Abstract
Human FGF1 (fibroblast growth factor 1) is a powerful signaling molecule with a short half-life in vivo and a denaturation temperature close to physiological. Binding to heparin increases the stability of FGF1 and is believed to be important in the formation of FGF1·fibroblast growth factor receptor (FGFR) active complex. In order to reveal the function of heparin in FGF1·FGFR complex formation and signaling, we constructed several FGF1 variants with reduced affinity for heparin and with diverse stability. We determined their biophysical properties and biological activities as well as their ability to translocate across cellular membranes. Our study showed that increased thermodynamic stability of FGF1 nicely compensates for decreased binding of heparin in FGFR activation, induction of DNA synthesis, and cell proliferation. By stepwise introduction of stabilizing mutations into the K118E (K132E) FGF1 variant that shows reduced affinity for heparin and is inactive in stimulation of DNA synthesis, we were able to restore the full mitogenic activity of this mutant. Our results indicate that the main role of heparin in FGF-induced signaling is to protect this naturally unstable protein against heat and/or proteolytic degradation and that heparin is not essential for a direct FGF1-FGFR interaction and receptor activation.
FGF1 (fibroblast growth factor 1) belongs to a family of polypeptide growth factors comprising in humans 22 structurally related proteins (1, 2). The signaling induced by the growth factor leads to a wide range of cellular responses during development as well as in adult life, such as growth regulation, differentiation, survival, stress response, migration, and proliferation of different cell types (3). The biological activity of FGF1 is exerted through binding to four high affinity cell surface receptors (FGFR1–4), resulting in receptor dimerization and transphosphorylation in its tyrosine kinase domain (4, 5). The activated FGFR3 induces cellular response by initiating several signaling cascades, including mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase/Akt, and phospholipase C-γ (PLC-γ) pathways (6).
In addition to FGFRs, FGF1 binds to heparan sulfates (HS) associated with proteoglycans at the cell surface and in the extracellular matrix (7). Among the physiological sugars, the highest affinity for FGF1 is shown by heparin, a widely used linear, highly sulfated polysaccharide composed of 2-O-sulfated iduronic acid and 6-O-sulfated, N-sulfated glucosamine units (8).
Despite many years of research, there is still controversy regarding the molecular role of heparin/HS in FGF1- and FGF2-induced signaling. Thus, the question of whether or not the linkage of two molecules of the growth factor by heparin/HS is an absolute prerequisite for induction of FGFR dimerization is still open. Numerous studies have concluded that the presence of heparin/HS is obligatory for FGF signaling. It is widely believed that heparin/HS is directly involved in receptor dimerization and is critical for mitogenic response stimulated by the growth factor (4, 6, 8–10).
On the other hand, several authors working on FGF1 and FGF2 have suggested that there is no mandatory requirement for heparin for the assembly and activation of the FGF·FGFR complex. They imply that heparin only plays a role in association of two molecules of the growth factor and therefore facilitates their binding to FGFR (11). It has been reported that FGF1 and FGF2 can interact with the FGFR and trigger phosphorylation of p42/44 MAPK and activation of other signaling pathways even in the absence of HS (12–16).
The accepted role of heparin/HS in FGF1 signaling is to prevent the degradation of the growth factor (17). The interaction with heparin or HS protects FGF1 against heat, acidic pH, and proteases (18, 19). HS also seems to regulate the activity of different FGFs by creating their local reservoir and generating a concentration gradient of the growth factor (6, 17).
The binding of FGF1 to heparin/HS is mediated by specific residues forming a positively charged patch on the protein surface (20, 21). The major contribution is made by Lys118 (Lys132 in the full-length numbering system), which was identified by Harper and Lobb (22), and Lys112 and Arg122 (23, 24). Additional residues of FGF1 involved in the interaction with heparin are the positively charged Lys113, Arg119, and Lys128 and the polar Asn18, Asn114, and Gln127 (20, 21). Site-directed mutagenesis and other studies have revealed the importance of Lys118 not only in heparin binding but also for the biological function of FGF1 (22, 25, 26). It was shown that the K118E (K132E) mutant is inactive in stimulation of DNA synthesis, although its affinity for FGFR and the ability to activate signaling cascades is not reduced (27, 28). Despite extensive research, the reason for the lack of mitogenic potential of K118E FGF1 is still not clear.
In this paper, we verified the function of heparin in FGF1·FGFR complex formation and signaling by constructing several FGF1 mutants with reduced affinity for heparin. To recover the stability of these variants, which could no longer be stabilized by heparin, we supplemented them stepwise with stabilizing mutations (29). We analyzed thoroughly their biological activity and their ability to translocate across cellular membranes (30–34). Interestingly, the full mitogenic activity of the K118E FGF1 variant was restored by the introduced stabilizing mutations.
Our results indicate that the main role of heparin in FGF-induced signaling is to protect this naturally unstable protein against heat denaturation and proteolytic degradation and that the increased stability of the growth factor can compensate for reduced heparin binding.
EXPERIMENTAL PROCEDURES
Materials
The site-directed mutagenesis kit (QuikChangeTM) was from Stratagene, and the DNA purification kit was from Qiagen. Escherichia coli strain BL21(DE3)pLysS was from New England Biolabs. Heparin-Sepharose CL-6B affinity resin was from Amersham Biosciences, and oligonucleotides for mutagenesis were from Sigma. The basic components of culture media were supplied by Merck, and guanidine hydrochloride was from ICN. [methyl-3H]Thymidine and [33P]phosphate were provided by Amersham Biosciences, and Na125I was from PerkinElmer Life Sciences. Complete protease inhibitor mixture was from Roche Applied Science. Phosphatase inhibitor mixture, lactacystin chloroquine, and trypsin were from Sigma, and bafilomycin A1 and leupeptin were from Calbiochem. Antibodies against MAPK (p42/p44) (rabbit), phospho-MAPK (p42/p44) (mouse), p38 MAPK (rabbit), Akt (rabbit), phospho-Akt (rabbit), PLC-γ (rabbit), FGFR (rabbit), and phospho-FGFR (mouse) were from Cell Signaling Technology. Anti-phospho-PLC-γ (rabbit), anti-FGF1 (goat), and anti-phospho-Tyr antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and anti-phospho-p38 MAPK antibody (mouse) was from BD Transduction Laboratory. Human antibody against EEA1 was a kind gift from Harald Stenmark. Horseradish peroxidase-conjugated and fluorescent secondary antibodies were from Jackson ImmunoResearch Laboratories. All other chemicals were from Sigma.
Cell Cultures
The NIH 3T3 cell line was obtained from ATCC. U2OS stably transfected with FGFR1 was a kind gift from Ellen M. Haugsten (35). NIH 3T3 cells were grown in Quantum 333 medium from PAA Laboratories GmbH containing 2% bovine serum (Invitrogen) and 100 units/ml penicillin and 100 μg/ml streptomycin. U2OS cells were grown in Dulbecco's modified Eagle's medium from Invitrogen containing 10% fetal calf serum (PAA Laboratories). Cells were seeded into tissue culture plates the day preceding the start of the experiments.
Protein Production and Purification
A construct comprising a truncated form (residues 21–154) of human FGF1 in the pET-3c vector was used. Mutagenesis and protein expression were performed as reported previously (29, 34, 36). In the case of mutants with significantly reduced heparin binding, a His6 tag was introduced at the N terminus of the protein to allow efficient purification. Recombinant proteins were expressed at 37 °C in E. coli BL21(DE3)pLysS strain and purified on a heparin-Sepharose CL-6B column or Ni2+-nitrilotriacetic acid column, followed by size exclusion chromatography on Superdex-75 using ÄKTA Explorer system (Amersham Biosciences). Protein purity was confirmed by silver-stained SDS-PAGE, and molecular masses were verified by electrospray ionization mass spectrometry. Native conformation of the variants was confirmed by circular dichroism and fluorescence spectra measurements. The numbering system used to identify amino acids in FGF1 has been reported previously (36).
Thermodynamic Studies
To analyze thermodynamic properties of the recombinant proteins we employed CD and fluorescence (29). To prevent both protein aggregation and accumulation of folding intermediates during heating, thermal denaturation was performed in the presence of a low concentration of a denaturant, 0.7 m guanidinium chloride, in phosphate buffer (37). The temperature-induced ellipticity changes at 227 nm were followed in cuvettes of 10-mm path length using an 8-s response time. Denaturation performed in the spectrofluorimeter was monitored by changes in fluorescence intensity at 353 nm of the single tryptophan residue of FGF1 excited at 280 nm (38). All denaturation data, irrespective of the method, were collected using a scan rate of 0.25 °C/min.
To analyze the impact of heparin on thermal denaturation of FGF1, we performed denaturation of the wild type and the mutants using the CD technique in the absence and presence of a 5-fold molar excess of heparin. The measurements were carried out in the presence of 1.5 m urea (equivalent of 0.7 m guanidinium chloride), which is a non-ionic denaturant and does not disturb the ionic interactions between heparin and growth factor (29).
The protein concentration was 2 × 10−6 m in all thermodynamic experiments. The data were analyzed using PeakFit software (Jandel Scientific Software), assuming a two-state reversible equilibrium transition, as described previously (29, 36).
Receptor Binding Assay
Binding of FGF1 to cell surface receptors was analyzed essentially as published (29, 34, 39). Wild type FGF1 in high salt buffer was dialyzed against PBS and iodinated using the Iodogen method. The labeled growth factor was then purified on a disposable PD-10 desalting column (Amersham Biosciences). Confluent NIH 3T3 cells were washed with ice-cold binding buffer (HEPES medium, 10 units/ml heparin) and incubated with 6 ng/ml 125I-labeled FGF1 and increasing concentrations of unlabeled FGF1 or its mutants for 3 h at 4 °C. Subsequently, the cells were washed twice with ice-cold binding buffer and once with ice-cold PBS, followed by the addition of ice-cold 1 m NaCl in PBS to dissociate the growth factor from heparans at the cell surface. After washing, the cells were lysed in 0.1 m KOH, and the solubilized radioactivity, representing the cell surface-bound 125I-FGF1, was measured with a γ-counter.
Microscopy
Serum-starved U2OS cells stably transfected with FGFR1 were incubated with 100 ng/ml selected FGF1 mutants for 15 min at 37 °C in the presence or absence of heparin (10 units/ml). The cells were then washed with PBS and fixed with 10% formalin solution (Sigma), permeabilized with 0.1% Triton X-100 in PBS, and stained with mouse anti-phospho-tyrosine antibody labeling the phosphorylated FGFR, human anti-EEA1 antibody (a specific marker for early endosomes), and DRAQ5 (a nuclear probe from Alexis Biochemicals). Finally, the cells were incubated with fluorescent secondary antibodies. The cells were examined with a Zeiss LSM Duo confocal microscope. Images of randomly chosen cells were taken.
Activation of Signaling Pathways
To detect phosphorylated forms of downstream signaling proteins, serum-starved NIH 3T3 cells were stimulated for 15 min with 5 and 50 ng/ml growth factor in the presence or absence of heparin (10 units/ml). The cells were lysed with SDS sample buffer, scraped from the plate, and sonicated. Total cell lysates were separated by SDS-PAGE, transferred onto Immobilon-P membrane, and subjected to immunoblot analysis for phosphorylated (activated) forms of the respective signaling proteins (29). After stripping, the membrane was reprobed with antibodies against total signaling proteins to ensure equal loading.
Measurements of DNA Synthesis
The mitogenic properties of FGF1 mutants were assessed by monitoring [3H]thymidine incorporation in response to the stimulation of NIH 3T3 cells (27, 29). The cells were starved in Dulbecco's modified Eagle's medium with 0.5% bovine serum for 24 h at 37 °C. Then increasing amounts (0.1–100 ng/ml) of FGF1 or its mutants were added, and the incubation was continued for 24 h at 37 °C. For the last 6 h, 1 μCi/ml [3H]thymidine was added to the cells. Finally, the cells were treated with 5% trichloroacetic acid and then solubilized in 0.1 m KOH, and the level of trichloroacetic acid-insoluble radioactivity was measured using a liquid scintillation β-counter. The relationship between the [3H]thymidine incorporation and log concentration of the added growth factor was fitted to a sigmoidal function. Specific mitogenic activity was expressed as the concentration giving 50% of maximal stimulation (EC50).
Proliferation Assay
Serum-starved NIH 3T3 cells were treated with 100 ng/ml wild type or mutant FGF1 in the presence or absence of heparin (10 units/ml) for 72 h and then counted.
Protease Digestion Assay
Digestion experiments were carried out at 37 °C by incubation of the wild type and mutants of FGF1 within trypsin at 1:80 and 1:20 molar ratios in a reaction buffer (100 mm Tris, 20 mm calcium chloride, pH 8.3). The proteolysis was stopped by the addition of SDS sample buffer. Products of the reaction were visualized by SDS-PAGE and Coomassie Blue staining.
In Vivo FGF1 Degradation Assay
Serum-starved NIH 3T3 cells were incubated with 10 ng/ml wild type or mutants of FGF1, in the presence or absence of heparin, for up to 24 h. Additionally, cells were incubated with 10 ng/ml wild type of FGF1 in the absence of heparin with or without pre-exposure for 30 min to lysosomal degradation inhibitors: bafilomycin A1 (10 nm), chloroquine (25 μm), leupeptin (30 μm), NH4Cl (20 mm), or lactacystin (10 μm), a proteasome inhibitor. Then the medium was removed, and the cells were lysed in lysis buffer at time points 0.1, 6, 9, 12, and 24 h. Lysates and medium fractions were adsorbed onto heparin-Sepharose or subjected to trichloroacetic acid precipitation. The amount of intact growth factor in the samples was analyzed by SDS-PAGE and immunoblotting using anti-FGF1 antibody.
In Vivo Phosphorylation of Externally Added FGF1
NIH 3T3 cells were starved for 24 h in Dulbecco's modified Eagle's medium with 1% bovine serum. Then the cells were incubated overnight in phosphate-free Dulbecco's modified Eagle's medium supplemented with 25 μCi/ml [33P]phosphate. Next the cells were treated with 10 units/ml heparin and 100 ng/ml recombinant FGF1 for 6 h. Subsequently, the cells were washed once with PBS containing phosphatase inhibitor mixture, lysed in lysis buffer (0.1 m NaCl, 10 mm Na2HPO4, 1% Triton X-100, 1 mm EDTA) supplemented with protease and phosphatase inhibitor mixtures, scraped, and sonicated. The lysates were adsorbed onto heparin-Sepharose for 2 h at 4 °C. To reduce the background, the beads were treated with 2 μg/ml tosylphenylalanyl chloromethyl ketone-treated trypsin for 30 min at room temperature. Finally, samples were subjected to SDS-PAGE, transferred onto Immobilon-P membrane (Millipore), and analyzed by immunoblotting (to detect the total amount of FGF1) and fluorography (to detect 33P-phosphorylated FGF1).
Digitonin Cell Fractionation
Cell fractionation was essentially performed as previously described (40). NIH 3T3 cells were starved for 24 h and then incubated with wild type FGF1 or different FGF1 mutants in the presence or absence of heparin for 6 h at 37 °C and washed with high salt/low pH buffer to remove FGF1 bound to the cell surface. The cells were then washed with PBS, and 20 μg/ml digitonin was added to permeabilize the cells. The cells were kept at room temperature for 5 min and on ice for an additional 30 min to allow the cytosol to leak into the buffer. Then the cells were scraped from the plastic and centrifuged. The supernatant was designated as the cytosol fraction. The cellular pellet was lysed with lysis buffer and centrifuged at 15,800 × g for 15 min. The supernatant was designated as the membrane fraction, and the nuclear pellet was sonicated in lysis buffer and centrifuged as above. The final supernatant was designated as the nuclear fraction. Growth factor from all fractions was precipitated with 50% trichloroacetic acid and solubilized in sample buffer. The samples were analyzed by SDS-PAGE and immunoblotting with FGF1-specific antibodies.
RESULTS
Preparation and Biophysical Properties of FGF1 Mutants
Design and Expression of FGF1 Mutants
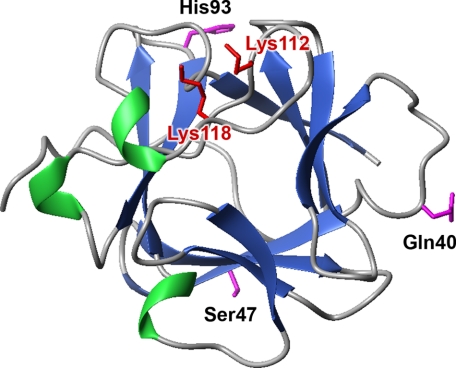
In a previous study (29), we showed that increased stability of FGF1 can prolong its half-life, increase protease resistance, and enhance the biological activities of the growth factor. Our results suggested that stabilizing mutations can compensate for the absence of heparin and allow full mitogenic activity of FGF1 in NIH 3T3 cells. To elucidate the significance of heparin in activation of the FGF1·FGFR complex and in FGF1-induced signaling, we constructed a series of 17 variants (six single, one double, and 10 multiple mutants) with reduced affinity for heparin/HS and with different stability (supplemental Tables S1 and S2). The mutations were designed based on the crystal structure of the FGF·FGFR complex (41, 42) and on our previous studies (29). Two crucial residues involved in heparin binding by the growth factor were selected for mutation (Lys112 and Lys118 in the 140-amino acid numbering system (38) corresponding to Lys126 and Lys132 in the full-length numbering system) and three residues known to stabilize the FGF1 molecule (29) (Fig. 1). All mutant proteins were expressed in the pET3c/BL21(DE3)pLysS system with a yield of 20–40 mg of pure protein/1 liter of culture.
FIGURE 1.
Spatial structure of FGF1 (Protein Data Bank code 1rg8). Amino acid residues mutated to lower FGF1 affinity for heparin and to increase its stability are marked in red and magenta, respectively.
Biophysical Characteristics of FGF1 Mutants
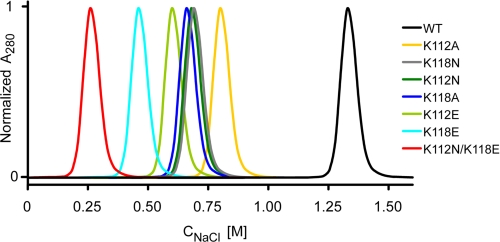
Native conformation of the FGF1 variants was confirmed by CD and fluorescence. There were no significant changes in fluorescence and in far- and near-CD spectra of the mutants as compared with the wild type growth factor, suggesting that the secondary and tertiary structures were preserved (data not shown). However, due to the lysine substitutions, individual mutants differed significantly in their affinity for heparin, as tested by elution from a heparin-Sepharose column with a linear NaCl gradient. Fig. 2 shows elution profiles of the six single and one double mutant with substitutions of Lys112 or/and Lys118 from the heparin-Sepharose column. Due to a reduced affinity for immobilized heparin, all variants were eluted at lower salt concentrations compared with the wild type FGF1, which was eluted at 1.33 m NaCl. The elution peak for the K118E variant was at 0.45 m NaCl, whereas the K112N mutant was eluted at 0.67 m. Mutations at position 112 reduced the affinity for heparin to a lesser extent than the substitutions of Lys118. Furthermore, mutations to alanine were less effective in lowering the heparin binding ability of FGF1 than the substitutions to asparagine and glutamic acid at the same positions.
FIGURE 2.
Elution profiles of FGF1 and its mutants from a heparin-Sepharose column.
In the case of the double lysine mutant (K112N/K118E), the affinity for heparin was very low, and the protein was eluted at 0.25 m NaCl. Because the heparan sulfates found on the cell surface are less sulfated than heparin, this mutant presumably does not exhibit any affinity for HS at physiological salt concentrations. The elution profiles of multiple mutants containing the K112N or/and K118E mutation and the stabilizing substitutions were the same as for the lysine mutants alone (data not shown).
Thermodynamic Stability of FGF1 Mutants
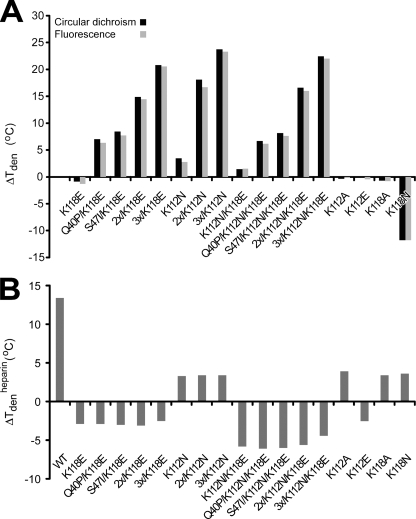
To measure accurately the conformational stability of the different FGF1 constructs, we employed thermal denaturation. Supplemental Table S1 summarizes the denaturation temperature (Tden) and the van't Hoff enthalpy data (ΔHden) obtained from denaturation experiments monitored by CD and fluorescence. The data demonstrate very good agreement between the two methods applied. The effects of the mutations on the conformational stability of FGF1 can be easily assessed by comparing the Tden values of the respective variant with the Tden value of the wild type (Fig. 3 A). Since very similar values were obtained for both measurement techniques, we discuss only the thermodynamic parameters based on the CD data.
FIGURE 3.
Thermal stability of FGF1 mutants. A, changes in denaturation temperature between the mutants and the wild type (ΔTden = Tden − Tden(WT)) monitored by fluorescence and circular dichroism in the presence of 0.7 m guanidinium chloride. B, changes in denaturation temperature of FGF1 mutants in the presence and absence of heparin (ΔTdenheparin = Tden+heparin − Tden−heparin) monitored by circular dichroism in the presence of 1.5 m urea.
The single mutants, with the exception of K112N and K118N, exhibited slightly lower thermal stability than the wild type. Introduction of the K112N substitution caused a marked increase of the denaturation temperature, by more than 3 °C. A similar replacement of Lys at position 118 (K118N) resulted in a strong reduction of the thermal stability of FGF1 by 12 °C. The double mutant K112N/K118E, containing the most thermodynamically favorable lysine mutation, K112N, and the mutation resulting in the lowest heparin affinity among the single mutants, K118E, exhibited a slightly elevated stability as compared with the wild type (ΔTden = 1.5 °C). The effects of these two point mutations sum up in the double mutant (supplemental Table S1 and Fig. 3A).
In this study, we made use of three previously described FGF1-stabilizing mutations, Q40P, S47I, and H93G (which did not affect any functional properties of the growth factor and stabilized it to the largest extent), and combined them with the substitutions that reduced the affinity of the growth factor to heparin in 10 multiple variants (29, 43). The introduction of the double stabilizing substitution (Q40P/S47I, denoted 2x) to the K112N, K118E, and K112N/K118E mutants increased the Tden of FGF1 by 18.2, 14.9, and 16.7 °C, respectively. Further, combination of all three stabilizing mutations (Q40P/S47I/H93G, denoted 3x) with the lysine substitutions yielded quadruple or quintuple mutants with an increase in Tden between 20.9 and 23.8 °C, thus providing a dramatic increase in thermal stability (supplemental Table S1 and Fig. 3A).
To test how the substitutions of lysines at position 112 and 118 influence the protective effect of heparin, we performed thermal unfolding of FGF1 and its mutants in the presence or absence of a 5-fold molar excess of heparin monitored by circular dichroism. To present the differences between the mutants, we calculated the change in denaturation temperature in the presence and absence of heparin (ΔTdenheparin = Tden+heparin − Tden−heparin) (supplemental Table S2 and Fig. 3B). Heparin stabilized the wild type FGF1 by 13.4 °C. Similar values were obtained for many other FGF1 mutants, available in the laboratory, with undisturbed affinity for heparin (data not shown). However, for all mutants with a decreased affinity for heparin presented in this study, we observed a significant reduction in ΔTdenheparin (supplemental Table S2 and Fig. 3B). Mutant K112N and all variants that contained this substitution exhibited ΔTdenheparin around 3.5 °C. Opposite charge substitutions, such as K112E and K118E, even caused a slight destabilization of the protein in the presence of heparin. For all the variants that carried the K118E mutation, the addition of heparin decreased the protein stability by close to 3 °C. In the case of the variants with double lysine substitutions (K112N/K118E), we observed a decrease in denaturation temperature in the presence of heparin in the range of 4–6.1 °C. These results confirmed that the selected lysine mutations strongly influenced the ability of FGF1 to bind properly to heparin and abolished its protective effect.
Biological Properties of FGF1 Mutants
Binding of the FGF1 Mutants to FGFR and Their Ability to Induce Its Activation
Using a competitive binding assay with 125I-labeled wild type FGF1, we tested the ability of the mutants to bind specifically to FGFRs in NIH 3T3 cells, which express mainly FGFR1, and displace the labeled FGF1 from the receptors. None of the single lysine substitutions that reduced the affinity of FGF1 for heparin altered the affinity of the growth factor for the receptor, since there were no differences in the ability of the mutants to compete with the wild type [125I]FGF1 (supplemental Fig. S1). This indicates that neither of the mutated residues is involved in the interactions with the high affinity receptor. However, variants containing the K112N/K118E double substitution were slightly less effective in competing out the labeled wild type FGF1. This discrepancy can be explained by the conditions of the experiment. For efficient 125I-labeling of FGF1, the presence of heparin is required, which causes effective dimerization of the growth factor in solution. Therefore, during the course of the experiment, the kinetics of receptor binding favors the dimerized labeled wild type FGF1 over the monomeric mutants with strongly reduced ability to bind free heparin.
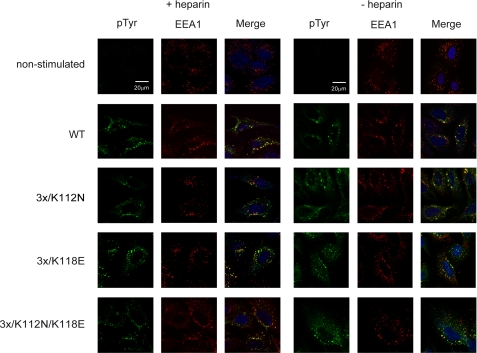
We also tested receptor dimerization monitored as autophoshorylation of the receptor upon stimulation by the wild type FGF1 or its mutants. For this purpose, we employed confocal microscopy. U2OS cells stably transfected with FGFR1 were incubated for 15 min with wild type FGF1 or selected mutants in the presence or absence of heparin. Then the cells were fixed and stained for the early endosome antigen (EEA1; red) and phosphotyrosines reflecting activated receptor (green), and then co-localization of the two markers in vesicular structures was examined. As shown in Fig. 4, both for the wild type FGF1 and the mutants, there was no detectable difference in the co-localization of EEA1 and activated FGFR1 in the absence or presence of heparin.
FIGURE 4.
Binding and internalization of FGF1 mutants. U2OS cells stably expressing FGFR1 were stimulated for 15 min with 100 ng/ml FGF1 or its selected variants in the presence or absence of heparin. Then the colocalization of EEA1 (red) and endocytosed FGFR1 (visualized by anti-phospho-Tyr (pTyr) antibody (green)) were examined by confocal microscopy.
Activation of Downstream Signaling Cascades
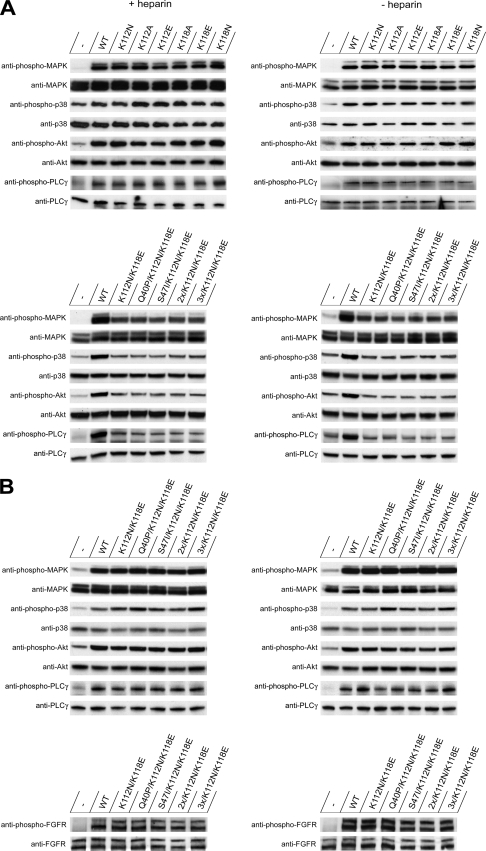
We next determined the ability of the mutants to activate the downstream signaling pathways in NIH 3T3 cells (Fig. 5, A and B, and supplemental Fig. S2A) and in U2OS cells stably transfected with FGFR1 (supplemental Fig. S2, B and C). We checked phosphorylation, reflecting the activation of FGFR as well as of MAPK, Akt, and p38 kinases and PLC-γ upon short term stimulation with the growth factor. The activation was detected by immunoblotting using specific antibodies that recognize only the phosphorylated form of the proteins.
FIGURE 5.
Activation of signaling cascades by wild type and mutants of FGF1 in the presence and absence of heparin. Serum-starved NIH 3T3 cells were treated with different concentrations of wild type FGF1 and its mutants (5 ng/ml (A) and 50 ng/ml (B)) for 15 min. To assay the activation of the downstream proteins, anti-phosphoprotein antibodies were used. To verify equal loading, stripped membranes were blotted with antibodies that recognized the total amount of each signaling protein.
To assure equal loading of the gels, the membranes were reprobed with antibodies recognizing the total amount of the signaling proteins whether or not they were phosphorylated. In non-treated cells, we detected only weak basal phosphorylation of the signaling proteins tested.
First, we tested the activation of the signaling cascades when the cells were treated with a low concentration (5 ng/ml) of FGF1. In the case of all (single and multiple) mutants with only one heparin-binding site mutated, there were no differences in FGFR stimulation and activation of the signaling pathways, as determined by monitoring the phosphorylation level of downstream kinases and PLC-γ in the presence or absence of heparin (Fig. 5A and supplemental Fig. S2, A and B). Single substitutions that lowered the affinity of FGF1 for heparin/HS had no effect on signal transduction as compared with the wild type, and the FGF1 mutants were functionally competent in activating signaling pathways even at a low concentration.
However, at 5 ng/ml FGF1, the double substitution in the heparin-binding site (K112N/K118E) significantly altered the efficiency of activation of the downstream signaling proteins (Fig. 5A and supplemental Fig. S2B), lowering their phosphorylation level in accordance with the FGFR binding experiments. Next, we checked if it was possible to compensate for the lower heparin binding of the K112N/K118E mutants using a higher concentration of the growth factors, and we studied the activation of the signaling cascades with 50 ng/ml FGF1 (Fig. 5B and supplemental Fig. S2C). In this case, the phosphorylation of the receptor as well as the activation of all tested pathways was on the level of the wild type for all mutants.
Mitogenic Activity of FGF1 Mutants
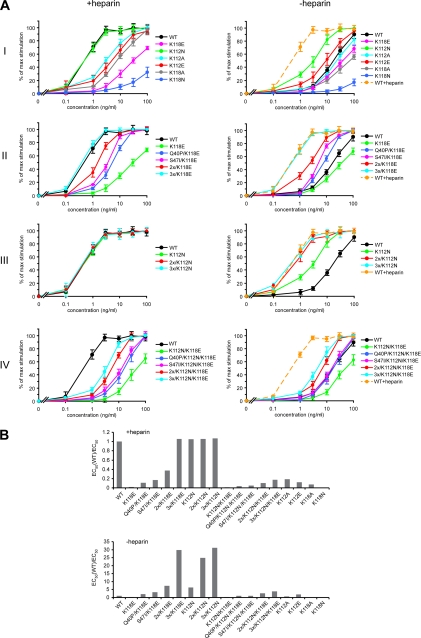
To check whether the inability to bind heparin can be compensated for by an increased stability of FGF1 with respect to its biological activities of the growth factor, we tested single and multiple mutants with reduced affinity for heparin and diverse stability for their mitogenic activity. Since the mitogenic potential of FGF1 is responsible for one of the crucial functions of the growth factor, stimulation of cell proliferation, we first checked the ability of the mutants to stimulate DNA synthesis in the presence and absence of heparin. This assay can be considered a long term response experiment, since it requires the survival of the growth factor in the medium for an extended period of time, over 12 h (44). Serum-starved NIH 3T3 cells were treated for 24 h with increasing concentrations of the wild type FGF1 or FGF1 mutants in the absence or presence of 10 units/ml heparin. Then the ability of the cells to incorporate [3H]thymidine during the last 6 h of the treatment was measured (Fig. 6A), and the specific mitogenic activity was calculated as described under “Experimental Procedures” (Fig. 6B).
FIGURE 6.
Mitogenic activity of wild type and mutants of FGF1. A, normalized mitogenic activity curves derived from a [3H]thymidine incorporation experiment in NIH 3T3 cells in the presence or absence of heparin. The data shown are mean values of five independent experiments ± S.D. B, relative mitogenic activity (in comparison with the wild type, EC50(WT)/EC50) of tested mutants in the presence and absence of heparin.
All but one single mutant with disturbed heparin binding exhibited reduced ability to stimulate DNA synthesis in the presence of heparin versus that of the wild type FGF1 (Fig. 6A, panel I). The K112N variant, which demonstrated the highest thermodynamic stability among the single mutants, was as mitogenically potent as the wild type in the presence of heparin. On the other hand, the K118N mutant, which was the least stable one, was also the least efficient in stimulating thymidine incorporation. The next in the rank was the K118E mutant, which had the most severely reduced affinity for heparin. In contrast to the wild type and the mutants that exhibited less affected heparin binding (K112N, K112A, K112E, and K118A), we did not observe any difference in the mitogenic activity in the absence and presence of heparin for the K118E mutant (Fig. 6A, panel I). These results suggest that the significantly reduced ability of the growth factor to stimulate DNA synthesis could be mainly caused by the loss of the heparin protection.
To test this possibility, we constructed series of mutants containing the K112N and/or K118E mutations and stabilizing substitutions in different combinations. Introduction of the stabilizing mutations into the K118E variant compensated for the lack of heparin protection and recovered the ability of the growth factor to stimulate mitogenesis regardless of the presence of heparin at the level of the wild type in the presence of heparin (Fig. 6A, panel II). All of the mutants with the K118E substitution behaved similarly in the presence and absence of heparin. We also observed a similar effect for a series of FGF1 variants containing the K112N mutation in the absence of heparin (Fig. 6A, panel III). This single mutation did not reduce heparin affinity as strongly as the K118E substitution did. Nevertheless, the introduction of the stabilizing mutations was also able to improve the mitogenic potential of the K112N variant. We also tested mutants of different stability containing the double substitution (K112N/K118E) in the heparin-binding site (Fig. 6A, panel IV). These mutants exhibited strongly reduced affinity for heparin and could be considered as unable to bind to the cell surface heparans. All of them behaved similarly with respect to the mitogenic properties regardless of the presence of heparin. We also observed a stepwise effect of the stabilizing mutations on the mitogenic activity. However, even the most potent combination of stabilizing substitutions (Q40P, S47I, and H93G) could not elicit a mitogenic response comparable with that of the wild type FGF1 in the presence of heparin. The mutant 3x/K112N/K118E exhibited an approximately 5 times lower mitogenic activity than the wild type in the presence of heparin.
A possible explanation of these data is that in the absence of heparin, cell surface heparans can still enable growth factor dimerization, despite the fact that they are not able to assure protection of FGF1. This is strongly supported by the behavior of the wild type FGF1 in the absence of heparin and its highly (but not completely) reduced mitogenic activity. For the mutants with the K112N/K118E substitution, even when they are very stable, the lack of affinity for heparans can change the kinetics of receptor binding. In such a case, higher concentrations of the growth factor are necessary to induce full mitogenic response.
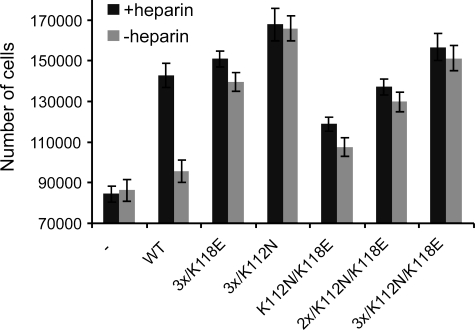
Proliferation of NIH 3T3 Cells in Response to Stimulation by FGF1 Mutants
As shown above, we observed considerable differences in the mitogenic activity of FGF1 mutants with the same heparin affinity but different stability. To further investigate the effect of stabilizing mutations on the biological activity of FGF1, we studied the ability of selected FGF1 variants to induce proliferation of NIH 3T3 cells. The cells were incubated for 72 h with 100 ng/ml FGF1 in the presence or absence of heparin; therefore, such an experiment can be considered a long term response assay. We found that the mutants with higher stability exhibited a prolonged ability to stimulate proliferation as compared with wild type FGF1, even when they had a reduced affinity for heparin (Fig. 7). The effect was particularly significant in the absence of heparin. The wild type induced almost no cell proliferation without heparin as compared with the control (non-stimulated cells). Introduction of the stabilizing mutations gradually enhanced the ability of FGF1 to stimulate cell proliferation. The most active variant in this assay was the 3x/K112N mutant, which stimulated the proliferation of NIH 3T3 cells without heparin almost twice as effectively as the wild type in the absence of heparin. The second in the rank among the mutants tested in the proliferation test was the 3x/K112N/K118E mutant, which was very stable but had an extremely reduced affinity for heparin. In this case, similarly to the 3x/K118E mutant, the extent of cell proliferation induced 72 h after the addition of the growth factor was the same in the presence and absence of heparin. Once again, for K112N/K118E, 2x/K112N/K118E, and 3x/K112N/K118E mutants, a stepwise compensation of the stabilizing mutations for the protective effect of heparin was visible.
FIGURE 7.
Stimulation of cell proliferation by FGF1 mutants. Serum-starved NIH 3T3 cells were treated with 100 ng/ml wild type or FGF1 mutant in the presence and absence of heparin. After 72 h, the number of cells was counted. Values are averages from three samples ± S.D.
Half-life of Mitogenic Activities
As discussed earlier, an important aspect of FGF1 activity, which is strongly affected by the presence of heparin (or heparans), is the duration of the action of the growth factor. To assess this parameter, we calculated for selected mutants the half-life of the active protein (i.e. the time required for the loss of one-half of the initial biological activity) (29). The wild type and two series of mutants containing the K112N or K118E mutations and the stabilizing ones were incubated for different periods of time at 37 °C in serum-free medium in the presence or absence of heparin before being added to NIH 3T3 cells. Then a standard experiment of [3H]thymidine incorporation in the absence of heparin was performed, the specific mitogenic activities were calculated, and the half-life values were derived (supplemental Fig. S3).
The activity of the K112N mutant was higher in the presence than in the absence of heparin. However, we did not observe such a tendency for the variants including the K118E substitution, considerably reducing the affinity of FGF1 to heparin. For these variants the data were similar irrespective of the presence of heparin.
All but one (the K118E) variants tested showed longer half-lives than the wild type when they were preincubated without heparin (supplemental Fig. S3). In contrast, in the presence of heparin, which protects the wild type against deactivation very effectively, only variants with the K112N mutation and the highly stable mutants with the K118E substitution exhibited longer half-life than the wild type (supplemental Fig. S3). The quadruple mutant 3x/K112N was the most resistant to deactivation, with a half-life over 36 h in the absence of heparin (i.e. more than 90-fold longer than that of the wild type (t½ = 24 min)). Among the mutants with the K118E mutation, the 3x/K118E variant exhibited the longest half-life (t½ = 10.5 h), being almost 30-fold more active than the wild type.
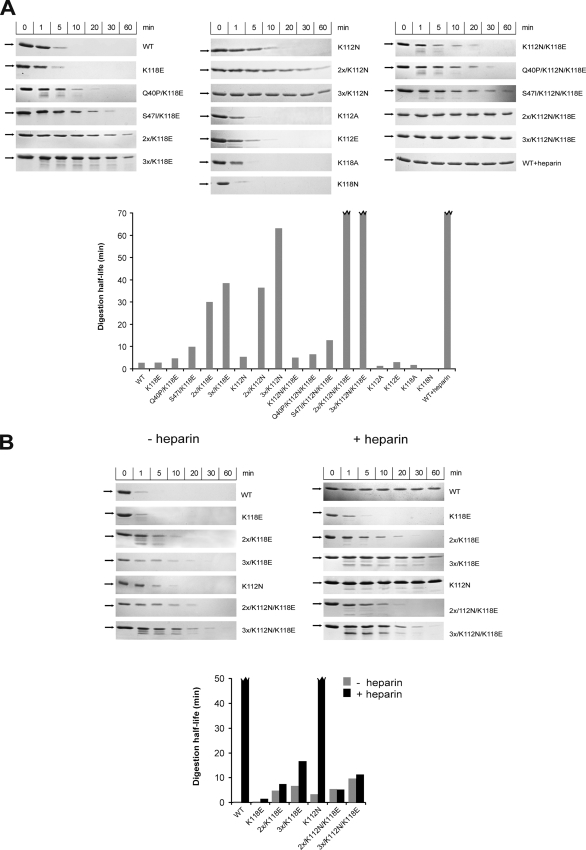
Susceptibility of FGF1 Mutants to Proteolysis
Our earlier study showed that the stabilizing mutations not only improved the thermodynamic stability of FGF1 but also increased its resistance to proteolytic degradation (29). To examine the relationship between proteolytic susceptibility, thermodynamic stability, and biological activity, we treated selected mutants with trypsin as a model protease at 37 °C. The proteolytic results obtained for the FGF1 mutants (Fig. 8) in the absence of heparin were in good agreement with the stability parameters reported in supplemental Table S1.
FIGURE 8.
Proteolytic degradation of wild type and mutants of FGF1. Growth factors were digested at 37 °C by trypsin at a molar ratio of 1:80 (A) and 1:20 in the presence and absence of heparin (B). The arrow indicates the position of intact FGF1. A broken line on the graph corresponds to much higher values than presented on the graph, which cannot be determined correctly within experimental conditions.
As shown in Fig. 8A, to digest wild type FGF1 completely, using trypsin at a molar ratio of 1:80, a period of 10 min was required, which corresponded to a protein half-life of slightly over 2.5 min. The mutant most susceptible to trypsin digestion was the K118N variant, which also exhibited the lowest denaturation temperature. The K112A, K112E, K118A, and K118E mutations did not seem to influence appreciably the susceptibility of FGF1 to proteolysis, similarly to its thermodynamic stability (Fig. 8A).
The K112N and K112N/K118E mutants turned out to be more resistant to trypsin digestion than the wild type, and only after 30 min of incubation with the protease at a molar ratio 1:80 were they totally digested. As expected, introduction of the stabilizing mutations to the K112N and K118E variants increased gradually their resistance to enzymatic degradation in the absence of heparin. Combination of Q40P, S47I, and H93G substitutions dramatically slowed the proteolytic processing extending the protein half-life 15 times in the case of the 3x/K118E variant and over 25 times for the 3x/K112N mutant. We could observe a synergistic effect of the stabilizing mutations for the 2x/K112N/K118E and 3x/K112N/K118E variants, for which even after 1 h of trypsin digestion (at a molar ratio of 1:80) there was no distinct fragmentation, and the intensity of the band corresponding to the intact form of FGF1 remained unchanged (Fig. 8A). For a higher trypsin concentration (ratio of 1:20), all mutants demonstrated again a clear correlation between the conformational stability and resistance to protease action and a cumulative, protective effect of the stabilizing mutations (Fig. 8B).
The presence of heparin strongly enhanced the resistance to proteolysis of the wild type and K112N FGF1, a mutant showing the least reduced heparin affinity of all of the analyzed mutants (Fig. 8B). For variants containing the K118E substitution, the presence of heparin only had a very slight enhancing effect on proteolytic resistance. Furthermore, all mutants with the both K118E and K112N substitutions had virtually identical protease susceptibility in the presence as well as in the absence of heparin (Fig. 8B). Because the binding of these mutants to heparin was strongly reduced, heparin did not exhibit any protective effect during the proteolytic degradation. This result fits very well with the thermodynamic data, since in the denaturation experiments, we observed a lack of protection by heparin during unfolding.
In Vivo FGF1 Degradation
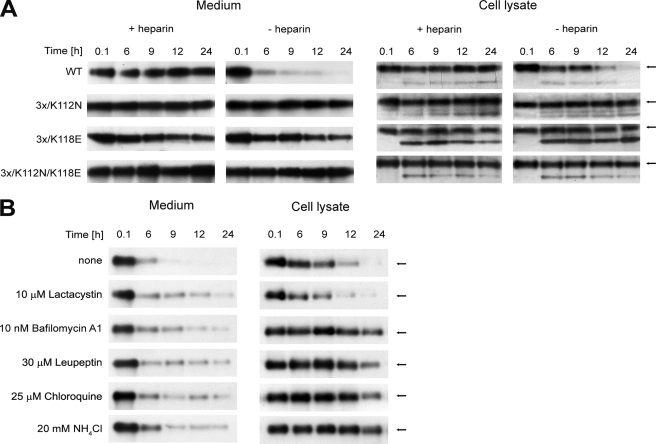
To test in vivo the results obtained in the in vitro proteolytic digestion assay, we checked the processing of selected FGF1 mutants in NIH 3T3 cell culture at 37 °C. We analyzed the amount of the growth factor in the medium and in cell lysates at different time points during the 24 h after FGF1 addition. The data in Fig. 9A demonstrate that in the presence of heparin, the wild type growth factor remained intact in both fractions throughout the experiment. However, in the absence of heparin, the wild type was much more efficiently degraded than the mutants with reduced heparin binding but increased stability.
FIGURE 9.
In vivo degradation of wild type and selected mutants of FGF1 during incubation with NIH 3T3 cells. A, presence of growth factors in medium and during the internalization process in cell lysates in the presence and absence of heparin. B, the effect of degradation inhibitors on extra- and intracellular processing of wild type FGF1 in the absence of heparin. The arrow indicates the position of intact FGF1.
After 6 h of incubation with the cells without heparin, the wild type growth factor was only faintly seen in the medium, whereas the 3x/K118E mutant was only degraded to a slight extent, and the remaining mutants were intact even after 24 h. The lack of degradation of the 3x/K112N/K118E mutant confirms the protective effect of the K112N mutation. Similar results were obtained for the cell lysates in the absence of heparin. There was no evident reduction in the amount of the mutants, whereas the degradation of the wild type protein was clearly visible after 12 h (Fig. 9A).
We observed almost no difference in in vivo processing in the presence and absence of heparin for the mutants tested. These results indicate that the stabilizing mutations significantly reduce in vivo degradation, slowing the rate of FGF1 proteolysis within the cell and in the medium to a similar extent as does the presence of heparin toward the wild type FGF1.
To further elucidate the mechanism of intracellular processing of FGF1, we performed an in vivo degradation experiment for the wild type without heparin in the absence or presence of several protein degradation inhibitors. Preincubation with lactacystin, a proteasome degradation inhibitor, showed no effect on degradation of internalized FGF1 in NIH 3T3 cells. In contrast, reduced degradation of FGF1 was observed when cells were pretreated with lysosomal degradation inhibitors, such as leupeptin, bafilomycin A1, chloroquine, and NH4Cl, which prevent endosomal acidification required for activity of lysomal proteases (Fig. 9B). These data suggest that the degradation of endocytosed FGF1 is controlled by the lysosomal pathway and that proteasome is not required for this process. In the presence of the inhibitors, we also observed slight reduction of FGF1 degradation in the medium, which could be caused by the reduced amount of proteases released from the cell.
The observed effect of inhibitors was similar to the effect of stabilizing mutations, suggesting that low stability of FGF1 makes the protein more susceptible to lysosomal degradation. This, again, confirms that increased stability protects FGF1 inside and outside the cell against proteolytic processing.
Translocation of FGF Variants into Cells
FGF1, similarly to FGF2, is able to cross the cellular membrane and to be translocated from the extracellular space into the cytosol and nucleus of target cells (33, 45). Therefore, we studied the ability of the mutants to enter the cells employing two previously described methods: in vivo phosphorylation of the externally added growth factor and digitonin fractionation of FGF1-treated cells (27, 40).
The in vivo phosphorylation assay takes advantage of the fact that FGF1 contains a single functional phosphorylation site for protein kinase C, which is a cytosolic and nuclear enzyme. Therefore, cells are only able to phosphorylate the growth factor that has crossed the cell membrane. However, because one of the experimental steps requires binding to heparin-Sepharose and since Lys118 is part of the site recognized by protein kinase C, this assay could only be used with heparin-binding site mutants containing the substitution of Lys112.
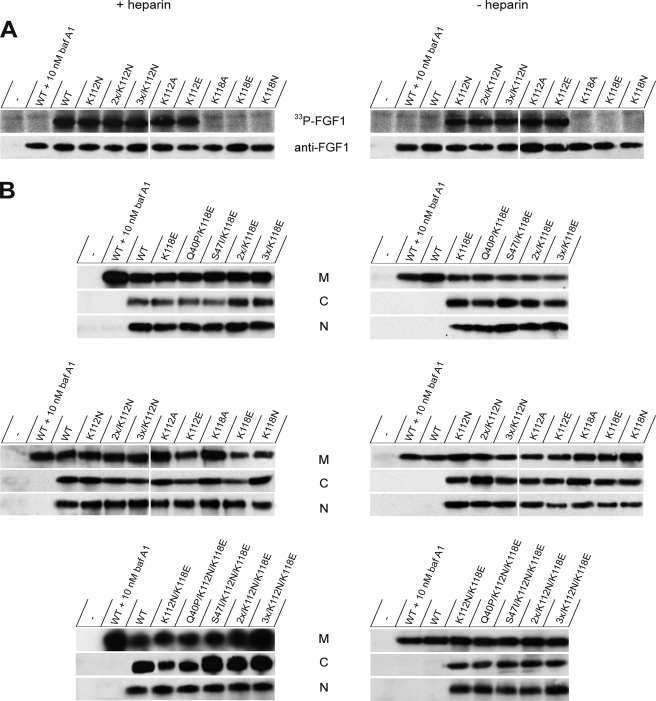
Serum-starved NIH 3T3 cells were incubated with FGF1 for 6 h in the presence of radioactive phosphate, and then the internalized growth factor was extracted from total cell lysates, run on SDS-PAGE, and electroblotted onto a membrane. FGF1 that had been radiolabeled in vivo was visualized by fluorography (Fig. 10A, top). The results clearly show that all of the variants containing a mutation at position 112 were effectively translocated into the cell, both in the presence and absence of heparin.
FIGURE 10.
Translocation of wild type and mutants of FGF1 in the presence and absence of heparin. Serum-starved NIH 3T3 cells were incubated with 100 ng/ml growth factor in the presence of 10 units/ml heparin for 6 h, and the translocation was monitored by in vivo phosphorylation (A) and digitonin-based fractionation (B) assays.
Mutants with substitution at position 118, which is involved in the phosphorylation site for protein kinase C, served as negative controls. As a control of total cellular uptake of FGF1, we used immunodetection with anti-FGF1 antibody (Fig. 10A, bottom).
To allow testing for the translocation of all of the mutants, we used a cell fractionation technique based on careful digitonin treatment of the cells under conditions where the plasma membrane is permeabilized while intracellular membranes remain intact (40, 46, 47). In this way, translocation of exogenously added FGF1 to the cytosol and nucleus can be monitored independently of its in vivo phosphorylation. NIH 3T3 cells were incubated with FGF1 variants for 6 h and then fractionated into cytosolic, nuclear, and membrane fractions, as described under “Experimental Procedures.” The membrane fraction, which represents growth factor from intracellular vesicles, can be considered as a loading control. To verify the quality of the fractionation procedure, we analyzed lysates from each fraction using antibodies against marker proteins for different compartments (data not shown).
Both in the presence and absence of heparin, all FGF1 mutants tested were detected in all three fractions (Fig. 10B). Interestingly, the wild type FGF1, which translocated nicely in the presence of heparin, was found neither in the cytosol nor in the nucleus in the absence of heparin, which was also seen using the phosphorylation method (Fig. 10, A and B). The presence of all of the mutants in the cytosolic and nuclear fractions indicates that reduced heparin binding not only does not disturb the translocation of the growth factor in the presence of heparin but also enables its entry into the cell in the absence of heparin.
In all of these experiments, translocation of FGF1 to the cytosol and nucleus could be blocked by bafilomycin A1 (Fig. 10, A and B), which was used as a negative control. This compound prevents growth factor translocation due to inhibition of the vesicular proton pumps essential to generate the vesicular membrane electric potential required for the process (31, 32).
DISCUSSION
Heparin and cell surface HS are considered to be important components of FGF-mediated signaling. The crystal structures of FGF1 and FGF2 complexes with FGFR solved in the presence of heparin clearly demonstrate its importance in complex formation (6, 20, 41, 48). Nevertheless, several reports question the direct requirement for heparin/HS in the assembly of a functional FGF·FGFR complex, stimulation of the signaling pathways, and generation of the mitogenic response (12, 14–16). Also, kinetic studies of FGF2·FGFR complex formation have revealed that in the presence of heparin, the dissociation rate constant is only 1 order of magnitude lower than in its absence (49).
In this study, we hypothesized that the main role of heparin is to protect FGF1 against heat and protease(s) under physiological conditions and that it is not required to form a functional FGF·FGFR complex. Free growth factor is a highly unstable protein with a denaturation temperature of about 40 °C at neutral pH, which means that a significant fraction of FGF1 is fully unfolded at physiological conditions and therefore prone to proteolysis. It is well documented that the addition of heparin effectively stabilizes the FGF1 molecule (17–19). Since the mitogenic potential of FGF1 is tested in assays taking at least 12 h, it is not clear whether the presence of heparin has a direct effect on the functional signaling complex or just a protective role. In our previous report, we showed that introduction of stabilizing mutation(s), either single or multiple, significantly improved the proteolytic resistance of free FGF1 and prolonged its biological action (29). Here, we applied the three most stabilizing mutations in combination, increasing the Tden of FGF1 by about 23 °C, to compensate for the absence of interaction with heparin, by providing the necessary protection against heat or protease(s).
We studied the biological and biophysical properties of a series of 17 variants of FGF1: six single mutants (substituting lysines at position 112 or 118 responsible for heparin binding with Ala, Glu, or Asn), one double lysine mutant (K112N/K118E), and 10 multiple mutants carrying mutations at position 112 and/or 118 together with different combinations of the three stabilizing mutations (Q40P, S47I, and H93G) (29) (Fig. 1). All FGF1 variants comprising mutation(s) at the heparin binding site exhibited reduced affinity for immobilized heparin as probed by the elution profile from a heparin-Sepharose column (Fig. 2). A stepwise introduction of the stabilizing mutations in these variants did not affect the respective elution profiles, in agreement with the crystal structure of the Q40P/S47I/H93G variant, which shows that the global conformation and the heparin binding site are not affected in the triple mutant (50). As expected, in the case of all variants, the introduction of stabilizing mutations resulted in a significant shift in Tden values, up to 64.1 °C (Fig. 3).
To verify the biological competence of the constructed mutants, we measured their activities in a series of experiments. Depending on the time scale of the test, we distinguish two classes of assays. In the short time response tests (time scale of minutes) we checked the ability of the mutants to bind to the receptor, activate it, and stimulate downstream signaling cascades. Long time response experiments (time scale of at least 12 h) included the stimulation of DNA synthesis, cell proliferation, and in vivo degradation. As shown earlier, sensitivity to heat denaturation and/or proteolysis can be revealed only in long time response experiments that are performed at 37 °C in cell culture containing extracellular proteases (29).
All of the mutants containing single substitutions that reduced heparin affinity (with or without the stabilizing mutations) showed no differences in competitive receptor binding, co-localization of phosphorylated receptors with early endosomes, and activation of FGF1 signaling pathways, as compared with the wild type (Figs. 4–6). These results indicate that none of the mutated residues are involved in the interactions with the high affinity receptor and that full short term activation of FGFR takes place even when the growth factor is defective in heparin binding.
However, in the case of the K112N/K118E mutants, the most defective ones in heparin binding (Fig. 2), a reduced signaling activation was observed at a low concentration (5 ng/ml) of the growth factor (Fig. 5). As expected, this effect could be overcome by using a high concentration of the FGF1 mutants (50 ng/ml). In this case, there was no difference in the level of signaling between the mutants containing the double substitution K112N/K118E and the wild type (Fig. 5).
For all variants, we tested the most crucial activity of the growth factor, its mitogenic potential. It was shown that a maximal mitogenic response requires the presence of a stable FGF1·FGFR complex over 12 h and that short term activation of the signaling pathways is not sufficient for stimulation of DNA synthesis by FGF1 (44, 51, 52). As is commonly done, we measured the ability of all of the mutants to stimulate cellular DNA synthesis after 24 h of incubation with the growth factor. All single mutants with reduced heparin affinity exhibited a significantly decreased ability to stimulate DNA synthesis. This could be due to the lack of protection of the growth factors provided by heparin and, in consequence, their higher susceptibility to proteases. Since we did not observe any difference in the activation of the signaling cascades in the case of the single mutants with reduced affinity for heparin, we concluded that the observed differences in the mitogenic potential of the variants were mainly due to differences in their stability and half-life.
This view is supported by the findings that full mitogenic activity could be restored to the K118E and K112N mutants when their stability was increased (Fig. 6). Introduction of the stabilizing mutations protected the growth factor with reduced heparin affinity sufficiently to compensate for the deficiency in heparin binding and ensured the survival of the proteins for a long time. Also, in the case of mutants containing a double substitution in the heparin-binding site (K112N/K118E), we observed a stepwise effect of the stabilizing mutations on the DNA synthesis (Fig. 6).
We also studied the effect of the heparin-binding site mutations in combination with the stabilizing substitutions on the ability of selected FGF1 variants to induce cell proliferation. In accordance with the effects on DNA synthesis, we found that increased stability provided prolonged ability to stimulate proliferation even when heparin affinity was strongly reduced (Fig. 7).
The compensating effect of the stabilizing mutations for the handicapped heparin protection was further demonstrated by testing the susceptibility of the FGF1 mutants to in vitro proteolysis and in vivo degradation (Figs. 8 and 9). The results of the trypsin digestion experiments were in excellent agreement with the thermodynamic stabilities and correlated nicely with the mitogenic activity assay and half-life experiments. This, again, confirms that the lack of mitogenic potential of the mutants with reduced heparin affinity reflects the lack of protection against heat/proteolysis and not their reduced functionality. We also observed that the degradation of endocytosed FGF1 was prevented by the lysosomal degradation inhibitors, which protect the growth factor in the absence of heparin to a similar extent as stabilizing mutations.
Our data showing a positive mitogenic effect of mutational stabilization of the K118E (K132E) variant are consistent with the results published by Klingenberg et al. (28). They showed that the double mutant C131S/K132E, with a cysteine substitution increasing significantly the half-life of FGF1, showed a higher mitogenic activity than the single K132E mutant (28, 53). Also, a recent paper of Imamura and co-workers (54) revealed that in BaF3 cells lacking the capacity to synthesize glycosaminoglycans, an FGF1:FGF2 chimeric protein (FGFC) was able to activate FGFR and induce cell proliferation in the absence of heparin. Similarly to our constructs, FGFC was more stable and resistant to protease digestion than FGF1 (54).
Our results are also in good agreement with the many studies on FGF2 suggesting that, in the absence of heparin, active FGF2·FGFR complex can still be formed. Fannon and Nugent (12) found that in Balb/c3T3 cells lacking HS due to complete inhibition of proteoglycan sulfation, FGF2 was able to bind to the receptor, induce the signaling, and stimulate DNA synthesis in a dose-dependent manner. They clearly showed that HS worked by decreasing the rate of dissociation from FGFR but not altering the rate of FGF2 association and therefore had a stabilizing effect on the FGF2·FGFR complex. Also, Delehedde et al. (15, 16) showed that in sulfated glycosaminoglycan-deficient cells generated by sodium chlorate treatment, FGF2 was unable to stimulate DNA synthesis; however, it was able to trigger a transient phosphorylation of p42/44 MAPK. Finally, several other studies have reported heparin/HS-independent binding of FGF2 to its receptor, showing that the expression of early genes upon FGF2 stimulation is induced to the same extent in the presence and absence of heparin (14). It was also found that mutations in the heparin-binding region of FGF2 had no effect on its affinity for the receptor (55) and that in heparan sulfate-deficient Chinese hamster ovary 677 cells, FGF2 was not completely dependent on heparin/HS for activation of FGFR1 tyrosine kinase (7).
The presented results are consistent with the symmetric “two-end” crystallographic model of FGF·FGFR interaction, which allows for receptor dimerization in the absence of heparin (48). In this model, heparin promotes coupling of two FGF·FGFR halves, but the formation of the FGF·FGFR dimer is dependent on protein-protein contacts (48).
The presence of HS on the cell surface can affect the efficiency of receptor-mediated translocation of FGF1 into cells (56–58). Analysis of the cellular localization of wild type FGF1 and its mutants with reduced affinity for heparin showed striking differences in their behavior (Fig. 10). In the presence of heparin, wild type FGF1 and all of its variants were effectively translocated into the cell (to the cytosol and nucleus). However, in the absence of heparin, the wild type did not cross the cellular membrane barrier, in clear contrast to all of the mutants studied here.
In the absence of heparin, wild type FGF1 binds to FGFR but also to the abundant heparan sulfates on the cell surface (17). Several reports show that in contrast to the binding of FGF1 to FGFR, its binding to cell surface heparans does not result in its functional translocation, instead directing the growth factor to the lysosome, where it is degraded (30, 58). The FGF1 mutants with low heparin affinity studied here can bind effectively only to FGFR and therefore can be efficiently translocated into the cytosol/nucleus. Because the affinity for heparin of wild type FGF1 is much higher than that for heparans, it seems that the increased level of heparin in specific stress conditions might be aiding efficient translocation of the growth factor. Such a mechanism may be important in regulating the intracellular fate of the growth factor. In this paper, we also confirmed that the translocation process is not influenced by FGF1 stability, which is in good agreement with our earlier studies. It was shown that stable, cross-linked FGF1 mutants were translocated to the cytosol and the nucleus equally well as the wild type FGF1 (46). On the other hand, we have reported that several mutants with decreased stability can also be effectively translocated across the cell membrane (34).
Surface heparans may play an important regulatory role by creating a local reservoir of FGF1 (6, 17). Since the concentration of heparin in normal blood is very low, it is likely that any free growth factor tends to become bound to surface heparans on the cells close to the site of production, which prevents its diffusion far away. This can be crucial, since growth factors may have biological activities that should be limited to a defined location. An example is FGF8, which ensures correct localization of asymmetric organs and tissues (59, 60). The fact that the growth factor is very sensitive to proteolysis when not bound to heparin or heparans may be a backup system ensuring that runaway molecules are destroyed before they can act at a wrong location. We conclude that an increased stability of FGF1 can compensate for reduced heparin binding and that the main role of heparin/HS is to protect FGF1 against proteolytic degradation and in this way prolong the signal from the FGF·FGFR complex.
Supplementary Material
Acknowledgment
We are grateful to Ellen M. Haugsten for help with the confocal experiments.
This work was supported by Polish Ministry of Science and Higher Education Grant N N301 4192 33, by the Norwegian Cancer Society, by Raquel and Chr. Bruuns legat, and by Torsteds legat.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- FGFR
- fibroblast growth factor receptor
- FGF
- fibroblast growth factor
- HS
- heparan sulfate(s)
- MAPK
- mitogen-activated protein kinase
- PLC-γ
- phospholipase C-γ
- PBS
- phosphate-buffered saline
- 2x
- substitution Q40P/S47I
- 3x
- substitution Q40P/S47I/H93G
- WT
- wild type.
REFERENCES
- 1.Ornitz D. M., Itoh N. (2001) Genome Biol. 2, REVIEWS3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh N., Ornitz D. M. (2004) Trends Genet. 20, 563–569 [DOI] [PubMed] [Google Scholar]
- 3.Eswarakumar V. P., Lax I., Schlessinger J. (2005) Cytokine Growth Factor Rev. 16, 139–149 [DOI] [PubMed] [Google Scholar]
- 4.Spivak-Kroizman T., Lemmon M. A., Dikic I., Ladbury J. E., Pinchasi D., Huang J., Jaye M., Crumley G., Schlessinger J., Lax I. (1994) Cell 79, 1015–1024 [DOI] [PubMed] [Google Scholar]
- 5.Ornitz D. M., Xu J., Colvin J. S., McEwen D. G., MacArthur C. A., Coulier F., Gao G., Goldfarb M. (1996) J. Biol. Chem. 271, 15292–15297 [DOI] [PubMed] [Google Scholar]
- 6.Mohammadi M., Olsen S. K., Ibrahimi O. A. (2005) Cytokine Growth Factor Rev. 16, 107–137 [DOI] [PubMed] [Google Scholar]
- 7.Lundin L., Rönnstrand L., Cross M., Hellberg C., Lindahl U., Claesson-Welsh L. (2003) Exp. Cell Res. 287, 190–198 [DOI] [PubMed] [Google Scholar]
- 8.Waksman G., Herr A. B. (1998) Nat. Struct. Biol. 5, 527–530 [DOI] [PubMed] [Google Scholar]
- 9.Wu Z. L., Zhang L., Yabe T., Kuberan B., Beeler D. L., Love A., Rosenberg R. D. (2003) J. Biol. Chem. 278, 17121–17129 [DOI] [PubMed] [Google Scholar]
- 10.Ornitz D. M., Herr A. B., Nilsson M., Westman J., Svahn C. M., Waksman G. (1995) Science 268, 432–436 [DOI] [PubMed] [Google Scholar]
- 11.de Paz J. L., Angulo J., Lassaletta J. M., Nieto P. M., Redondo-Horcajo M., Lozano R. M., Giménez-Gallego G., Martín-Lomas M. (2001) ChemBioChem 2, 673–685 [DOI] [PubMed] [Google Scholar]
- 12.Fannon M., Nugent M. A. (1996) J. Biol. Chem. 271, 17949–17956 [DOI] [PubMed] [Google Scholar]
- 13.Nugent M. A., Edelman E. R. (1992) Biochemistry 31, 8876–8883 [DOI] [PubMed] [Google Scholar]
- 14.Roghani M., Mansukhani A., Dell'Era P., Bellosta P., Basilico C., Rifkin D. B., Moscatelli D. (1994) J. Biol. Chem. 269, 3976–3984 [PubMed] [Google Scholar]
- 15.Delehedde M., Seve M., Sergeant N., Wartelle I., Lyon M., Rudland P. S., Fernig D. G. (2000) J. Biol. Chem. 275, 33905–33910 [DOI] [PubMed] [Google Scholar]
- 16.Delehedde M., Lyon M., Gallagher J. T., Rudland P. S., Fernig D. G. (2002) Biochem. J. 366, 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powers C. J., McLeskey S. W., Wellstein A. (2000) Endocr. Relat. Cancer 7, 165–197 [DOI] [PubMed] [Google Scholar]
- 18.Gospodarowicz D., Cheng J. (1986) J. Cell. Physiol. 128, 475–484 [DOI] [PubMed] [Google Scholar]
- 19.Damon D. H., Lobb R. R., D'Amore P. A., Wagner J. A. (1989) J. Cell. Physiol. 138, 221–226 [DOI] [PubMed] [Google Scholar]
- 20.Pellegrini L., Burke D. F., von Delft F., Mulloy B., Blundell T. L. (2000) Nature 407, 1029–1034 [DOI] [PubMed] [Google Scholar]
- 21.DiGabriele A. D., Lax I., Chen D. I., Svahn C. M., Jaye M., Schlessinger J., Hendrickson W. A. (1998) Nature 393, 812–817 [DOI] [PubMed] [Google Scholar]
- 22.Harper J. W., Lobb R. R. (1988) Biochemistry 27, 671–678 [DOI] [PubMed] [Google Scholar]
- 23.Zhu X., Komiya H., Chirino A., Faham S., Fox G. M., Arakawa T., Hsu B. T., Rees D. C. (1991) Science 251, 90–93 [DOI] [PubMed] [Google Scholar]
- 24.Volkin D. B., Tsai P. K., Dabora J. M., Gress J. O., Burke C. J., Linhardt R. J., Middaugh C. R. (1993) Arch. Biochem. Biophys. 300, 30–41 [DOI] [PubMed] [Google Scholar]
- 25.Burgess W. H., Shaheen A. M., Ravera M., Jaye M., Donohue P. J., Winkles J. A. (1990) J. Cell Biol. 111, 2129–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong P., Hampton B., Szylobryt E., Gallagher A. M., Jaye M., Burgess W. H. (1995) J. Biol. Chem. 270, 25805–25811 [DOI] [PubMed] [Google Scholar]
- 27.Klingenberg O., Wiedlocha A., Rapak A., Muñoz R., Falnes P., Olsnes S. (1998) J. Biol. Chem. 273, 11164–11172 [DOI] [PubMed] [Google Scholar]
- 28.Klingenberg O., Wiedlocha A., Olsnes S. (1999) J. Biol. Chem. 274, 18081–18086 [DOI] [PubMed] [Google Scholar]
- 29.Zakrzewska M., Krowarsch D., Wiedlocha A., Olsnes S., Otlewski J. (2005) J. Mol. Biol. 352, 860–875 [DOI] [PubMed] [Google Scholar]
- 30.Wiedłocha A., Falnes P. O., Rapak A., Klingenberg O., Muñoz R., Olsnes S. (1995) J. Biol. Chem. 270, 30680–30685 [DOI] [PubMed] [Google Scholar]
- 31.Małecki J., Wiedłocha A., Wesche J., Olsnes S. (2002) EMBO J. 21, 4480–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Małecki J., Wesche J., Skjerpen C. S., Wiedłocha A., Olsnes S. (2004) Mol. Biol. Cell 15, 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiedłuocha A., Sørensen V. (2004) Curr. Top. Microbiol. Immunol. 286, 45–79 [DOI] [PubMed] [Google Scholar]
- 34.Zakrzewska M., Krowarsch D., Wiedlocha A., Olsnes S., Otlewski J. (2006) Biochemistry 45, 15338–15348 [DOI] [PubMed] [Google Scholar]
- 35.Haugsten E. M., Malecki J., Bjørklund S. M., Olsnes S., Wesche J. (2008) Mol. Biol. Cell 19, 3390–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zakrzewska M., Krowarsch D., Wiedlocha A., Otlewski J. (2004) Protein Eng. Des. Sel. 17, 603–611 [DOI] [PubMed] [Google Scholar]
- 37.Blaber S. I., Culajay J. F., Khurana A., Blaber M. (1999) Biophys. J. 77, 470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gimenez-Gallego G., Conn G., Hatcher V. B., Thomas K. A. (1986) Biochem. Biophys. Res. Commun. 138, 611–617 [DOI] [PubMed] [Google Scholar]
- 39.Wiedłocha A., Falnes P. O., Rapak A., Muñoz R., Klingenberg O., Olsnes S. (1996) Mol. Cell. Biol. 16, 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiedłocha A., Nilsen T., Wesche J., Sçrensen V., Maùecki J., Marcinkowska E., Olsnes S. (2005) Mol. Biol. Cell 16, 794–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. (2000) Mol. Cell 6, 743–750 [DOI] [PubMed] [Google Scholar]
- 42.Plotnikov A. N., Hubbard S. R., Schlessinger J., Mohammadi M. (2000) Cell 101, 413–424 [DOI] [PubMed] [Google Scholar]
- 43.Culajay J. F., Blaber S. I., Khurana A., Blaber M. (2000) Biochemistry 39, 7153–7158 [DOI] [PubMed] [Google Scholar]
- 44.Zhan X., Hu X., Friesel R., Maciag T. (1993) J. Biol. Chem. 268, 9611–9620 [PubMed] [Google Scholar]
- 45.Olsnes S., Klingenberg O., Wiedłocha A. (2003) Physiol Rev. 83, 163–182 [DOI] [PubMed] [Google Scholar]
- 46.Wesche J., Wiedłocha A., Falnes P. O., Choe S., Olsnes S. (2000) Biochemistry 39, 15091–15100 [DOI] [PubMed] [Google Scholar]
- 47.Sørensen V., Zhen Y., Zakrzewska M., Haugsten E. M., Wälchli S., Nilsen T., Olsnes S., Wiedlocha A. (2008) Mol. Cell. Biol. 28, 4129–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohammadi M., Olsen S. K., Goetz R. (2005) Curr. Opin. Struct. Biol. 15, 506–516 [DOI] [PubMed] [Google Scholar]
- 49.Ibrahimi O. A., Zhang F., Hrstka S. C., Mohammadi M., Linhardt R. J. (2004) Biochemistry 43, 4724–4730 [DOI] [PubMed] [Google Scholar]
- 50.Szlachcic A., Zakrzewska M., Krowarsch D., Os V., Helland R., Smalås A. O., Otlewski J. (2009) Acta Crystallogr. D Biol. Crystallogr. 65, 67–73 [DOI] [PubMed] [Google Scholar]
- 51.Imamura T., Oka S., Tanahashi T., Okita Y. (1994) Exp. Cell Res. 215, 363–372 [DOI] [PubMed] [Google Scholar]
- 52.Ornitz D. M. (2000) BioEssays 22, 108–112 [DOI] [PubMed] [Google Scholar]
- 53.Ortega S., Schaeffer M. T., Soderman D., DiSalvo J., Linemeyer D. L., Gimenez-Gallego G., Thomas K. A. (1991) J. Biol. Chem. 266, 5842–5846 [PubMed] [Google Scholar]
- 54.Motomura K., Hagiwara A., Komi-Kuramochi A., Hanyu Y., Honda E., Suzuki M., Kimura M., Oki J., Asada M., Sakaguchi N., Nakayama F., Akashi M., Imamura T. (2008) Biochim. Biophys. Acta 1780, 1432–1440 [DOI] [PubMed] [Google Scholar]
- 55.Presta M., Statuto M., Isacchi A., Caccia P., Pozzi A., Gualandris A., Rusnati M., Bergonzoni L., Sarmientos P. (1992) Biochem. Biophys. Res. Commun. 185, 1098–1107 [DOI] [PubMed] [Google Scholar]
- 56.Roghani M., Moscatelli D. (1992) J. Biol. Chem. 267, 22156–22162 [PubMed] [Google Scholar]
- 57.Gleizes P. E., Noaillac-Depeyre J., Amalric F., Gas N. (1995) Eur. J. Cell Biol. 66, 47–59 [PubMed] [Google Scholar]
- 58.Citores L., Khnykin D., Sçrensen V., Wesche J., Klingenberg O., Wiedùocha A., Olsnes S. (2001) J. Cell Sci. 114, 1677–1689 [DOI] [PubMed] [Google Scholar]
- 59.Regan J. C., Concha M. L., Roussigne M., Russell C., Wilson S. W. (2009) Neuron 61, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hong S. K., Dawid I. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2230–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.