Abstract
Human FBP21 (formin-binding protein 21) contains a matrin-type zinc finger and two tandem WW domains. It is a component of the spliceosomes and interacts with several established splicing factors. Here we demonstrate for the first time that FBP21 is an activator of pre-mRNA splicing in vivo and that its splicing activation function and interaction with the splicing factor SIPP1 (splicing factor that interacts with PQBP1 and PP1) are both mediated by the two tandem WW domains of group III. We determined the solution structure of the tandem WW domains of FBP21 and found that the WW domains recognize peptide ligands containing either group II (PPLP) or group III (PPR) motifs. The binding interfaces involve both the XP and XP2 grooves of the two WW domains. Significantly, the tandem WW domains of FBP21 are connected by a highly flexible region, enabling their simultaneous interaction with two proline-rich motifs of SIPP1. The strong interaction between SIPP1 and FBP21 can be explained by the conjugation of two low affinity interactions with the tandem WW domains. Our study provides a structural basis for understanding the molecular mechanism underlying the functional implication of FBP21 and the biological specificity of tandem WW domains.
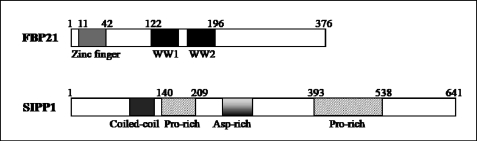
Gene expression in eukaryotic cells involves several steps, including transcription, mRNA processing, and export. Pre-mRNA splicing takes place in the spliceosome, a highly dynamic ribonucleoprotein particle that consists of five small nuclear RNAs and at least 150 proteins. Small nuclear ribonucleoproteins (snRNPs)3 and numerous protein factors are essential for the formation of the active spliceosome (1, 2). In budding yeast, the splicing factor Prp40 participates in cross-intron bridging by interacting with the branch point-binding protein (BBP) and the U5 snRNP component Prp8. Prp40 contacts the 5′ splice site and interacts with BBP, bringing the 5′ splice site and the branch point in spatial proximity. These interactions are believed to be conserved in mammals (3–5). FBP21 (formin-binding protein 21), the mammalian Prp40-like protein, colocalizes with splicing factors in nuclear storage sites for pre-mRNA splicing factors. In addition, FBP21 is a component of the mammalian spliceosomal A/B complex and is associated with U2 snRNPs (6). FBP21 interacts directly with the splicing factors U1 snRNP protein U1C, the core snRNP proteins SmB and SmB′, and the branch point-binding protein SF1/mBBP, suggesting that it may also play a role in cross-intron bridging of U1 and U2 snRNPs in the spliceosomes. FBP21 contains a matrin-type zinc finger and two group III WW domains (Fig. 1) that are structurally related to those of the established splicing factors U1C and Prp40, respectively (6, 7). The binding of FBP21 to splicing factors is mediated by its tandem WW domains, which represent interaction modules for proline-rich ligands (4, 8, 9). Although the above data strongly suggest that FBP21 has a role in pre-mRNA splicing, there are no in vivo data to support this contention.
FIGURE 1.
Domain structure of human FBP21 and SIPP1, as predicted by PROSITE.
The splicing factor SIPP1 (splicing factor that interacts with PQBP-1 and PP1) contains two proline-rich regions (Fig. 1) that are capable of binding to the WW domain of PQBP1 (polyglutamine tract-binding protein 1) and functions as a pre-mRNA splicing activator in intact cells (10). SIPP1 is also present in the sub-spliceosomal complex containing FBP21, suggesting that it is also a candidate interactor of FBP21. A yeast two-hybrid screening suggested that FBP21 and SIPP1 interact with each other (6). However, the interaction still needs to be confirmed by independent approaches, and the involved interaction sites and its functional relevance remain to be examined.
Many proteins contain multiple arrays of WW domains (11), which may increase the specificity and affinity for ligands (12, 13) or enhance their functional diversity by their ability to bind more than one ligand (14, 15). As the number and spatial arrangement of WW domains can be variable (14, 16), it is likely that the length and structure of the interdomain linkers are of crucial significance. Little is currently known about the functional importance of the interdomain linker in proteins with multiple WW domains. Although a number of structures of individual WW domains have been solved (17–22), only two structures of tandem WW domains, namely those of yeast Prp40 and Drosophila Su(dx), have been determined, and their structures are markedly different (7, 14). So far, the relationship between the functional diversity and the structural organization of multiple WW domains remains poorly characterized.
In this study, we provide the first data showing that human spliceosomal protein FBP21 stimulates pre-mRNA splicing in vivo. We also show that FBP21 interacts directly with SIPP1 in vitro as well as in vivo. Both the splicing function and the association with splicing factors, including SIPP1, are mediated by its tandem WW domains. Furthermore, we have solved the solution structure of the two tandem WW domains of FBP21 using NMR spectroscopy, which represents the first available structure of WW domains of group III. The ligand specificity and binding interface of the tandem WW domains, as well as the importance of having two WW domains, have been explored by NMR chemical shift perturbation, isothermal titration calorimetry (ITC), and site-directed mutagenesis. Our data reveal a cooperation between the two tandem WW domains in ligand binding, which results from the conjugation of two low affinity binding sites and the presence of a highly flexible linker region between them.
MATERIALS AND METHODS
Preparation of Recombinant Proteins
The nucleotide sequence encoding the tandem WW domains (amino acids 122–196) of human FBP21 was amplified by PCR from a human brain cDNA library (BD Biosciences) using specific primers (Takara) that generate flanking NdeI and XhoI sites for insertion into pET-22b (+) (Novagen) as a fusion with a C-terminal His tag (LEH6). Full-length FBP21 was cloned in the pET-28a (+) vector between the NcoI and XhoI sites. Wild type FBP21 and the indicated FBP21 mutants and fragments were cloned in the pEGFP-C1 vector, in-frame with the EGFP tag. Point mutations were made according to the QuikChangeTM site-directed mutagenesis protocol of Stratagene, using the appropriate primers and templates. The sequences of the DNA constructs were verified by DNA sequencing. The constructs GST-SIPP1-(1–372), GST-SIPP1-(180–372), GST-SIPP1-(253–641), and GST-SIPP1-(358–641) were obtained as described (10).
Expression, Purification, and Isotope Labeling of Tandem WW Domains of FBP21
The recombinant plasmid encoding the tandem WW domains of FBP21 was transformed into the bacterial BL21 (DE3) strain. Bacteria were grown at 37 °C in minimal medium (M9), and the target protein expression was induced at the mid-log phase (A600, 0.6–0.8) by the addition of 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. Uniformly 13C- and/or 15N-labeled recombinant protein was produced by the same way in M9 with d-[13C]glucose and/or 15NH4Cl as the sole sources of carbon and nitrogen, respectively. The protein was purified by HiTrap chelating column chromatography (GE Healthcare) and size-exclusion chromatography on Superdex 16/60 column (GE Healthcare). The NMR samples had a protein concentration of 1.0–1.2 mm, estimated by BCA kits (Pierce), and dissolved in 20 mm Tris buffer, 50 mm NaCl, 1 mm EDTA in 90% H2O, 10% 2H2O or 100% 2H2O, pH 6.5. Uniformly 15N-labeled mutant proteins were produced in the same way. The integrity of FBP21 tandem WW domains mutants was assessed by their 15N-HSQC spectra, which were all similar to that of the wild type protein and could be reassigned.
Peptide Synthesis and Purification
The proline-rich peptides from FBP21 ligands (Table 2) were chemically synthesized using a standard Fmoc (N-9-fluorenyl methoxycarbonyl) strategy at Shanghai Sangon Ltd. The synthetic peptides were purified by reverse-phase high pressure liquid chromatography using a C18 column and eluted with a linear gradient, 15–30%, of acetonitrile. Pooled fractions of the pure peptides were lyophilized and verified by matrix-assisted laser desorption ionization time of-flight mass spectrometry.
TABLE 2.
Interactions between FBP21 tandem WW domains and proline-rich peptides
| Motif | Peptide | Protein | Sequence | Bindinga |
|
|---|---|---|---|---|---|
| WW1 | WW2 | ||||
| Group III | P1 (13AA) | SmB′-(221–233) | RGPPPPGMRPPRP | +b | + |
| PPR/PGMc | P2 (10AA) | SIPP1-(481–490) | GPPPRGPPPR | + | + |
| P3 (10AA) | SIPP1-(501–510) | PPRPGMMRPP | + | + | |
| Group II | P4 (9AA) | Leukocyte formin-(541–549) | APPPPPPLP | + | + |
| PPLP/PPPPc | P5 (10AA) | DIAPH3-(589–598) | PPPPPPPPPP | + | + |
| P6 (10AA) | SIPP1-(491–500) | LPPPAPPGIP | + | + | |
| P7 (20AA) | SIPP1-(481–500)(p2+p6) | GPPPRGPPPRLPPPAPPGIP | + | + | |
a The interactions between FBP21 tandem WW domains and proline-rich peptides were studied using NMR chemical shift perturbation experiments.
b The proline-rich peptides bind to the WW domains.
c The proline-rich motifs in peptides are shown in boldface.
Antibodies
Polyclonal antibodies against SIPP1 were raised in rabbits, as described previously (10). Anti-EGFP and anti-rabbit secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Dako (Denmark), respectively. Anti-SC35 was ordered from BD Biosciences, and goat anti-mouse red secondary antibodies (goat anti-mouse IgG (H + L), DyLight 549-conjugated) were from Thermo Scientific-Pierce. Anti-His (H3), anti-GST antibodies, and IRDye 680 secondary antibodies were purchased from Santa Cruz Biotechnology and Odyssey. Polyclonal antibodies against FBP21 were a kind gift from Dr. Mark Bedford (Department of Cell Biology, Harvard Medical School, Boston) (6).
Cell Cultures and Immunoprecipitation
HEK293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal calf serum. 48 h after transfection, the cells were washed twice with ice-cold phosphate-buffered saline and lysed in 50 mm Tris at pH 7.5, 0.3 m NaCl, 0.5% (v/v) Triton X-100, 0.5 mm phenylmethanesulfonyl fluoride, 0.5 mm benzamidine, and 5 μm leupeptin. After sonication, the cell lysates were cleared by centrifugation (10 min at 10,000 × g), and the supernatants were used for immunoprecipitations. The cleared cell lysates were incubated with anti-EGFP antibodies coupled to protein A-TSK-Sepharose for 3 h at 10 °C. After one wash with Tris-buffered saline supplemented with 0.1 m LiCl and three washes with Tris-buffered saline plus 0.1% (v/v) Nonidet P-40, the beads were resuspended in 25 mm Tris, pH 7.5, and used for immunoblotting with anti-FBP21 and anti-SIPP1 antibodies.
GST Pulldown Assays
For the in vitro interaction of His-tagged FBP21 with GST-SIPP1-(1–372), GST-SIPP1-(180–372), GST-SIPP1-(253–641), and GST-SIPP1-(358–641), recombinant GST and GST fusion proteins were immobilized on 100 μl of glutathione-agarose beads in GST binding buffer (50 mm Tris/HCl, pH 7.5, 50 mm NaCl, and 1 mm EDTA). After washing with 3 ml of GST binding buffer, the beads were incubated with an equivalent amount of His-tagged FBP21. After incubation for 1 h at 4 °C, the beads were washed five times (1 ml each time) with GST binding buffer and then the same buffer supplemented with different concentrations of NaCl (2 ml of each buffer). Finally, the beads were eluted by GST binding buffer, supplemented with 15 mm glutathione. The bound proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. The bound FBP21 was further detected by Western blot with anti-His antibodies and visualized by Odyssey Infrared Imaging System.
Immunofluorescence and Confocal Microscopy
24 h after transfection the cells were washed twice with PBS and fixed for 10 min with formaldehyde (2–5% (v/v) of the 30% stock solution in PBS containing 2% sucrose). Cell permeabilization was performed by a 10-min incubation in PBS solution containing 0.5% Nonidet P-40 and 10% sucrose. Primary antibodies were diluted in PBS containing 2% fetal calf serum. After incubation at room temperature for 1 h, cells were washed at least three times with PBS plus 2% fetal calf serum and then incubated for 1 h with secondary antibodies. Finally, the cells were washed three times for 10 min in PBS. Confocal images were obtained with a Zeiss LSM-510 laser-scanning confocal microscope (Jena, Germany), equipped with the Zeiss Axiovert 100 M.
In Vivo Splicing Assay
This was performed essentially as described by Nasim et al. (23). Briefly, HEK293 cells were transfected with the reporter gene TN24 and the indicated plasmids. 48 h after transfection the cells were lysed using the passive lysis buffer (Promega Corp., Madison, WI). An aliquot of the lysate was used to measure luciferase activity using the assay system from Promega. The β-galactosidase activity was measured with o-nitrophenyl-β-d-galactopyranoside as a substrate. The expression of the constructs in the lysates was verified by Western blot analysis with anti-FBP21 antibodies.
Reverse Transcription (RT)-PCR
RNA isolated from HEK293 cells using the mammalian total RNA miniprep kit (Sigma) was reverse-transcribed with oligo(dT) primer (Sigma) and the Moloney murine leukemia virus reverse transcriptase (Fermentas). 1.5% of this cDNA was analyzed in duplicate by real time PCR, using the Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen) in a Rotorgene detection system (Corbett Research). Primers used were forward primer 5′-AACATCAGCCGCTACAGTCAA-3′ and reverse primer 5′-ACGTGATGTTCTCCTCGATAT-3′ (57). The PCR products were separated by 2.5% agarose gel electrophoresis.
NMR Spectroscopy
NMR experiments for the structure determination of the FBP21 tandem WW domains were carried out at 293 K on Bruker DMX-500 and DMX-600 NMR spectrometers equipped with cryoprobes. For the backbone and aliphatic side chain resonance assignments, two-dimensional 1H,15N-HSQC, three-dimensional CBCANH, CBCA(CO)NH, HNCO, HN(CA)CO, (H)C(CO)NH-TOCSY, HBHA(CBCACO)NH, H(C)(CO)NH-TOCSY, HCCH-TOCSY, HCCH-COSY, 15N- and 13C-edited NOESY-HSQC spectra were used. Aromatic side-chain assignments were obtained from two-dimensional homonuclear TOCSY and NOESY and three-dimensional 13C-edited NOESY-HSQC spectra. Inter-proton distance restraints were obtained from three-dimensional 15N- and 13C-edited NOESY-HSQC experiments using mixing times of 110 ms. The chemical shift index (24) and TALOS programs (25) were used together to obtain the backbone dihedral angles (Φ and Ψ) in secondary structures on the basis of chemical shift information. There was no hydrogen bond obtained from slow exchanging amide protons identified after exchanging the H2O buffer for D2O. NMR data were processed by NMRPipe and NMRDraw (26) and assigned with Sparky.
Structure Calculations
Structure calculation for FBP21 tandem WW domains was performed on the basis of proton-proton NOE restraints and dihedral angle restraints (Φ and Ψ) with a torsion angle dynamics simulated annealing protocol using the CNS version 1.1 program (27). The NOE-derived distance restraints were classified into four groups with the upper boundaries of 3.0, 4.0, 5.0, and 6.0 Å and lower boundary of 1.80 Å on the basis of NOE intensity measurements. TALOS predictions, only “good” with 9 or 10 matches in agreement, were used and converted into restraints on Φ and Ψ angles. 20 models were selected on the basis of energetic criteria (low total energy, using the accept.inp routine) to form a representative ensemble of the 200 calculated structures. The quality of the final structures was assessed using the program PROCHECK-NMR (28).
NMR Backbone Relaxation Experiments
15N relaxation experiments were carried out at 293 K on Bruker DMX500 NMR spectrometer. Heteronuclear 1H-15N NOEs as well as longitudinal (R1) and transverse (R2) 15N relaxation rates were measured using standard two-dimensional methods (29, 30). The relaxation delays were set to 11, 61, 142, and 242 (run twice), 362, 523, and 753 ms, and 1.144 s for T1 measurements, and 17.6, 35.2, and 52.8 (run twice) and 70.4, 105.6, and 140.8 ms for T2 measurements. A recycle delay of 1 s was used for measurement of R1 and R2 relaxation rates. The heteronuclear NOE experiments were run twice in an interleaved mode with and without (reference experiment) proton saturation during the recovery delay. The exponential curve fitting and extract of T1 and T2 were processed by Sparky. The steady-state 1H-15N NOE enhancements were calculated as the ratio of peak intensity in spectra recorded with and without proton saturation.
1DNH RDCs
1DNH RDCs were measured on a 0.5 mm 15N-labeled protein sample in the NMR buffer containing 25 mg/ml Pf1 phage (Pf1 magnetic resonance cosolvent) (31) at 293 K using the in-phase/anti-phase method of spectra recording (32). The RDC values were obtained by subtracting the reference value in isotropic solution.
Chemical Shift Perturbation
Chemical shift perturbation experiments were performed on Bruker DMX500 at 298 K using two-dimensional 1H-15N-HSQC experiments (33). Samples for NMR titrations contained 0.5 mm 15N-labeled protein in the same NMR buffer as described above. The unlabeled peptide stock solutions were added stepwise to give the peptide/protein molar ratios ranging from 0:1 to 4:1 or 6:1; each step was monitored by two-dimensional 1H-15N HSQC spectrum. The combined chemical shift changes were calculated using Equation 1.
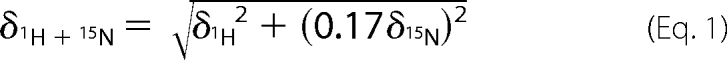 |
ITC
Calorimetry experiments were performed with a VP-ITC microcalorimeter (MicroCal, Inc.) at 298 K. The protein and peptide samples were dissolved in 20 mm Tris buffer, 50 mm NaCl, pH 7.4. The 2.0–3.0 mm peptide solution was titrated into the sample cell containing the FBP21 tandem WW domains or its mutants (80–100 μm). Control experiments were performed by making identical injection of peptide into the sample cell containing only buffer to correct the heat effects not directly related to the binding reaction. Data analysis was performed using the Microcal Origin software supplied with the instruments. The stoichiometry (N), association constant (K), standard enthalpy change (ΔH), and standard entropy change (ΔS) for the interactions were obtained from fitting of the experimental titration curve.
RESULTS
WW Domains of FBP21 Are Essential for Its Pre-mRNA Splicing Activation Function
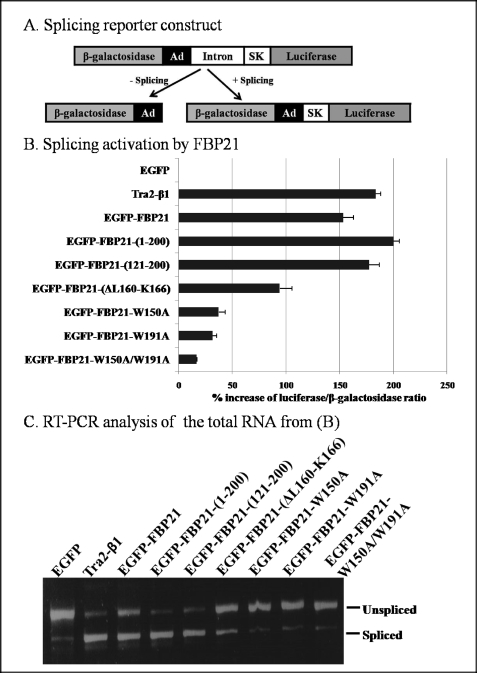
FBP21 is a component of the U2 snRNP complex of the spliceosomes (6). To examine whether FBP21 is essential for pre-mRNA splicing, we performed in vivo splicing assays following the transient expression of EGFP-tagged variants of FBP21 in HEK293T cells. The splicing efficiency was quantified using the pTN24 reporter (23). Unspliced transcripts from this reporter gene yield only β-galactosidase, whereas spliced transcripts yield a fusion protein with both β-galactosidase and luciferase activities (Fig. 2A). Thus, the luciferase/β-galactosidase ratio provides a measure of splicing efficiency. The ratio after the expression of EGFP was used as a negative control and set at 100% (Fig. 2B). Tra2-β1, an established splicing factor (34), was used as a positive control. The expression of Tra2-β1 increased the luciferase/β-galactosidase ratio by some 180%, whereas wild type EGFP-FBP21 increased the splicing efficiency by 150%. Interestingly, the fragments FBP21-(1–200) and FBP21-(121–200), which both contain the tandem WW domains, stimulated the splicing efficiency by about 200%, but mutation of either of the two WW domains, as in FBP21-W150A or FBP21-W195A, nearly abolished the stimulatory effect on splicing. EGFP-FBP21-(ΔL160-K166), which lacked 7 of the 12 residues of the interdomain linker, stimulated the splicing efficiency by about 90%, much less than that of full-length EGFP-FBP21. It was verified that all EGFP-FBP21 constructs were expressed to a similar extent (supplemental Fig. S1).
FIGURE 2.
FBP21 is an activator of pre-mRNA splicing in vivo. A, structure of the reporter construct. The β-galactosidase and luciferase genes are fused in-frame but separated by an intronic sequence derived from the adenovirus (Ad) and the skeletal muscle isoform of human tropomyosin (SK). The intron contains three translation stop signals. In the absence of splicing only β-galactosidase is generated, whereas an active fusion of β-galactosidase and luciferase is generated after splicing of the primary transcript. B, HEK293T cells were transiently transfected with the reporter plasmid (pTN24) and the indicated expression plasmids. The graph shows the % stimulation of splicing, based on the luciferase/β-galactosidase ratio, as compared with the ratio obtained with EGFP. The data represent the means ± S.E. (n = 3) and are representative for five different experiments. The standard deviations of the normalized ratios are indicated by error bars. C, total RNA from B was isolated and analyzed by RT-PCR. The two DNA bands were derived from spliced and unspliced RNA as indicated.
To confirm that the luciferase/β-galactosidase ratio was correlated with the proportion of spliced mRNA, the experiments were repeated, and the RNA was analyzed by RT-PCR. The DNA bands corresponding to spliced and unspliced RNA are shown in Fig. 2C. The results from RT-PCR analysis agreed well with the results obtained using the double reporter activities. Wild type EGFP-FBP21, EGFP-FBP21-(1–200), and EGFP-FBP21-(121–200) promoted splicing to a similar extent as did the positive control Tra2-β1, whereas there were almost no stimulation with mutants of the WW domain. Similarly, EGFP-FBP21-(ΔL160-K166) was a less efficient splicing stimulator than was full-length EGFP-FBP21. Obviously, there were parallel changes in the apparent level of splicing by both assays. Thus, our data indicate that FBP21 is a stimulator of pre-mRNA splicing by a mechanism that requires both WW domains and the interdomain linker.
WW Domains Mediate the Interaction of FBP21 with Pre-mRNA Splicing Factors
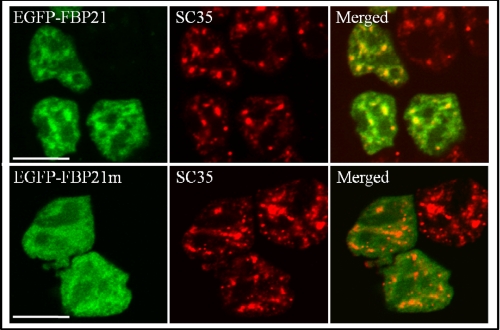
Because WW domains are protein interaction domains, we have subsequently examined whether they mediate the targeting of FBP21 to splicing factors. Endogenous FBP21 colocalizes with the essential splicing factor SC35 in the nuclear speckles (6), which contain numerous pre-mRNA splicing factors (10, 35, 36). EGFP-tagged FBP21 also partially colocalized with SC35 in the nuclear speckles in both HEK293T cells (Fig. 3) and HeLa cells (supplemental Fig. S2). However, mutation of the WW domains, as in FBP21-W150A, FBP21-W195A, and FBP21-W150A/W195A, abolished the speckled distribution and colocalization with SC35 (Fig. 3 and supplemental Fig. S2). These data are consistent with the notion that the WW domains target FBP21 to splicing factors.
FIGURE 3.
Subcellular localization of EGFP-FBP21. EGFP-FBP21 (upper panel) or EGFP-FBP21m (EGFP-FBP21-W150A/W191A, lower panel) was transiently expressed in HEK293T cells. After 24 h the cells were fixed and analyzed for the presence of the EGFP fusions by fluorescence microscopy (left column) and endogenous SC35 by immunocytochemistry with anti-SC35 antibodies and DyLight 549-conjugated goat anti-mouse secondary antibodies (middle column). The right column shows the overlay pictures. Bars, 10 μm. Similar results were obtained in HeLa cells (supplemental Fig. S2).
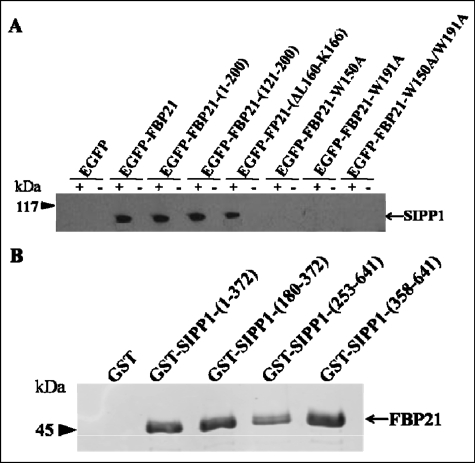
WW domains represent docking sites for proline-rich sequences (4, 9). Because FBP21 is present in a spliceosomal subcomplex together with the protein SIPP1 (6), which contains two proline-rich regions, we have examined whether FBB21 and SIPP1 interact with each other. First, we performed coimmunoprecipitation experiments following the transient expression of EGFP fusions of FBP21 variants in HEK293T cells. EGFP-FBP21, EGFP-FBP2-(1–200), and EGFP-FBP21-(121–200), as well as EGFP-FBP21-(ΔL160-K166), all interacted with endogenous SIPP1 (Fig. 4A). However, EGFP as well as EGFP-tagged fusions of FBP21-W150A, FBP21-W195A, and FBP21-W150A/W195A did not coimmunoprecipitate with SIPP1. The immunoprecipitated EGFP-FBP21 fusions were verified by Western blotting with anti-FBP21 antibodies (supplemental Fig. S3A). To distinguish between direct and indirect interactions, we subsequently performed pulldown experiments with bacterially expressed GST-tagged SIPP1 fragments and His-tagged FBP21 (Fig. 4B). GST-SIPP1-(1–372) and GST-SIPP1-(180–372), which contain the N-terminal proline-rich region, as well as GST-SIPP1-(253–641) and GST-SIPP1-(358–641), which harbor the C-terminal proline-rich region, all interacted with His-FBP21. The expression of GST-SIPP1 fusions is shown in supplemental Fig. S3B and is further verified by Western blotting with anti-GST antibodies (supplemental Fig. S3C). The SIPP1 fragments were readily degraded, which is often seen for PP1-interacting proteins/domains, and can be explained by their disordered structure, making them accessible to proteases (37). Collectively, our data strongly indicate that SIPP1 is a direct interactor of the WW domains of FBP21.
FIGURE 4.
Interaction of FBP21 and SIPP1. A, endogenous SIPP1 coimmunoprecipitates with EGFP-FBP21. EGFP and EGFP-FBP21 fusions were transfected into HEK293T cells. After lysis of the cells, FBP21 was immunoprecipitated from the lysates with anti-EGFP antibodies. The coimmunoprecipitated SIPP1s were detected by Western blot analysis with anti-SIPP1 antibodies. The immunoprecipitates were obtained with (+) and without (−) primary anti-EGFP antibodies. The immunoprecipitation of EGFP-FBP21 (variants) is shown in supplemental Fig. S3A. B, interaction of purified, recombinant FBP21 and GST-SIPP1. The interactions in vitro were examined by GST pulldown assay and further detected by Western blot analysis with anti-His (H-3) antibodies, as visualized by the Odyssey Infrared Imaging System. Four GST fusion SIPP1 fragments (1–372, 180–372, 253–641, and 358–641) were examined, the former two fragments contain the first proline-rich region and the latter two contain the second proline-rich region of SIPP1.GST alone was used as a negative control. The expression of GST-SIPP1 fusions is shown in supplemental Fig. S3B and further verified by Western blotting with anti-GST antibodies (supplemental Fig. S3C).
Structure of the Tandem WW Domains of FBP21
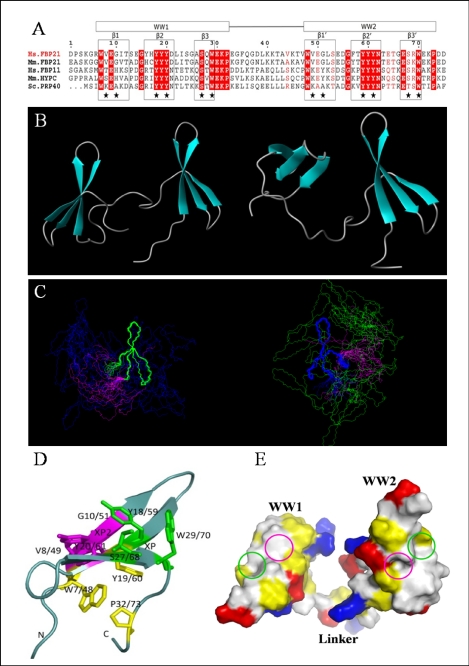
Because the WW domains of FBP21 are crucial for its splicing function, probably because they mediate the interaction with the proline-rich regions of the splicing factor SIPP1, we have determined the solution structure of the tandem WW domains of FBP21. The recombinant fragment used for these structural studies, i.e. human FBP21-(122–196), includes two group III WW domains and a 12-residue linker (Fig. 5A). The tandem WW domains share a 53% sequence identity, and they both contain a conserved “YYY” motif, which is found in nearly all group II/III WW domains. The solution structure of the FBP21 tandem WW domains was determined by multidimensional heteronuclear and homonuclear NMR spectroscopy.
FIGURE 5.
Solution structure of the tandem WW domains of FBP21. A, sequence alignment of FBP21 tandem WW domains with its orthologues as follows: Hs.FBP21 and Hs.FBP11 from Homo sapiens; Mm.FBP21 and Mm.HYPC from Mus musculus; and Sc.PRP40 from Saccharomyces cerevisiae. The WW domain boundaries are indicated by boxes at the top, and the secondary structure elements are boxed and indicated above the alignment. Residues identical in all five sequences are shown in red columns, and conserved residues are shown in red letters. Stars indicate residues forming binding interface. The sequence alignment was made using ClustalW, and the panel was generated by ESPript 2.2. B, ribbon representation of two structures in different interdomain orientations with secondary elements of WW domains shown as turquoise arrows. C, stereoview of the 20 lowest energy NMR structures of FBP21 tandem WW domains, superimposed on backbone atoms of WW1 (blue) and WW2 (green), respectively, and the flexible linker is shown in magenta. The structures cannot be superimposed over the entire molecule due to the flexible interdomain motion. D, view of the binding surface on the tandem WW domains of FBP21. Residues that form the XP and XP2 grooves are shown as green and purple sticks, respectively. The side chains of residues forming a hydrophobic core that stabilizes the fold are shown in yellow. The figures were made using MOLMOL and PyMol. E, space-filling structure of the WW domains of FBP21. The molecular surface is colored according to electric and hydrophobic potential. Blue colors represent positive charges; red colors are negative charges; neutral areas are gray, and yellow represents hydrophobic residues. The green and purple circles represent the XP and XP2 grooves, respectively.
Table 1 lists the structural statistics for the 20 deposited NMR structures. A total of 200 conformers was calculated, and the 20 conformers with the lowest energy were selected for analysis and presentation. Fig. 5, B and C, shows the ribbon representation of the energy-minimized average structure and superposition of 20 NMR structures of human FBP21 tandem WW domains by MOLMOL (38). The coordinates of these 20 NMR structures have been deposited into the Protein Data Bank (code 2JXW). Because of the linker flexibility and interdomain dynamics, the overall structure of the FBP21 tandem WW domains does not converge to one conformation. The calculation of an overall r.m.s.d. is meaningless for structures that cannot be superimposed over the entire molecule. In contrast to the whole tandem WW domains of FBP21, the structure for the individual WW domains converged well, which is reflected in the average r.m.s.d. values of 0.78 Å (all heavy atoms) and 0.29 Å (backbone atoms) at WW1, and 0.86 Å (all heavy atoms) and 0.35 Å (backbone atoms) at WW2. A PROCHECK analysis of the 20 NMR structures indicated that >95% of the residues lie in the most favored region and the additionally allowed region of the Ramachandran plot. The residues in the disallowed regions were those located in the N- and C-terminal parts and the flexible linker because of the paucity of inter-residual NOEs.
TABLE 1.
Summary of structure statistics
Structural statistics for the 20 NMR structures of FBP21 tandem WW domains. None of the structure exhibits distance violations >0.5 Å or dihedral angle violations >5°.
| NMR restraints in the structure calculation | |
| Distance restraints | 1404 |
| Intra-residue | 392 |
| Sequential (|i − j| = 1) | 404 |
| Medium range (|i − j| < 5) | 172 |
| Long range (|i − j| ≥ 5) | 436 |
| Dihedral angle restraints | 56 |
| Total | 1460 |
| Statistics for the 20 lowest energy structures | |
| Lennard-Jones potential energy (kcal mol−1) | −239.15 ± 11.07 |
| Mean r.m.s.d. from idealized covalent geometry | |
| Bonds | 0.0008 ± 0.00003 Å |
| Angles | 0.2780 ± 0.0020° |
| Impropers | 0.1004 ± 0.0080° |
| Mean r.m.s.d. from experimental restraints | |
| Distance | 0.0018 ± 0.0005 Å |
| cdih | 0.2980 ± 0.0590° |
| Coordinate r.m.s.d. of N, Cα, C′/all heavy atoms (Å)a | |
| Residues 6–32 (WW1) | 0.29/0.78 |
| Residues 47–73 (WW2) | 0.35/0.86 |
| Ramachandran plot (% residues)b | |
| Residues in most favored regions | 89.9 |
| Residues in additional allowed regions | 10 |
| Residues in generously allowed regions | 0 |
| Residues in disallowed regions | 0.1 |
a The calculation of an overall r.m.s.d. is meaningless because the flexible linker caused interdomain motion.
b Data exclude glycine, proline, and flexible residues 1–5, 13–14, 33–46, 54–55, and 74–75.
The two WW domains of FBP21 share 30–60% sequence identity with its homologues (Fig. 5A). The structures of the individual FBP21 WW domains are very similar to those of previously solved WW domain structures, and the pairwise r.m.s.d. of the β-sheet backbone atoms to the solution structures of WW domains from scPrp40 (7), hsFBP11 (22), mmFBP28 (39) are between 1.0 and 1.6 Å.
The FBP21 tandem WW domains each show the typical WW domain fold, consisting of a triple-stranded antiparallel β-sheet (Fig. 5B). The triple-stranded β-sheet structure of WW1 and WW2 domains has two sides (Fig. 5D). On one side, the highly conserved residues Trp-7/48, Tyr-19/60, and Pro-32/73 form a hydrophobic core that brings together the N and C termini and maintains the structure of WW domain. On the other side the conserved XP groove, formed by Trp-29/70, Tyr-18/59, and Ser-27/68, together with the XP2 groove, formed by Val-8/49, Glu-9/50, Gly-10/51, Tyr-18/59, and Tyr-20/61, make the binding surface for the proline-rich ligand. The two XP grooves transverse the low ridge between Tyr-18/59 and Tyr-20/61 nearly merge to a larger continuous groove (Fig. 5, D–E).
Dynamic Properties of the Tandem WW Domains of FBP21
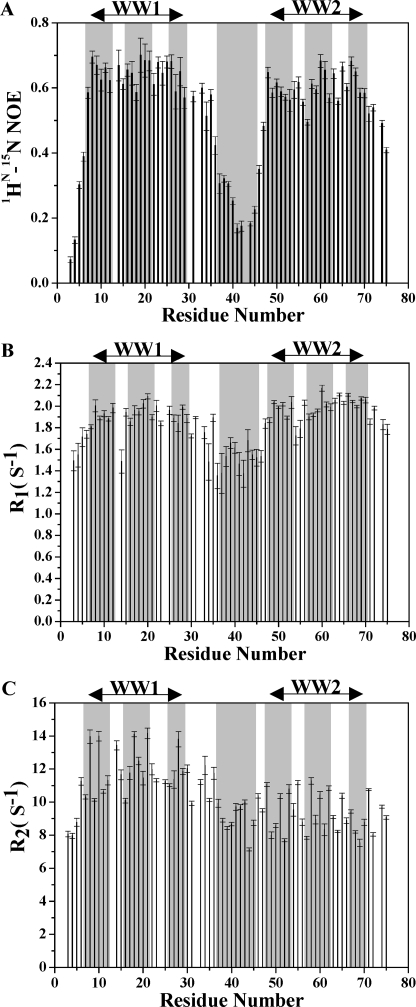
The structure of the tandem WW domains of FBP21 suggests a considerable flexibility in the relative orientations of WW1 and WW2 toward each other. To determine whether this reflects a true conformational flexibility or is rather because of the lack of observed NOEs, we further investigated the dynamic properties of FBP21 tandem WW domains using 15N relaxation measurements. For this purpose, we recorded the longitudinal relaxation times T1, the transverse relaxation times T2, as well as the heteronuclear {1H}-15N NOE values. Fig. 6 presents the 15N relaxation data of the tandem WW domain backbone amide nitrogens, including heteronuclear {1H}-15N NOEs, 15N R1 and R2, which were obtained for 90% of the residues. The steady-state heteronuclear {1H}-15N NOE, a sensitive indicator of internal motions on a sub-nanosecond timescale, revealed that the WW1 and WW2 domains of FBP21 have 1H-15N NOE values of 0.65 and 0.61, respectively, consistent with a relatively rigid domain structure. In contrast, the interdomain linker displayed a 1H-15N NOE value of only 0.27, indicating that this region is highly flexible and lacks a well ordered structure. The R1 plot also shows relatively uniform values of the two WW domains, whereas residues in the interdomain linker display somewhat smaller R1 values. Small R1 values are likely to arise from fast mobility (40, 41), in agreement with the small NOEs in the same region. Neither the NOEs nor the R1 values show significant systematic differences between the two domains, despite the apparent high mobility of interdomain linker. However, the R2 values turned out to be different between the two WW domains. The WW1 domain has an average R2 of 11.62 ± 0.22 s−1, whereas the WW2 domain has an average R2 of 9.86 ± 0.16 s−1. The similar R1 and the larger R2 values of the WW1 domain compared with those of the WW2 domain imply that the former domain being affected by extended millisecond-microsecond mobility (41).
FIGURE 6.
Backbone dynamics of the tandem WW domains of FBP21. Plots of 1HN-15N NOE (A), 15N R1 (B), and R2 (C) versus residue number of WW1–2 are shown. Secondary structure elements and linker region are boxed in gray. Similar data were recorded in ligand-bound state (supplemental Fig. S7).
Additional information on the interdomain orientation and motion of FBP21 tandem WW domains was obtained by the measurement of RDCs. As shown in supplemental Fig. S4A, the measured 1H-15N RDCs were markedly different for WW1 and WW2. The RDCs in the β-strands of WW1 are mostly negative, whereas they are positive for the corresponding residues in WW2. The opposite sign of RDCs suggests that there is no colinearity between the tandem WW domains, and the interdomain orientation between the two WW domains is not fixed (14, 42). However, the similar R2/R1 ratio of the two WW domains obtained from 15N relaxation experiments indicates that the two domains, on average, tumble together in solution. Thus, we conclude that although the linker flexibility leads to some orientation independence between the tandem WW domains, there remains limitations to this flexibility.
Ligand Specificity Analyses by Chemical Shift Perturbation
NMR is an extraordinarily powerful method to analyze the binding properties of individual domains in a multidomain protein, because of its unique ability to identify residues that participate in the binding in a direct manner.
To delineate whether these WW domains recognize proline-rich motifs of both group II and group III, and to map the binding interface of a proline-rich ligand on the two WW domains of FBP21, 1H and 15N chemical shift changes were measured upon ligand binding to FBP21-(122–196) (Table 2). Among the ligands, three peptides of different length were derived from SIPP1, namely P2 (residues 481–490), P6 (residues 491–500), and P7 (residues 481–500). Two additional ligand peptides, namely P1 and P3, were chosen from the splicing factors SmB′ (residues 221–233) and SIPP1 (residues 501–510), respectively (6, 9). P1, P2, and P3 contain PGM/PPR motifs that were postulated to bind to the WW domain of group III. Other peptides, derived from leukocyte formin (P4, residues 541–549) (43), DIAPH3 (P5, residues 589–598) (44), and the SIPP1-derived peptide P6 (see above) contain PPLP or PPP motifs that were previously reported to interact with WW domains of group II (21, 22). The data presented in Table 2 and Table 3 indicates that both WW domains of FBP21 recognize the group III PPR/PGM-binding motif. The tandem WW domains of FBP21 also interact with the group II PPLP and PPPP motifs. WW1 and WW2 of FBP21 behave similarly in terms of ligand selectivity.
TABLE 3.
Dissociation constants and binding stoichiometry for FBP21 tandem WW domains-peptide interactions
| WW1–2 | P2 (GPPPRGPPPR) |
P6 (LPPPAPPGIP) |
P7 (GPPPRGPPPRLPPPAPPGIP) |
|||
|---|---|---|---|---|---|---|
| Na | Kdb | N | Kd | N | Kd | |
| mm | mm | mm | ||||
| Wild type | 2 | 1.15 (±0.03) | 2 | 2.42 (±0.40) | 1 | 0.039 (±0.001) |
| W29A | 1 | 3.50 (±0.33) | /c | / | 1 | 0.122 (±0.005) |
| W70A | 1 | 2.07 (±0.09) | / | / | 1 | 0.187 (±0.012) |
a N indicates stoichiometry of the FBP21 tandem WW domains (mutant)-ligand interaction.
b Equilibrium dissociation constant was determined by ITC at 25 °C.
c / indicates no data acquired.
Ligand Binding Interfaces on the Two WW Domains of FBP21
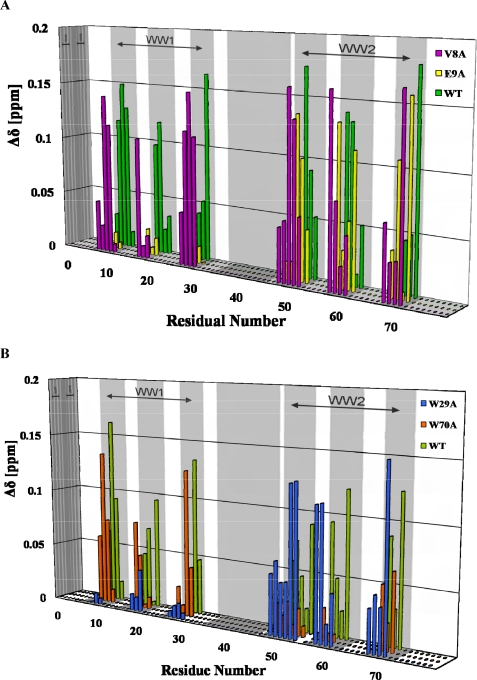
Representative chemical shift changes upon titration of proline-rich peptides are shown in Fig. 7 and supplemental Fig. S5. Most of the residues that experienced significant chemical shift changes were located in the three β-strands of each WW domain, namely residues 8–10 in β1, 18–20 in β2, and 27–30 in β3 of WW1. The corresponding residues of WW2 are 49–51, 59–60, and 68–71, respectively. Therefore, the NMR titration results can be used to map the ligand binding interface on the tandem WW domains of FBP21 (Fig. 5, D and E). The canonical XP groove formed by residues Trp-29/70, Ser-27/68, and Tyr-18/59 is highly conserved in most WW domains (22). The second groove, the XP2 groove, is formed by residues Val-8/49, Glu-9/50, Gly-10/51, Tyr-18/59, and Tyr-20/61. The XP and XP2 grooves, Fig. 5E, highlighted with green and purple circles, respectively, constitute the binding surface of both WW domains of FBP21.
FIGURE 7.
Chemical shift changes upon ligand binding versus residue number of FBP21 tandem WW domains. The combined chemical shift changes in parts/million are calculated using Equation 1. FBP21 tandem WW domains are titrated by peptides of different length as follows: P2 (10 amino acids, residues 481–490 in blue), P6 (10 amino acids, residues 491–500 in red), and P7 (20 amino acids, residues 481–500 in green) from the same protein SIPP1 to the same peptide/protein molar ratio 6:1. Secondary structure elements and linker region are represented by gray boxes.
The XP groove residues of the two WW domains of FBP21 are highly conserved in almost all WW domains, whereas the residues forming the XP2 groove are more variable, particularly in residues Val-8/49, Glu-9/50, and Gly-10/51 in the β1-strand. The contribution of the latter residues to ligand binding of WW domains of groups II and III and their role in the conjugation of tandem WW domains on ligand binding are still unknown. We have used site-directed mutagenesis (V8A, E9A, W29A, and W70A) to explore these issues in more detail (Fig. 8). The V8A mutation did not significantly affect the ligand affinity, whereas the E9A mutation abolished the binding of WW1 to ligand, indicating that Glu-9 is essential for ligand binding of the XP2 groove, whereas Val-8 does not contribute directly to ligand binding (Fig. 8A). The W29A and W70A mutations in the XP groove abolished the ability of WW1 and WW2, respectively, to bind ligands (Fig. 8B), confirming the importance of these residues for ligand binding.
FIGURE 8.
Mutational analysis of the FBP21 tandem WW domains. Combined chemical shift changes upon ligand binding are calculated as in Equation 1; the mutants V8A, E9A, W29A, and W70A are colored as shown, secondary structure elements are represented by gray boxes. A, peptide P1 (RGPPPPGMRPPRP) added to the FBP21 tandem WW domains and indicated mutants. B, peptide P2 (GPPPRGPPPR) added to the FBP21 tandem WW domains and indicated mutants. WT, wild type.
Tandem WW Domains Confer Higher Ligand Affinity
As direct interactions between FBP21 tandem WW domains and ligand motifs of groups II and III have been identified by NMR chemical shift perturbation, ITC was used to quantitate binding affinity and stoichiometry between the WW domains and peptides. The peptide sequences tested in ITC experiments are shown in Table 3, and representative integrated heat data are shown in Fig. 9. The two single motif ligands P2 and P6 both interacted with wild type tandem WW domains with a binding stoichiometry of 2. This is consistent with the result from NMR chemical shift perturbation, which shows that the single motif ligands bind to both WW domains of FBP21. Titrating the W29A or W70A mutants of FBP21 tandem WW domains with single motif peptides P2 also yielded consistent results with NMR chemical shift perturbation experiments (Fig. 8B). The binding stoichiometry for the W29A or W70A mutants and single motif ligands are 1, corroborating that the mutation abolished the binding of the corresponding WW domain to ligand. The binding stoichiometry between the tandem WW domains and the tandem motifs ligand P7 is 1, and the NMR chemical shift perturbation experiments showed that the two WW domains had equivalent chemical shift changes upon ligand binding (Fig. 7). Furthermore, the W29A or W70A mutants bind to P7 with the same stoichiometry as the wild type FBP21 tandem WW domains. Because the result of NMR chemical shift perturbation (Fig. 8B) and ITC (Table 3) both showed that the tryptophan to alanine mutation abolished the binding of the corresponding WW domain to ligand, steric hindrance probably impedes the binding of another wild type WW domain to the other available motif in P7. So the W29A or W70A mutants and tandem motif peptide P7 remained in 1:1 interaction. Thus, results from ITC and NMR together indicated that the FBP21 tandem WW domains interact with the two motifs in P7 simultaneously. A schematic figure of the interactions between FBP21 tandem WW domains and single/tandem motif peptides is shown in supplemental Fig. S6.
FIGURE 9.
Representative ITC profile of FBP21 tandem WW domains and peptide interactions. The upper panel represents raw data obtained after injections of peptide P7 (GPPPRGPPPRLPPPAPPGIP) into FBP21 tandem WW domains solution. The bottom panel showing the fit of the data with a one set of sites model. The Stoichiometry (N) and Kd values are given with the binding curve for the interaction.
Remarkably, compared with individual P2 or P6, the tandem motif peptide P7, which actually represents a combination of the peptides P2 and P6, shows much stronger association with FBP21 tandem WW domains. As shown in Table 3, peptide P7 (Kd ∼ 10−5 m) has 30–60-fold higher affinity than P2 (Kd ∼ 10−3 m) or P6 (Kd ∼ 10−3 m). This implies that the tandem WW domains bind to two consecutive proline-rich motif ligands with 30–60-fold higher affinity than to peptide containing only one motif. Moreover, because both the W29A and W70A mutants bind to P7 with lower affinity than the wild type further confirmed that the high affinity between the tandem WW domains and ligand P7 need the two WW domains to work together. The pairs of equivalent residues in the two WW domains of FBP21, i.e. Trp-29 and Trp-70, Ser-27 and Ser-68, Tyr-18 and Tyr-59, Tyr-20 and Tyr-61, Val-8 and Val-49, Glu-9 and Glu-50, Gly-10 and Gly-51, all behaved in a similar manner concerning chemical shift changes upon the three ligands binding (Fig. 7). However, no obvious chemical shift changes in the linker region were observed upon binding of the peptides P2 and P6, whereas several residues in this region, including Phe-35, Gln-36, Gly-37, Asp-38, Lys-41, Thr-42, Val-44, and Thr-46, were obviously affected by P7 binding (Fig. 7). Taken together, our data suggest that the linker region is sufficiently flexible to enable simultaneous binding of the two WW domains to two distinct proline-rich motifs in a single ligand, conferring increased affinity of FBP21 to ligands with multiple docking motifs.
DISCUSSION
Structure of the Tandem WW Domains of FBP21
Based on their ligand specificity, WW domains have been classified into four groups (45–47). Group I recognizes PPXY motifs, where X can be any residue; group II recognizes PPLP motifs; group III binds so-called PPR motifs, which represent stretches of prolines C-terminally flanked by an arginine; and group IV recognizes phospho-Ser or phospho-Thr followed by a proline. Of these four groups, WW domains of group III are the least characterized, and their differentiation from group II domains remains ambiguous (45, 48). We report here for the first time the solution structure of WW domains of group III and studied their ligand specificity using synthetic peptides (Table 2). Our results show that the two WW domains of FBP21 both recognize peptides containing PPR, PPLP, as well as PPPP motifs. We also found that the binding interfaces on both WW domains involve both the XP and XP2 grooves, similar to what has been reported for group II domains (49, 50). In conclusion, WW domains of group II and group III cannot be clearly differentiated based on their three-dimensional structure, binding interface, and ligand specificity and should be viewed as a single group.
We have provided unequivocal evidence that the 12-residue linker between the WW domains of FBP21 is highly flexible. This linker flexibility leads to some interdomain motion between the two WW domains. A comparison of dynamics data between the free and bound states of FBP21 tandem WW domains is shown in supplemental Fig. S4 and supplemental Fig. S7. Comparing the dynamics data in free and bound states, we found that R2 values of the two WW domains are much larger, and there are no systematic differences anymore between them in the bound state (supplemental Fig. S7). These results indicated that the tandem WW domains do possess some interdomain motion, and this interdomain mobility is much restricted when the two WW domains simultaneously interact with the two motifs in peptide P7.
High sequence similarity of the two WW domains, broader resonance lines caused by intense interdomain motion, and extensive spectral overlap together make resolving the structure of tandem WW domains a big challenge. Only two tandem WW structures have been reported (7, 14), one of which represents the two tandem WW domains of yeast splicing factor Prp40 (7). The WW domains of the Prp40 have a defined interdomain orientation, whereas the tandem WW domains of the mammalian FBP21 are connected by a much more flexible linker. The second previously determined tandem WW domains structure is represented by WW domains 3 and 4 of Su(dx) (14). WW4 of Su(dx) contains a tryptophan to phenylalanine substitution and only folds well upon ligand binding. When WW4 is paired with WW3, interdomain association impedes proper folding of WW4. This mode of autoinhibition is relieved by binding of a ligand to WW3, enabling ligand binding to WW4. This cooperativity was proposed to facilitate the transient regulatory interactions between Su(dx) and Notch in the endocytic pathway (51). In contrast, the WW domains of FBP21 are already well folded in their free state and the two WW domains behave equivalently in ligand binding. Moreover, the two WW domains of FBP21 together confer higher binding affinity. As for the third solved tandem WW domains structure, our study reveals that even with high sequence identity and similar domain array, the mechanism of tandem WW domains binding to ligand, the relative domain orientation, and dynamics can be very different and context-dependent.
FBP21 as a Pre-mRNA Splicing Activator
We have shown here for the first time that FBP21 functions as a splicing activator in vivo (Fig. 2). FBP21 enhanced the splicing efficiency of a reporter gene up to 2.5-fold, and a similar effect was obtained with FBP21-(121–200) and FBP21-(1–200), both of which contain the two tandem WW domains. However, this splicing activation effect was lost by mutation of the WW domains, attesting to the key role of these protein interaction domains. This conclusion is in accordance with our observation that the WW domains of FBP21 mediate its interaction with the splicing factor in nuclear speckles (Fig. 3) and with its ligand SIPP1 (Fig. 4). Importantly, the splicing activation effect of FBP21 was nearly completely abolished by mutation of only one of its two WW domains (Fig. 2, B and C). This can be explained by our observation that each of the WW domains of FBP21 represents a relatively weak binding site and that the high affinity binding of SIPP1 requires its simultaneous interaction with both WW domains (Table 3 and Fig. 4A). In this respect it is worth recalling that so many proteins contain tandem, multiple arrays of WW domains (11). It has been reported that the mere conjugation of two low affinity docking sites can create a high affinity interaction (12, 13, 15). However, it is not well established how these tandem domains function together. We obtained evidence for the cooperation between the two WW domains of FBP21 and illustrated the importance of the flexible interdomain linker for the first time. As shown in Fig. 7, the simultaneous interaction of the two WW domains of FBP21 with the two binding motifs in ligand is probably helped by the flexibility of the intervening linker region, which enables the tandem WW domains to adopt a variable spatial orientation toward each other. Fig. 2, B and C, also shows the significance of the interdomain linker. Shortening the linker between the two WW domains substantially decreased the splicing efficiency of FBP21. Unexpectedly, EGFP-FBP21-(ΔL160-K166), which has a shortened linker between the two WW domains, was still targeted to the nuclear speckles (supplemental Fig. S2) and maintained the interaction with SIPP1, although the interaction is much weaker (Fig. 4A). One interpretation is that the interaction between FBP21 and a certain splicing factor only requires the two WW domains and some limited interdomain motion, whereas more complicated processes such as pre-mRNA splicing need a higher conformational flexibility. Furthermore, as shown in Fig. 5A, from human to yeast, the linker between two WW domains are not as conserved as the key residues within the WW domains. However, no insertion or deletion occurred in the linker region. Indeed, the length of the linker is fixed, implying that the space separation between the two WW domains is important. Because little is known about the functional importance of the linker region in proteins with multiple WW domains until now, it will be very interesting to explore this in further detail.
Increasing evidence supports the notion that pre-mRNA synthesis and its subsequent processing is coordinated (52–55). WW domains were suggested to act as a platform that links transcription and pre-mRNA splicing (11, 56). Indeed, WW domain-containing proteins not only bind to the phosphorylated C-terminal domain of the largest subunit of RNA polymerase II but also recruit splicing factors such as 17 S U2 snRNP, SF1, and U2AF to facilitate 3′ splice site recognition of emerging mRNA precursors and/or to dictate splice site selection during alternative splicing (53, 56). In the dynamic spliceosomal complex, the WW domains of FBP21 associate with U2 snRNPs and interact directly with the U1 snRNP protein U1C, the core snRNP proteins SmB and SmB′, and the branch point-binding protein SF1/mBBP (6). We have shown here that the tandem WW domains of FBP21 also interact directly with splicing factor SIPP1. Intriguingly, the PQBP1 (polyglutamine tract-binding protein 1), which can bind to the C-terminal domain of the largest subunit of RNA polymerase II, also interacts with SIPP1 via its WW domain. This suggests that FBP21 may also be implicated in the coordination of transcription and pre-mRNA splicing via its tandem WW domains. The variable spatial relationships between the two WW domains of FBP21 may facilitate its function in this dynamic cotranscriptional pre-mRNA splicing process.
Supplementary Material
Acknowledgments
We thank Dr. M. Bedford (Boston) for the generous gift of the anti-FBP21 antibodies and Prof. Xuebiao Yao (University of Science and Technology of China) for the vectors. We are grateful to Dr. Yinshan Yang for help with the structure determination, Dr. Chao Xu for stimulating discussions, and Nicole Sente for expert technical assistance.
This work was supported by Chinese National Natural Science Foundation Grant 30670426, Chinese National Fundamental Research Project Grants 2006CB806507 and 2006CB910201, the Chinese National High Tech R & D Program Grant 2006AA02A315, and the Fund for Scientific Research-Flanders Grant G.0670.09N.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
The atomic coordinates and structure factors (code 2JXW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- snRNP
- small nuclear ribonucleoprotein
- SIPP1
- splicing factor that interacts with PQBP-1 and PP1
- NOE
- nuclear Overhauser effect
- EGFP
- enhanced green fluorescent protein
- GST
- glutathione S-transferase
- RDCs
- residual dipolar couplings
- RT
- reverse transcription
- ITC
- isothermal titration calorimetry
- NOESY
- nuclear Overhauser effect spectroscopy
- PBS
- phosphate-buffered saline
- BBB
- the branch point-binding protein
- HSQC
- heteronuclear single quantum coherence
- r.m.s.d
- root mean square deviation.
REFERENCES
- 1.Krämer A. (1996) Annu. Rev. Biochem. 65, 367–409 [DOI] [PubMed] [Google Scholar]
- 2.Will C. L., Lührmann R. (1997) Curr. Opin. Cell Biol. 9, 320–328 [DOI] [PubMed] [Google Scholar]
- 3.Abovich N., Rosbash M. (1997) Cell 89, 403–412 [DOI] [PubMed] [Google Scholar]
- 4.Bedford M. T., Chan D. C., Leder P. (1997) EMBO J. 16, 2376–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berglund J. A., Chua K., Abovich N., Reed R., Rosbash M. (1997) Cell 89, 781–787 [DOI] [PubMed] [Google Scholar]
- 6.Bedford M. T., Reed R., Leder P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10602–10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiesner S., Stier G., Sattler M., Macias M. J. (2002) J. Mol. Biol. 324, 807–822 [DOI] [PubMed] [Google Scholar]
- 8.Sudol M., Chen H. I., Bougeret C., Einbond A., Bork P. (1995) FEBS Lett. 369, 67–71 [DOI] [PubMed] [Google Scholar]
- 9.Bedford M. T., Sarbassova D., Xu J., Leder P., Yaffe M. B. (2000) J. Biol. Chem. 275, 10359–10369 [DOI] [PubMed] [Google Scholar]
- 10.Llorian M., Beullens M., Andrés I., Ortiz J. M., Bollen M. (2004) Biochem. J. 378, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingham R. J., Colwill K., Howard C., Dettwiler S., Lim C. S., Yu J., Hersi K., Raaijmakers J., Gish G., Mbamalu G., Taylor L., Yeung B., Vassilovski G., Amin M., Chen F., Matskova L., Winberg G., Ernberg I., Linding R., O'donnell P., Starostine A., Keller W., Metalnikov P., Stark C., Pawson T. (2005) Mol. Cell. Biol. 25, 7092–7106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eck M. J., Pluskey S., Trüb T., Harrison S. C., Shoelson S. E. (1996) Nature 379, 277–280 [DOI] [PubMed] [Google Scholar]
- 13.Ottinger E. A., Botfield M. C., Shoelson S. E. (1998) J. Biol. Chem. 273, 729–735 [DOI] [PubMed] [Google Scholar]
- 14.Fedoroff O. Y., Townson S. A., Golovanov A. P., Baron M., Avis J. M. (2004) J. Biol. Chem. 279, 34991–35000 [DOI] [PubMed] [Google Scholar]
- 15.Ramirez-Espain X., Ruiz L., Martin-Malpartida P., Oschkinat H., Macias M. J. (2007) J. Mol. Biol. 373, 1255–1268 [DOI] [PubMed] [Google Scholar]
- 16.Itani O. A., Campbell J. R., Herrero J., Snyder P. M., Thomas C. P. (2003) Am. J. Physiol. Renal Physiol. 285, F916–F929 [DOI] [PubMed] [Google Scholar]
- 17.Huang X., Poy F., Zhang R., Joachimiak A., Sudol M., Eck M. J. (2000) Nat. Struct. Biol. 7, 634–638 [DOI] [PubMed] [Google Scholar]
- 18.Macias M. J., Hyvönen M., Baraldi E., Schultz J., Sudol M., Saraste M., Oschkinat H. (1996) Nature 382, 646–649 [DOI] [PubMed] [Google Scholar]
- 19.Kanelis V., Rotin D., Forman-Kay J. D. (2001) Nat. Struct. Biol. 8, 407–412 [DOI] [PubMed] [Google Scholar]
- 20.Pires J. R., Taha-Nejad F., Toepert F., Ast T., Hoffmüller U., Schneider-Mergener J., Kühne R., Macias M. J., Oschkinat H. (2001) J. Mol. Biol. 314, 1147–1156 [DOI] [PubMed] [Google Scholar]
- 21.Meiyappan M., Birrane G., Ladias J. A. (2007) J. Mol. Biol. 372, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pires J. R., Parthier C., Aido-Machado R., Wiedemann U., Otte L., Böhm G., Rudolph R., Oschkinat H. (2005) J. Mol. Biol. 348, 399–408 [DOI] [PubMed] [Google Scholar]
- 23.Nasim M. T., Chowdhury H. M., Eperon I. C. (2002) Nucleic Acids Res. 30, e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart D. S., Sykes B. D. (1994) J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 25.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 27.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 28.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 29.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 30.Opella S. J., Kim Y., McDonnell P. (1994) Methods Enzymol. 239, 536–560 [DOI] [PubMed] [Google Scholar]
- 31.Hansen M. R., Mueller L., Pardi A. (1998) Nat. Struct. Biol. 5, 1065–1074 [DOI] [PubMed] [Google Scholar]
- 32.Ottiger M., Delaglio F., Bax A. (1998) J. Magn. Reson. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 33.Kim S., Cullis D. N., Feig L. A., Baleja J. D. (2001) Biochemistry 40, 6776–6785 [DOI] [PubMed] [Google Scholar]
- 34.Stoilov P., Daoud R., Nayler O., Stamm S. (2004) Hum. Mol. Genet. 13, 509–524 [DOI] [PubMed] [Google Scholar]
- 35.Zhou Z., Licklider L. J., Gygi S. P., Reed R. (2002) Nature 419, 182–185 [DOI] [PubMed] [Google Scholar]
- 36.Rappsilber J., Ryder U., Lamond A. I., Mann M. (2002) Genome Res. 12, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dancheck B., Nairn A. C., Peti W. (2008) Biochemistry 47, 12346–12356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 39.Macias M. J., Gervais V., Civera C., Oschkinat H. (2000) Nat. Struct. Biol. 7, 375–379 [DOI] [PubMed] [Google Scholar]
- 40.Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A. (1992) Biochemistry 31, 5269–5278 [DOI] [PubMed] [Google Scholar]
- 41.Babini E., Bertini I., Capozzi F., Chirivino E., Luchinat C. (2006) Structure 14, 1029–1038 [DOI] [PubMed] [Google Scholar]
- 42.Braddock D. T., Cai M., Baber J. L., Huang Y., Clore G. M. (2001) J. Am. Chem. Soc. 123, 8634–8635 [DOI] [PubMed] [Google Scholar]
- 43.Favaro P. M., de Souza Medina S., Traina F., Bassères D. S., Costa F. F., Saad S. T. (2003) Biochem. Biophys. Res. Commun. 311, 365–371 [DOI] [PubMed] [Google Scholar]
- 44.Katoh M., Katoh M. (2004) Int. J. Mol. Med. 13, 473–478 [PubMed] [Google Scholar]
- 45.Sudol M., Hunter T. (2000) Cell 103, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 46.Sudol M., Sliwa K., Russo T. (2001) FEBS Lett. 490, 190–195 [DOI] [PubMed] [Google Scholar]
- 47.Otte L., Wiedemann U., Schlegel B., Pires J. R., Beyermann M., Schmieder P., Krause G., Volkmer-Engert R., Schneider-Mergener J., Oschkinat H. (2003) Protein Sci. 12, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ilsley J. L., Sudol M., Winder S. J. (2002) Cell. Signal. 14, 183–189 [DOI] [PubMed] [Google Scholar]
- 49.Kato Y., Miyakawa T., Kurita J., Tanokura M. (2006) J. Biol. Chem. 281, 40321–40329 [DOI] [PubMed] [Google Scholar]
- 50.Kato Y., Hino Y., Nagata K., Tanokura M. (2006) Proteins 63, 227–234 [DOI] [PubMed] [Google Scholar]
- 51.Jennings M. D., Blankley R. T., Baron M., Golovanov A. P., Avis J. M. (2007) J. Biol. Chem. 282, 29032–29042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Proudfoot N. J., Furger A., Dye M. J. (2002) Cell 108, 501–512 [DOI] [PubMed] [Google Scholar]
- 53.Goldstrohm A. C., Greenleaf A. L., Garcia-Blanco M. A. (2001) Gene 277, 31–47 [DOI] [PubMed] [Google Scholar]
- 54.Cramer P., Srebrow A., Kadener S., Werbajh S., de la Mata M., Melen G., Nogués G., Kornblihtt A. R. (2001) FEBS Lett. 498, 179–182 [DOI] [PubMed] [Google Scholar]
- 55.Bentley D. (2002) Curr. Opin. Cell Biol. 14, 336–342 [DOI] [PubMed] [Google Scholar]
- 56.Lin K. T., Lu R. M., Tarn W. Y. (2004) Mol. Cell. Biol. 24, 9176–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nasim M. T., Eperon I. C. (2006) Nat. Protoc. 1, 1022–1028 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.