenBankTM revealed no other obvious homologs of the GAPM family outside the Apicomplexan phylum.
GAPM sequence alignment revealed interesting patterns of amino acid conservation across the entire GAPM family as well as within the three groups (Fig. 1 and supplemental Fig. S1). Inspection of the residues forming the five short loops separating the six transmembrane domains reveals that loops two and four are highly conserved across all Apicomplexa. Comparatively, loops 1, 3, and 5 are less conserved. Such a high level of conservation in loops 2 and 4 suggests that these residues may assist in maintaining overall GAPM integrity or facilitating interactions with other proteins. Interestingly, the N and C termini appear to be highly conserved within but not between each of the three GAPM groups and may potentially determine group specific functions.
GAPMs Localize to the IMC
To facilitate fluorescent and immunodetection of GAPMs in the absence of antibodies to the endogenous proteins, members from each of the three groups in both P. falciparum and T. gondii were tagged at their C terminus with either green fluorescent protein (GFP), cherry fluorescent protein (ChFP) (31) and/or a hemagglutinin epitope tag (HA). For tagging the GAPMs in P. falciparum, cDNA sequences were ligated in front and in-frame with the sequences of GFP or a double HA tag. The gene fusions were inserted into a modified version of the pTGFP-GPI plasmid (21) (supplemental Fig. S2). In this plasmid system the expression of gene fusions can be suppressed by the addition of the tetracycline analogue anhydrotetracycline and induced by removing the drug. Once induced, the gene fusions come under transcriptional control of a transactivator protein (TATi2) whose expression is in turn regulated by a schizont blood stage merozoite surface protein 2 promoter (20, 21). Examination of the microarray gene transcription profile of the Pfgapms confirmed the endogenous genes were also expressed during schizogony (32, 33). Once parasite lines transfected with the Pfgapm gene fusion plasmids were established, anhydrotetracycline was removed and those expressing the GFP fusion proteins were examined by microscopy. GFP fluorescence was only observed for the PfGAPM1-GFP and PfGAPM2-GFP fusions and initially only in a low percentage of schizont and merozoite stage parasites. Over several weeks however, this percentage increased greatly even in the presence of anhydrotetracycline, which should have silenced expression. This indicated that the plasmids containing the PfGAPM-GFP fusion may have recombined into the Pfgapm chromosomal locus tagging the endogenous gene with GFP. Southern blot analysis of PfGAMP2-GFP parasites confirmed this (supplemental Fig. S2). The PfGAPM1-GFP parasites grew poorly and no further work was done with this line. Our attention instead turned to the HA-tagged PfGAPM transfectants. These were examined by immunofluorescence microscopy with a HA monoclonal antibody (12CA5 mAb; used for all subsequent experiments) after several weeks in culture. Most schizont/merozoites expressed the PfGAPM1-HA fusion protein and Southern blot analysis also confirmed integration into the endogenous locus (supplemental Fig. S2). PfGAPM3-HA-expressing parasites were similarly produced, but integration was not confirmed by Southern blot analysis. Although transfected lines containing the PfGAPM2-HA and PfGAPM3-GFP plasmids were generated, the resultant lines did not express detectable levels of fusion proteins possibly due to some unforeseen deleterious effects.
To determine in which cellular membrane the GAPMs resided, P. falciparum schizonts were examined by immunofluorescence microscopy. In Plasmodium asexual blood stages, cell replication occurs by schizogony where several rounds of nuclear division and associated organelle development occur within a common cytoplasm bound by a single plasma membrane. Daughter merozoites are formed late in schizogony when cytokinesis pinches off the individual merozoites. We microscopically examined immature PfGAPM1-HA and PfGAPM3-HA transgenic schizonts, because the multiple forming IMCs are distinct from the single bounding plasma membrane. Parasites were labeled with anti-HA and either the IMC-specific PfGAP45 rabbit antibody or parasite plasma membrane-specific rabbit antibody for the C-terminal domain of merozoite surface protein 1 (MSP119). The localization pattern of PfGAPM1-HA and PfGAPM3-HA was indistinguishable from that of PfGAP45 and markedly distinct from that of known plasma membrane protein MSP1 (Fig. 2A and supplemental Fig. S4). Additionally, live fluorescence microscopy of PfGAPM1-GFP and PfGAPM2-GFP transgenic schizonts demonstrated an identical IMC expression pattern to that of PfGAPM1-HA and PfGAPM3-HA throughout schizogony and a developmental series of images is shown in supplemental Fig. S3.
FIGURE 2.
GAPM proteins localize to the IMC. A, immunofluorescence images of PfGAPM1-HA and PfGAPM3-HA expressing P. falciparum schizonts probed with an anti-HA antibody and antibodies to either the IMC-resident protein PfGAP45 or the MSP119 subunit of the plasma membrane protein MSP1. PfGAPM1-HA and PfGAPM3-HA co-localize with PfGAP45 but not with MSP119. B, TgGAPM-ChFP-HA-expressing parasites were treated with C. septicum α-toxin to swell the parasite membrane away from the IMC. Probing fixed parasites with anti-HA and the parasite membrane marker SAG1 validates that TgGAPM proteins reside in the IMC. Scale bar, 1 μm.
To ascertain if the GAPMs localize to the IMC in other Apicomplexa, the GAPMs of T. gondii were investigated by live fluorescence microscopy. The cDNA sequences of Tggapm1a, Tggapm2b, and Tggapm3 were N-terminally joined to a fusion of both cherry fluorescent protein and the HA epitopes (ChFP-HA). The gene fusions were ligated into the pCTCh3HA plasmid under the control of the α-tubulin promoter expressed during the tachyzoite stage (supplemental Fig. S2). After transfection, ectopic expression of the fusion proteins was observed near the surface of tachyzoites developing within mammalian host cells (supplemental Fig. S5). The GAPMs were particularly concentrated in membranous organelles of the developing daughter cells suggestive of IMC localization (supplemental Fig. S5). To confirm this, T. gondii GAPM-ChFP-HA transgenic parasites were treated with Clostridium septicum α-toxin, which swells the parasite membrane away from the underlying IMC (27). Treated transgenic parasites were fixed and labeled with antibodies specific to the parasite plasma membrane (SAG1) and the TgGAPM-ChFP-HA fusion protein (anti-HA). Upon fluorescence microscopy, treated parasites exhibited markedly swollen outer membranes, whereas TgGAPM-ChFP-HA labeling was only apparent in the normal crescent shape of the parasite confirming IMC localization (Fig. 2B).
GAPM Proteins Are Present in Large and Extremely Stable Complexes
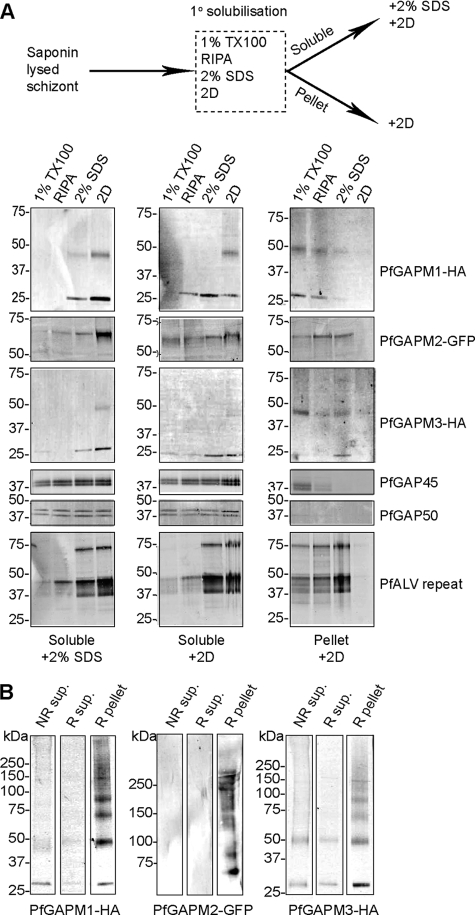
To ensure that the GAPM fusion proteins were full-length and had been tagged as predicted, the P. falciparum schizonts expressing these proteins were examined by Western blot analysis. The parasitized red blood cells were firstly treated with saponin to remove host cell proteins, and the remaining schizonts were then solubilized in standard SDS-PAGE protein sample buffer containing 2% SDS and the reducing agent dithiothreitol. Western blots of the proteins revealed monomeric bands slightly smaller than the sizes expected (PfGAPM1-HA: 36.6 kDa, PfGAPM2-GFP: 65.2 kDa, and PfGAPM3-HA: 34 kDa) as well as high molecular mass species particularly for PfGAPM1-HA (data not shown). The resistance of these proteins to SDS dissociation prompted further investigation. PfGAPM1-HA-, PfGAPM2-GFP-, and PfGAPM3-HA-expressing schizonts were saponin treated as described above and were then solubilized in four detergent/denaturant mixes of differing strengths. From weakest to strongest the mixes were; 1) 1% of the non-ionic detergent Triton X-100; 2) RIPA buffer containing 1% Triton X-100, 1% of the weak ionic detergent deoxycholate, and 0.1% of the strong ionic detergent SDS; 3) 2% SDS; and 4) two-dimensional sample buffer containing the denaturant urea. Soluble and insoluble material were separated by centrifugation and prior to fractionation by SDS-PAGE, the soluble fraction was mixed with standard 2% SDS sample buffer or two-dimensional sample buffer (Fig. 3A). The insoluble pellet material was treated with two-dimensional sample buffer only. Immunoblots of the proteins probed with their corresponding antibodies indicated the solubility of PfGAPM1-HA was poor in Triton X-100, better in RIPA buffer, and substantially increased with the stronger 2% SDS and two-dimensional buffers (Fig. 3A). PfGAPM2-GFP was the most easily extracted and was mostly solubilized in all buffers (Fig. 3A). PfGAPM3-HA was also solubilized by all the buffers; however, the stronger 2% SDS and two-dimensional buffers were required for best extraction (Fig. 3A). The solubility of other IMC-associated proteins was also investigated. Actin-myosin motor components PfGAP45 and PfGAP50 were easily solubilized by all detergent treatments and only a small amount of these proteins remained in the insoluble pellet fractions in Triton X-100 and RIPA (Fig. 3A).
FIGURE 3.
P. falciparum GAPM proteins are present in oligomeric complexes resistant to disassociation. A, Western blots of schizont stage transgenic parasites solubilized with detergent mixes of different strengths (1% TX-100 < RIPA < 2% SDS < 2D) indicate that PfGAPM1-HA is the most resistant to solubilisation, PfGAPM3-HA is intermediate and PfGAPM2-GFP the most easily solubilized. Other known IMC proteins, PfGAP45 and PfGAP50, are easily solubilized by all detergent treatments. Cytoskeletal ALVs are most readily solubilized by the harsher detergent treatments. B, cross-linking parasite proteins with DSP indicated that the PfGAPMs form insoluble high molecular weight oligomers. Mono- and dimeric forms of the PfGAPMs are more soluble, and the majority of the GAPM protein remained in the insoluble pellet fraction. NR, non-reduced; R, reduced.
To investigate the solubility of the IMC-associated cytoskeleton, antisera were raised to a repeat region common to the alveolin family of cytoskeletal proteins characteristic of the alveolates, a vast group of organisms that include apicomplexa, dinoflagellates, and ciliates (34). The Plasmodium alveolin antibody (PfALV) recognized three major bands that migrated at 75, 42, and 37 kDa, which probably correspond to the alveolins PfALV2, PfALV4, and PfALV5, respectively (34), that microarray data predict are expressed in blood stages (35, 32). The predicted masses of these blood-stage alveolins are 62, 34, and 30 kDa, respectively, so the proteins migrate more slowly than expected, which has been reported previously for the alveolins of other species (34). Interestingly, the solubility of the blood-stage alveolins was similar to PfGAPM1 and -3 in that they were only partially solubilized with the weaker Triton X-100 and RIPA detergent mixes and required the harsher 2% SDS and two-dimensional sample buffers for better extraction (Fig. 3A).
The resistance of some of the PfGAPM proteins to solubilization in mild detergent mixes and to complete dissociation in SDS-containing buffers suggested that the proteins could be components of large, stable complexes within the IMC. To investigate this further, live PfGAPM schizont stage parasites were cross-linked with the reduction sensitive, cell-permeable cross-linker dithiobis succinimidyl propionate (DSP) after host cell proteins had been removed by saponin lysis. The parasites were then solubilized in two-dimensional urea sample buffer without a reducing agent, and the insoluble material was pelleted by centrifugation. Dithiothreitol was then added to partially reduce the DSP cross-links in the insoluble pellet and also to half of the soluble fraction. Western blots of the material probed with anti-HA and anti-GFP indicated that DSP cross-linking had caused most PfGAPM proteins that had previously been soluble to become insoluble (Fig. 3B). PfGAPM1-HA and PfGAPM3-HA, which previously had been efficiently reduced to monomer and dimer in two-dimensional urea sample buffer, now contained many larger bands that appeared to increase in increments of 28 kDa suggesting that the proteins were derived from partially reduced homo-oligomeric complexes of up to 150 kDa (pentameric) (Fig. 3B). PfGAPM2-GFP was also only present in DSP-treated pellet samples as a high molecular weight smear suggesting that it too forms poorly soluble macromolecular complexes when cross-linked (Fig. 3B). The only PfGAPM proteins that were present in the soluble fractions were mostly low molecular weight monomers and dimers. These could have been reduced and then dissociated from large complexes or might have been derived from newly synthesized proteins that had not yet assembled into the IMC complexes.
Similar experiments were completed to investigate the solubility of GAPM proteins in T. gondii (supplemental Fig. S6). These data revealed that the Toxoplasma proteins follow similar trends to those of Plasmodium in not only migrating more quickly than expected but also in their respective resistance to breakdown into monomers. TgGAPM1, like PfGAPM1, is the most resistant, TgGAPM2 and PfGAPM2 are the least resistant, and TgGAPM3 and PfGAPM3 are intermediate (Fig. 3A and supplemental Fig. S6). Overall, however, the Toxoplasma GAPM aggregates appeared more resistant to breakdown than their Plasmodium counterparts (supplemental Fig. S6).
GAPM Proteins Interact with Components of the Actin-myosin Motor and Cytoskeletal Network
The stability of GAPM protein complexes, in addition to their large size, suggested that they may play a structural role within the IMC such as pinning the membranes together, anchoring the actin-myosin motor or linking the IMC to the cytoskeleton. To explore potential interactions between the GAPM proteins and other parasite proteins, PfGAPM1-HA was immunoprecipitated from schizont-stage parasites in RIPA buffer rather than 1% Triton X-100, because it solubilized the GAPM protein yet still supported interaction between the HA-epitope and the commercial anti-HA beads. Parallel control experiments were performed with wild-type 3D7 parasites, and the immunoprecipitates were resolved by SDS-PAGE (supplemental Fig. S7). Protein bands unique to PfGAPM1-HA but not 3D7 were excised, and their contents identified by mass spectroscopy-based protein sequencing (supplemental Fig. S7). Apart from PfGAPM1, about a third of the proteins identified were known IMC-associated proteins that included GAP50, actin, PfGAPM2, and ALV4. Another protein identified was 6-phosphofructokinase. Although this abundant glycolytic enzyme could be a contaminant, it has recently been shown that, in Toxoplasma, glycolytic enzymes specifically associate with the IMC prior to host cell egress to presumably provide ATP to the actin-myosin motor (16). The same could be true in Plasmodium. Of the remaining proteins precipitating with PfGAPM1-HA, about a third were involved in vesicle-mediated trafficking and could have been involved in IMC formation. The remaining parasite proteins were highly expressed probably nonspecific contaminants.
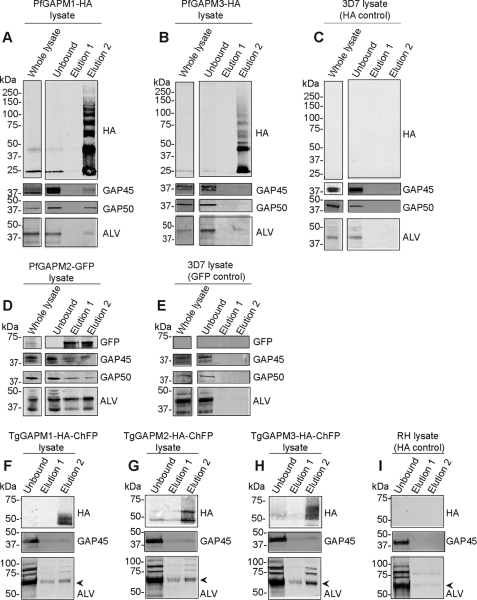
To validate the interactions of the GAPMs with the actin-myosin motor and alveolin network, Western blots of GAPM immunoprecipitates were probed with protein-specific antibodies. For P. falciparum, all precipitations were performed with anti-HA beads except for PfGAPM2-GFP in which anti-GFP beads were used (Fig. 4, A–E). Western blots probed with either the anti-HA or the rabbit anti-GFP antibody confirmed that the immunoprecipitations were successful in PfGAPM parasites but not in 3D7 control parasites (Fig. 4, A–E). To detect if other IMC proteins were co-precipitating, the blots were probed with rabbit anti-PfGAP45 and anti-PfGAP50. These antibodies produced positive signals in the PfGAPM precipitations but not in the 3D7 control reactions (Fig. 4, A–E). The strongest GAP45 and GAP50 co-immunoprecipitations were in the PfGAPM1-HA and PfGAPM2-GFP samples and the weakest in PfGAPM3-HA (Fig. 4A, B and D). It is worth noting that, whereas >90% of the GAPM proteins were often precipitated from the sample, usually <10% of the GAP45/50 proteins were precipitated. This suggested that only a minor proportion of the actin-myosin motor components might be interacting with the PfGAPMs or that the interactions between the proteins might have been destabilized by the detergents used.
FIGURE 4.
GAPM proteins interact with components of the actin-myosin motor and subpellicular network. PfGAPM1-HA (A) and PfGAPM3-HA (B) were immunoprecipitated with anti-HA beads from parasites expressing the tagged proteins. To ensure the precipitations worked, Western blots were probed with anti-HA. Probing the precipitates with rabbit anti-GAP45, anti-GAP50, and anti-ALV repeat indicated that these proteins were binding to PfGAPM1-HA and to a lesser extent to PfGAPM3-HA. D, PfGAPM2-GFP-expressing parasites were immunoprecipitated with anti-GFP beads and Western blots were probed as previously described. GAP45 and -50 and particularly ALV4 appeared to bind to PfGAPM2-GFP. C and E, the validity of these interactions was supported by failure to precipitate any of these proteins from wild type 3D7 parasites with anti-HA and anti-GFP beads. F–H, anti-HA immunoprecipitations were performed on tachyzoites expressing TgGAPM-1/2/3-HA-ChFP and Western blots of these probed with anti-HA certified the precipitations worked. The Western blots were also probed with anti-GAP45 and anti-ALV repeat, which indicated these proteins co-immunoprecipitated with the TgGAPMs. I, control immunoprecipitations with wild-type RH parasites and anti-HA beads were unsuccessful for GAP45 but small amounts of a protein that bound the ALV-repeat antibody was precipitated. The arrowhead indicates a non-specifically binding ALV band.
Interactions of PfGAPM with the alveolins were also investigated by probing with anti-ALV. This antibody detected a 42-kDa band in precipitates probably corresponding to PfALV4 in all three GAPMs (Fig. 4, A–E). An additional ∼35-kDa band was observed in the PfGAPM2-GFP precipitation that could be ALV5 (Fig. 4D). PfGAPM1, PfGAPM2, and PfGAPM3, respectively, precipitated ∼17%, 26%, and 16% of the total ALV detected in the sample. A degree of specificity was therefore observed between the PfGAPMs with PfGAPM2-GFP precipitating the most ALV4, as well as the additional alveolin, ALV5 (Fig. 4D).
When probed with anti-HA, a ladder of proteins ranging from 28 kDa to 150 kDa was seen in the second elution for both PfGAPM1-HA and PfGAPM3-HA immunoprecipitations (Fig. 4, A and B). The origins of these ladders are unclear because the two-dimensional sample buffer used to solubilize the eluted proteins should have dissociated the complexes. We speculate that the high concentration of PfGAPM proteins collecting on the anti-HA agarose beads during the immunoprecipitation may have caused the hydrophobic PfGAPM proteins to aggregate, further increasing their resistance to complete denaturation into monomers and dimers.
To investigate whether T. gondii GAPMs also co-immunoprecipitated members of the actin-myosin motor, transgenic T. gondii parasites expressing C-terminally ChFP/HA-tagged GAPM proteins were used in immunoprecipitation assays as above. These samples were subsequently analyzed by Western blotting that revealed each TgGAPM investigated also co-immunoprecipitated TgGAP45 while RH-strain control parasites did not. Additionally, ALV-specific bands were detected at 62, 74, 82, and 92 kDa for each TgGAPM (Fig. 4, F–H); however, because minor amounts of the 62-kDa band were also detected in RH-strain control parasites, this might not represent a genuine TgGAPM-specific interaction. Toxoplasma encodes nearly twice as many alveolins as P. falciparum (12 versus 7) so it is not possible to speculate which specific alveolins were interacting with the TgGAPM1/2/3-ChFP-HA proteins. We note however that similar sized alveolins were previously detected with a TgALV3 antibody (34).
As was seen for PfGAPM immunoprecipitations, only a small fraction of the total TgGAP45 and ALV proteins were detected in the immunoprecipitated material. We were unable to investigate proposed TgGAPM interactions with TgGAP50 due to a lack of antibody to this protein.
To validate the proposed interactions between the GAPM proteins and the actin-myosin motor, reciprocal immunoprecipitation experiments were performed. Anti-ALV and anti-GAP45 antibodies were found to interact poorly with their antigens in RIPA buffer (data not shown), therefore reciprocal immunoprecipitations were performed in 1% Triton X-100. Tagged P. falciparum GAPM schizonts were solubilized and incubated with either rabbit anti-ALV or rabbit anti-PfGAP45. Immune complexes were precipitated with anti-rabbit conjugated Dynal beads. Antibodies specific to PfGAP45 co-precipitated small amounts of PfGAPM2 and PfGAPM3 (Fig. 5, B and C). Slightly larger amounts of these proteins were co-precipitated with anti-ALV suggestive of a more extensive interaction between the GAPMs and the ALVs than between the GAPMs and PfGAP45 (Fig. 5, B and C). PfGAPM1 was co-immunoprecipitated poorly with ALV and GAP45 probably due to its poor solubility in Triton X-100 (Fig. 5A).
FIGURE 5.
Reciprocal immunoprecipitation assays indicate that the alveolins and GAP45 interact with GAPM proteins. Western blots of alveolin and GAP45 immunoprecipitation assays on lysates from P. falciparum schizonts expressing either PfGAPM1-HA (A), PfGAPM2-GFP (B), or PfGAPM3-HA (C) were probed with antibodies listed in individual panels. Probing with anti-ALV or anti-GAP45 confirmed that immunoprecipitations were successful. Probing with either anti-HA (A and C) or anti-GFP (B) indicated that each GAPM co-purified with the alveolins and GAP45. The arrow represents the anti-ALV IgG band.
DISCUSSION
The GAPMs identified in this study constitute a three-member family of six-pass transmembrane proteins present in all sequenced Apicomplexa. In the groups tested here, namely Plasmodium blood-stage merozoites and Toxoplasma tachyzoites, the GAPMs localize to the IMC implying that the IMCs of all motile Apicomplexan zoite stages will contain these proteins. No related proteins are present outside of this phylum suggesting that GAPMs perform vital, Apicomplexan-specific functions.
The tagged GAPMs interact with the cytoskeletal alveolins and to a slightly lesser degree the actin-myosin motor components GAP45/50. This suggests that the GAPMs might function as protein “rivets” that attach the IMC to the cytoskeleton or even as protein tethers that span the IMC to link and immobilize the myosin complex onto the cytoskeleton (Fig. 6). If true, then why have previous immunoprecipitation-based attempts to identify anchors of the GAP45/50/myosin complex failed to identify the GAPMs (11, 12)? We believe these attempts may have failed because the interactions of the GAPMs with GAP45/50 do not survive the immunoprecipitation process well and the GAPMs are difficult to resolve in monomeric form by SDS-PAGE and to detect without specific reagents. With six transmembrane-spanning domains and the size of the complexes they form (>200 kDa) the GAPMs are probably firmly embedded in the IMC membrane. For immunoprecipitation to work the GAPMs have to be solubilized, which in their case requires extensive sonication in the stringent RIPA-detergent mix. Both of these treatments can disrupt protein-protein interactions, and it is no surprise that little (<10%) of the GAP45/50 and GAPM interaction survives co-immunoprecipitation. The sonication is probably necessary to shear the cytoskeleton and detach the IMC, which is also why only ∼20% of the available alveolins co-precipitate with the GAPMs.
FIGURE 6.
Proposed model of GAPM interaction. GAPM proteins reside within the IMC and have been shown to interact with components of the actin-myosin motor and subpellicular network. The plus signs represent the solubility of each of the proteins in each buffer as judged by Western blotting: +, proteins readily soluble in listed buffer; ++++, proteins poorly soluble in listed buffer.
To validate interaction of the GAPMs with the alveolins and GAP45/50 we attempted reciprocal immunoprecipitations with anti-PfGAP45 in P. falciparum, anti-TgGAP45 in Toxoplasma, and anti-ALV in both species. We found these immunoprecipitations worked poorly in RIPA, because the stringent buffer reduced interaction of the antibodies with their ligands. These precipitations were therefore repeated in 1% Triton X-100. In P. falciparum this detergent easily extracted GAP45/50 but only poorly the PfGAPMs, especially PfGAPM1. This probably accounts for why the GAPMs were only poorly co-precipitated with GAP45 and alveolin, with much of the GAPMs remaining in the pellet after detergent extraction. Previous attempts to identify GAP45/50/myosin-tethering proteins in Toxoplasma were probably unsuccessful because they were performed with Triton X-100, and the GAPMs in this parasite are even more insoluble than in P. falciparum (11). Chemical cross-linkers were also used to identify proteins attaching to the GAP45/50/myosin complex (12) but as we have shown they cause the GAPMs to become extremely insoluble.
Even if the GAPM proteins could be co-immunoprecipitated in appreciable amounts with GAP45/50, they may have avoided detection after SDS-PAGE, because they migrated as indistinct high molecular weight aggregates. We found it necessary to treat the immunoprecipitates with two-dimensional sample buffer to dissociate the GAPMs into monomeric form. We note that, although PfGAPM2-GFP was the most easily extracted protein, this may have been assisted by the highly soluble GFP moiety that is almost as large as PfGAPM2 itself.
All of our P. falciparum immunoprecipitations with the GAPs, GAPMs, and alveolins successfully co-precipitated each other suggesting they all physically bind to each other though not necessarily in equimolar ratios. Another interpretation of our data is that small fragments of IMC containing all of these proteins are being nonspecifically precipitated, because they are resistant to mechanical and detergent disruption. These fragments would, however, have to be small, because the parasite lysates were cleared by centrifugation prior to immunoprecipitation. We also observed a certain degree of specificity with our immunoprecipitations that argues against the GAPMs pulling down IMC fragments with attached cytoskeleton. For example, PfGAPM2 specifically precipitates two alveolins, whereas the others only precipitate a single cytoskeletal protein. To absolutely resolve the IMC fragment issue would require the use of a resident IMC protein known not to bind to the proteins mentioned above as a negative control in the immunoprecipitation reactions, but unfortunately no such protein is known. In conclusion, based on the numerous times we have repeated these precipitations and the fact that some work in the stringent RIPA buffer, we are confident that GAPMs do contact both the alveolin cytoskeleton on one side of the IMC and the GAP-myosin complex on the other side (Fig. 6).
In each of the immunoprecipitation assays shown here, the strongest interactions were seen between the GAPMs and the alveolins of the subpellicular network. This network resides on the cytoplasmic face of the IMC suggesting that the GAPMs may also localize to this face. In addition, the organization of GAPMs into highly stable oligomers makes these proteins prime candidates for being constituents of the uncharacterized family of 9 nm intramembranous particles (IMPs) seen throughout the IMC in freeze-fracture studies (6, 13–15). It has been observed that IMPs localize to different parts of the IMC. A single line of IMPs overlay the cytoskeletal filaments (probably 10 nm alveolin containing filaments) (15) and a double row lays over microtubules (13). Furthermore, other types of IMPs might form pores in the IMC or may suture the edges of the flattened vesicles that comprise the IMC (14). This degree of specialization hints that if the GAPMs are indeed true IMP proteins, then each type of IMP might contain a different GAPM. To establish whether the GAPMs bind to each other we attempted to make specific antibodies to the N- and C-terminal tails of PfGAPM1 to determine if the protein co-immunoprecipitates with the GFP- and HA-tagged PfGAPM2 and PfGAPM3 proteins. Unfortunately, the PfGAPM1 antibodies performed very poorly (data not shown), so specific antibodies will have to be raised to the other GAPMs. It is worth noting, though, that when partially cross-linked, the PfGAPM1-HA and -3HA proteins migrated in precise increments of their own molecular weights hinting that they might be homo-oligomers. Sizes up to the pentamer (∼150 kDa) have been detected by immunoblot analysis of partially cross-linked material, although to form a 9 nm particle the protein complexes would probably have to be ∼500 kDa.
We have attempted immunoelectron microscopy to determine if the GAPMs are components of these 9 nm IMPs. If present, immunogold particles specific for the HA-tagged GAPMs might label the IMC with a periodicity corresponding to IMPs observed by freeze fracture microscopy. Unfortunately, the immunoelectron microscopy experiments were not successful because we could not achieve specific labeling of the developing IMC in gluteraldehyde-fixed schizont stage P. falciparum parasites (data not shown). It is possible that the gluteraldehyde used for electron microscopy may have cross-linked the GAPMs to such an extent that access to the HA epitope was greatly reduced, resulting in poor labeling. Supporting this is our finding that GAPM labeling with anti-HA or anti-GFP is greatly reduced in immunofluorescence microscopy of cells fixed in paraformaldehyde when compared with methanol/acetone (data not shown). Successful immunoelectron microscopy of the GAPMs may therefore require alternative cryo-preservation methods.
The GAPM proteins appear to form complexes, possibly homo-oligomers, though how large they are remains to be ascertained. Unless reduced to mono- or dimeric forms the GAPM proteins when in complexes appear very insoluble. The presence of GAPMs in IMPs would explain why the GAPMs poorly precipitate GAPs and alveolins. The large membrane particles may only bind to a small number of neighboring actin-myosin complexes and alveolin subunits particularly if the latter were forming a filament. Data presented here suggest that the GAPM proteins form complexes on the cytoplasmic face of the IMC where they interact with components of the subpellicular network. This localization would permit interactions with the GAP45/50 complex, because it is believed that the majority of GAP50 resides within the IMC (11) and would explain why only a small proportion of the GAP45/50 proteins can be co-immunoprecipitated with the GAPM proteins (Fig. 6). Establishing where the GAPMs reside within the IMC and what each truly interacts with will do much to determine the structural organization of this important apicomplexan organelle of motility.
Supplementary Material
Acknowledgments
We thank Jake Baum for the GAP45 and -50 antibodies, Con Beckers for the TgGAP45 antibody, and Paul Sanders and Thomas Nebl for technical advice. We also thank Giel van Dooren of the University of Georgia, Athens, for his gift of the T. gondii transfection plasmid pCTG and J. I. Rood, D. Lyras, and C. L. Kennedy of Monash University, Victoria, Australia for the kind provision of the C. septicum α-toxin. We also thank the Walter and Eliza Hall Institute's antibody production facility and the Australian Red Cross for supplying blood.
This work was supported, in part, by the National Health and Medical Research Council of Australia and Wellcome Trust, UK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1 and S2.
- IMC
- inner membrane complex
- DRM
- detergent-resistant membrane
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- RIPA
- radioimmune precipitation assay
- bis-tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- mAb
- monoclonal antibody
- ChFP
- cherry fluorescent protein
- DSP
- dithiobis succinimidyl propionate
- IMP
- intramembranous particle
- ALV
- alveolin
- GPI
- glycosylphosphatidylinositol.
REFERENCES
- 1.Snow R. W., Guerra C. A., Noor A. M., Myint H. Y., Hay S. I. (2005) Nature 434, 214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum J., Gilberger T. W., Frischknecht F., Meissner M. (2008) Trends Parasitol. 24, 557–563 [DOI] [PubMed] [Google Scholar]
- 3.Cowman A. F., Crabb B. S. (2006) Cell 124, 755–766 [DOI] [PubMed] [Google Scholar]
- 4.Tardieux I., Ménard R. (2008) Traffic 9, 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keeley A., Soldati D. (2004) Trends Cell Biol. 14, 528–532 [DOI] [PubMed] [Google Scholar]
- 6.Soldati D., Meissner M. (2004) Curr. Opin Cell Biol. 16, 32–40 [DOI] [PubMed] [Google Scholar]
- 7.Bergman L. W., Kaiser K., Fujioka H., Coppens I., Daly T. M., Fox S., Matuschewski K., Nussenzweig V., Kappe S. H. (2003) J. Cell Sci. 116, 39–49 [DOI] [PubMed] [Google Scholar]
- 8.Pinder J. C., Fowler R. E., Dluzewski A. R., Bannister L. H., Lavin F. M., Mitchell G. H., Wilson R. J., Gratzer W. B. (1998) J. Cell Sci. 111, 1831–1839 [DOI] [PubMed] [Google Scholar]
- 9.Bannister L. H., Hopkins J. M., Fowler R. E., Krishna S., Mitchell G. H. (2000) Parasitology 121, 273–287 [DOI] [PubMed] [Google Scholar]
- 10.Pinder J., Fowler R., Bannister L., Dluzewski A., Mitchell G. H. (2000) Parasitol Today 16, 240–245 [DOI] [PubMed] [Google Scholar]
- 11.Gaskins E., Gilk S., DeVore N., Mann T., Ward G., Beckers C. (2004) J. Cell Biol. 165, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson T. M., Rajfur Z., Jacobson K., Beckers C. J. (2007) Mol. Biol. Cell 18, 3039–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrissette N. S., Murray J. M., Roos D. S. (1997) J. Cell Sci. 110, 35–42 [DOI] [PubMed] [Google Scholar]
- 14.Raibaud A., Lupetti P., Paul R. E., Mercati D., Brey P. T., Sinden R. E., Heuser J. E., Dallai R. (2001) J. Struct. Biol. 135, 47–57 [DOI] [PubMed] [Google Scholar]
- 15.Mann T., Beckers C. (2001) Mol. Biochem. Parasitol. 115, 257–268 [DOI] [PubMed] [Google Scholar]
- 16.Pomel S., Luk F. C., Beckers C. J. (2008) PLoS Pathog. 4, e1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders P. R., Gilson P. R., Cantin G. T., Greenbaum D. C., Nebl T., Carucci D. J., McConville M. J., Schofield L., Hodder A. N., Yates J. R., 3rd, Crabb B. S. (2005) J. Biol. Chem. 280, 40169–40176 [DOI] [PubMed] [Google Scholar]
- 18.Sanders P. R., Cantin G. T., Greenbaum D. C., Gilson P. R., Nebl T., Moritz R. L., Yates J. R., 3rd, Hodder A. N., Crabb B. S. (2007) Mol. Biochem. Parasitol. 154, 148–157 [DOI] [PubMed] [Google Scholar]
- 19.Edgar R. C. (2004) BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilson P. R., O'Donnell R. A., Nebl T., Sanders P. R., Wickham M. E., McElwain T. F., de Koning-Ward T. F., Crabb B. S. (2008) Mol. Microbiol. 68, 124–138 [DOI] [PubMed] [Google Scholar]
- 21.Meissner M., Krejany E., Gilson P. R., de Koning-Ward T. F., Soldati D., Crabb B. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2980–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Dooren G. G., Tomova C., Agrawal S., Humbel B. M., Striepen B. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 13574–13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trager W., Jensen J. B. (1976) Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 24.Crabb B. S., Rug M., Gilberger T. W., Thompson J. K., Triglia T., Maier A. G., Cowman A. F. (2004) Methods Mol. Biol. 270, 263–276 [DOI] [PubMed] [Google Scholar]
- 25.Striepen B., He C. Y., Matrajt M., Soldati D., Roos D. S. (1998) Mol. Biochem. Parasitol. 92, 325–338 [DOI] [PubMed] [Google Scholar]
- 26.Baum J., Richard D., Healer J., Rug M., Krnajski Z., Gilberger T. W., Green J. L., Holder A. A., Cowman A. F. (2006) J. Biol. Chem. 281, 5197–5208 [DOI] [PubMed] [Google Scholar]
- 27.Wichroski M. J., Melton J. A., Donahue C. G., Tweten R. K., Ward G. E. (2002) Infect. Immun. 70, 4353–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aurrecoechea C., Brestelli J., Brunk B. P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Miller J. A., Nayak V., Pennington C., Pinney D. F., Roos D. S., Ross C., Stoeckert C. J., Jr., Treatman C., Wang H. (2009) Nucleic Acids Res. 37, D539–D543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 30.Aurrecoechea C., Heiges M., Wang H., Wang Z., Fischer S., Rhodes P., Miller J., Kraemer E., Stoeckert C. J., Jr., Roos D. S., Kissinger J. C. (2007) Nucleic Acids Res. 35, D427–D430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giepmans B. N., Adams S. R., Ellisman M. H., Tsien R. Y. (2006) Science 312, 217–224 [DOI] [PubMed] [Google Scholar]
- 32.Bozdech Z., Llinás M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. (2003) PLoS Biol. 1, E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Roch K. G., Johnson J. R., Florens L., Zhou Y., Santrosyan A., Grainger M., Yan S. F., Williamson K. C., Holder A. A., Carucci D. J., Yates J. R., 3rd, Winzeler E. A. (2004) Genome Res. 14, 2308–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gould S. B., Tham W. H., Cowman A. F., McFadden G. I., Waller R. F. (2008) Mol. Biol. Evol. 25, 1219–1230 [DOI] [PubMed] [Google Scholar]
- 35.Le Roch K. G., Zhou Y., Blair P. L., Grainger M., Moch J. K., Haynes J. D., De La Vega P., Holder A. A., Batalov S., Carucci D. J., Winzeler E. A. (2003) Science 301, 1503–1508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.